Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Methods
2.1. Isolation of Pseudomonas aeruginosa
2.2. Antimicrobial Susceptibility by the Automated Method
2.3. Antimicrobial Susceptibility Testing by Disk Diffusion
2.4. Total DNA Extraction
2.5. Molecular Detection of Genes Related to Virulence and Antimicrobial Resistance
2.6. Sequencing of PCR Products
2.7. Genotyping Using Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR
2.8. Statistical Analysis of the Data
2.9. Ethical Aspects
3. Results
3.1. Characterization of Samples and Drug Susceptibility Profile
3.2. Identification and Detection by Multiplex-PCR of Virulence Genes in Pseudomonas aeruginosa Isolates
3.3. Molecular Detection of Genes That Confer Antimicrobial Resistance in Pseudomonas aeruginosa Isolates
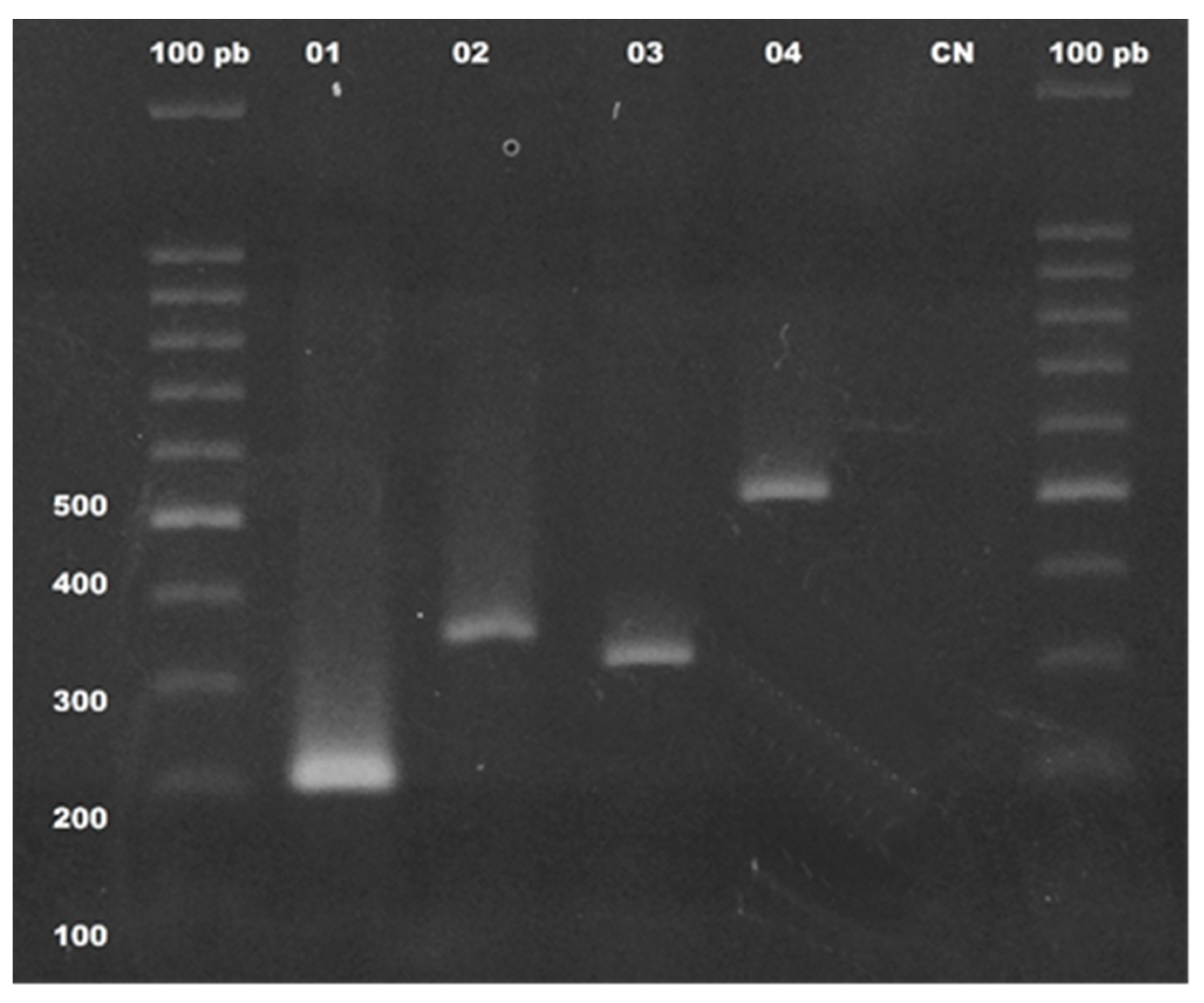
3.4. Sequencing Analysis of blaOXA-23, blaOXA-51, and mcr-1 Genes and the 16S Ribosomal Region
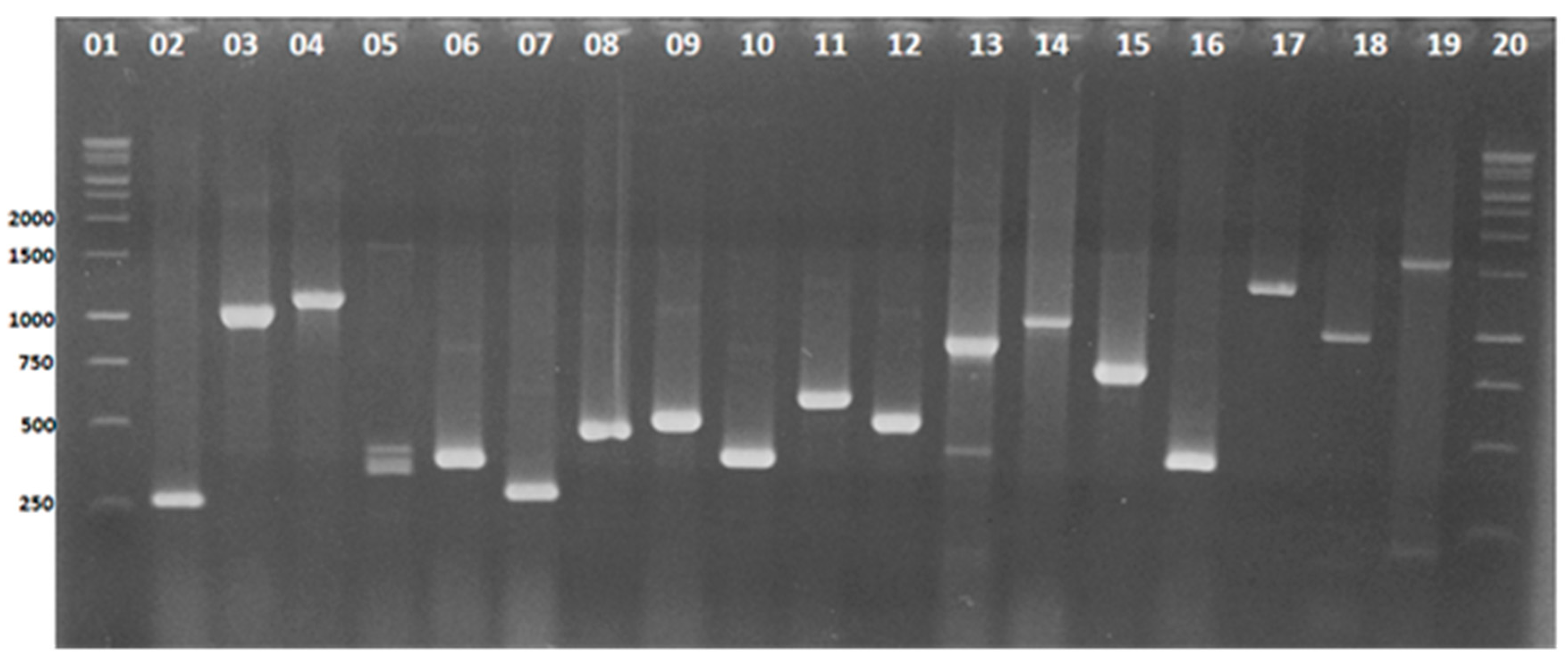
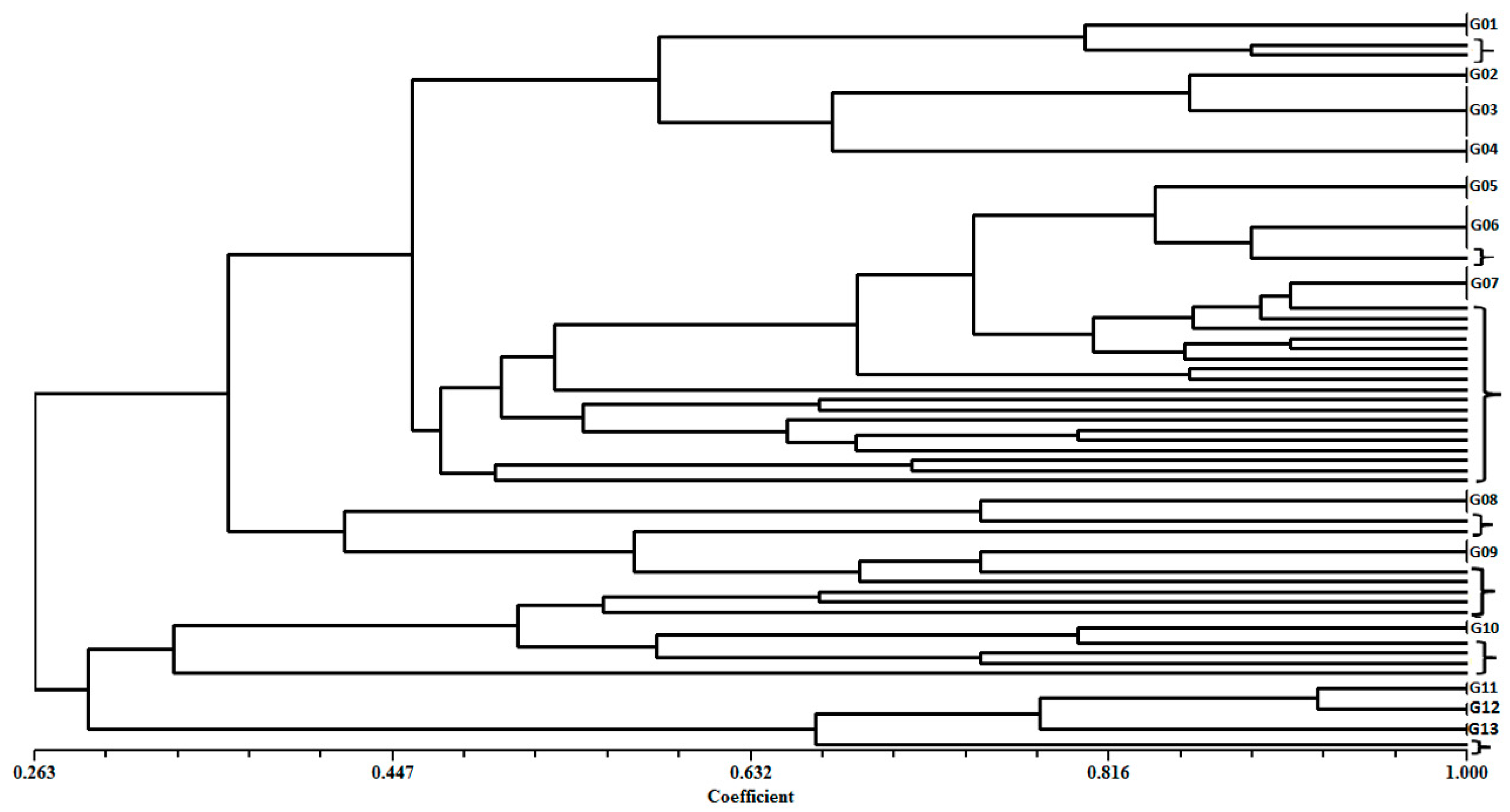
3.5. Clonal Profile of Pseudomonas aeruginosa Isolates by ERIC-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fazeli, N.; Momtaz, H. Virulence Gene Profiles of Multidrug-Resistant Pseudomonas aeruginosa Isolated from Iranian Hospital Infections. Iran. Red Crescent Med. J. 2014, 16, e15722. [Google Scholar] [CrossRef]
- Gomila, M.; Pena, A.; Mulet, M.; Lalucat, J.; Garcia-Valdes, E. Phylogenomics and systematics in Pseudomona. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef]
- Fujitani, S.; Moffett, K.S.; Yu, V. Pseudomonas aeruginosa. Antimicrobe: Infectious Disease & Antimicrobial Agents. Available online: http://www.antimicrobe.org/new/b112 (accessed on 1 March 2017).
- Vale de Macedo, G.H.R.; Costa, G.D.E.; Oliveira, E.R.; Damasceno, G.V.; Mendonca, J.S.P.; Silva, L.D.S.; Chagas, V.L.; Bazan, J.M.N.; Alianca, A.; Miranda, R.C.M.; et al. Interplay between ESKAPE Pathogens and Immunity in Skin Infections: An Overview of the Major Determinants of Virulence and Antibiotic Resistance. Pathogens 2021, 10, 148. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Badamchi, A.; Masoumi, H.; Javadinia, S.; Asgarian, R.; Tabatabaee, A. Molecular detection of six virulence genes in Pseudomonas aeruginosa isolates detected in children with urinary tract infection. Microb. Pathog. 2017, 107, 44–47. [Google Scholar] [CrossRef]
- Benie, C.K.; Dadie, A.; Guessennd, N.; N’Gbesso-Kouadio, N.A.; Kouame, N.D.; N’Golo, D.C.; Aka, S.; Dako, E.; Dje, K.M.; Dosso, M. Characterization of Virulence Potential of Pseudomonas aeruginosa Isolated from Bovine Meat, Fresh Fish, and Smoked Fish. Eur. J. Microbiol. Immunol. 2017, 7, 55–64. [Google Scholar] [CrossRef]
- Schiavano, G.F.; Carloni, E.; Andreoni, F.; Magi, S.; Chironna, M.; Brandi, G.; Amagliani, G. Prevalence and antibiotic resistance of Pseudomonas aeruginosa in water samples in central Italy and molecular characterization of oprD in imipenem resistant isolates. PLOS ONE 2017, 12, e0189172. [Google Scholar] [CrossRef] [PubMed]
- Ruffin, M.; Brochiero, E. Repair Process Impairment by Pseudomonas aeruginosa in Epithelial Tissues: Major Features and Potential Therapeutic Avenues. Front Cell Infect. Microbiol. 2019, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: A formidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef]
- Mittal, R.; Aggarwal, S.; Sharma, S.; Chhibber, S.; Harjai, K. Urinary tract infections caused by Pseudomonas aeruginosa: A minireview. J. Infect. Public Health 2009, 2, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.M.; Flaws, M.L. Antimicrobial resistance mechanisms in Pseudomonas aeruginosa. Am. Soc. Clin. Lab. Sci. 2011, 24, 47–51. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, R.J.; Roque, F.; Rodrigues, A.T.; Herdeiro, M.T.; Ramalheira, E. O uso de antibióticos e as resistências bacterianas: Breves notas sobre a sua evolução. Rev. Port. Saúde Pública 2016, 34, 77–84. [Google Scholar] [CrossRef]
- Nóbrega, M.d.S.; do Carmo Filho, J.R.; Pereira, M.S. Drug resistance evolution of Pseudomonas aeruginosa and Acinetobacter baumannii in intensive care units. Rev. Eletrônica Enferm. 2013, 15, 696–703. [Google Scholar]
- Kritsotakis, E.I.; Kontopidou, F.; Astrinaki, E.; Roumbelaki, M.; Ioannidou, E.; Gikas, A. Prevalence, incidence burden, and clinical impact of healthcare-associated infections and antimicrobial resistance: A national prevalent cohort study in acute care hospitals in Greece. Infect. Drug Resist. 2017, 10, 317–328. [Google Scholar] [CrossRef]
- Matos, E.C.O.d.; Matos, H.J.d.; Conceição, M.L.; Rodrigues, Y.C.; Carneiro, I.C.d.R.S.; Lima, K.V.B. Clinical and microbiological features of infections caused by Pseudomonas aeruginosa in patients hospitalized in intensive care units. Rev. Soc. Bras. Med. Trop. 2016, 49, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 20th Informational Supplement. CLSI Document M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Lanotte, P.; Watt, S.; Mereghetti, L.; Dartiguelongue, N.; Rastegar-Lari, A.; Goudeau, A.; Quentin, R. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J. Med. Microbiol. 2004, 53, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489. [Google Scholar] [CrossRef]
- De Vos, D.; Lim, A., Jr.; Pirnay, J.P.; Struelens, M.; Vandenvelde, C.; Duinslaeger, L.; Vanderkelen, A.; Cornelis, P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol. 1997, 35, 1295–1299. [Google Scholar] [CrossRef]
- Wozniak, D.J.; Ohman, D. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 1994, 176, 6007–6014. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, N.; Dhall, S.; Chhibber, S.; Harjai, K. Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. Int. J. Mol. Epidemiol. Genet. 2014, 5, 125–134. [Google Scholar]
- Khan, A.A.; Cerniglia, C.E. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl. Environ. Microbiol. 1994, 60, 3739–3745. [Google Scholar] [CrossRef]
- Smith, L.; Rose, B.; Tingpej, P.; Zhu, H.; Conibear, T.; Manos, J.; Bye, P.; Elkins, M.; Willcox, M.; Bell, S.; et al. Protease IV production in Pseudomonas aeruginosa from the lungs of adults with cystic fibrosis. J. Med. Microbiol. 2006, 55, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Paulyn, A.T.; Umeh, E.; Nna, E. Detection of virulence genes in urinary Pseudomonas aeruginosa from pregnant women attending Antenatal clinic in Makurdi, Central Nigeria. Sci. Res. J. 2016, 4, 9. [Google Scholar]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; Ostholm-Balkhed, A.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Devereux, R.; Willis, S.G. Amplification of ribosomal RNA sequences. In Molecular Microbial Ecology Manual; Akkermans, A.D.L., Van Elsas, J.D., De Bruijn, F.J., Eds.; Springer: Berlin, Germany, 1995; pp. 277–287. [Google Scholar]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Mesaros, N.; Nordmann, P.; Plésiat, P.; Roussel-Delvallez, M.; Van Eldere, J.; Glupczynski, Y.; Van Laethem, Y.; Jacobs, F.; Lebecque, P.; Malfroot, A.; et al. Pseudomonas aeruginosa: Resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect 2007, 13, 560–578. [Google Scholar] [CrossRef] [PubMed]
- Haidar, G.; Philips, N.J.; Shields, R.K.; Snyder, D.; Cheng, S.; Potoski, B.A.; Hao, B.; Press, E.G.; Cooper, V.S.; Clancy, C.J. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: Clinical effectiveness and evolution of resistance. Clin. Infect. Dis. 2017, 65, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Azimi, A.; Peymani, A.; Pour, P.K. Phenotypic and molecular detection of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from patients with burns in Tehran, Iran. Rev. Soc. Bras. Med. Trop. 2018, 51, 610–615. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Chung, E.S.; Lee, J.Y.; Rhee, J.Y.; Ko, K.S. Colistin resistance in Pseudomonas aeruginosa that is not linked to arnB. J. Med. Microbiol. 2017, 66, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Ocampo-Sosa, A.A.; Domínguez, M.A.; Gago, J.F.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; Peña, C.; Martínez-Martínez, L. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012, 56, 6349–6357. [Google Scholar] [CrossRef] [PubMed]
- Paluchowska, P.; Skałkowska, M.; Spelak, A.; Budak, A. Occurrence of alert pathogens in hospital environment. Part II. Multidrug-resistant non-fermenting bacilli. Med. Dośw. I Mikrobiol. 2012, 64, 45–53. [Google Scholar]
- Michalska, A.D.; Sacha, P.T.; Ojdana, D.; Wieczorek, A.; Tryniszewska, E. Prevalence of resistance to aminoglycosides and fluoroquinolones among Pseudomonas aeruginosa strains in a University Hospital in Northeastern Poland. Braz. J. Microbiol. 2014, 45, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Pathmanathan, S.G.; Samat, N.A.; Mohamed, R. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from a Malaysian Hospital. Malays. J. Med. Sci. 2009, 16, 27–32. [Google Scholar] [PubMed]
- Bhandari, S.; Banjara, M.R.; Lekhak, B.; Bhatta, D.R.; Regmi, S.R. Multi-drug and pan-drug resistant Pseudomonas aeruginosa: A challenge in post-antibiotic era. Nepal J. Sci. Technol. 2013, 13, 197–202. [Google Scholar] [CrossRef]
- Moazami, G.S.; Eftekhar, F. Assessment of carbapenem susceptibility and multidrug-resistance in Pseudomonas aeruginosa burn isolates in Tehran. Jundishapur J. Microbiol. 2013, 6, 162–165. [Google Scholar]
- Saderi, H.; Owlia, P. Detection of multidrug resistant (MDR) and extremely drug resistant (XDR) P. aeruginosa isolated from patients in Tehran, Iran. Iran. J. Pathol. 2015, 10, 265–271. [Google Scholar]
- Regenbogen, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and consistent evolution of colistin resistance in XDR Pseudomonas aeruginosa during morbidostat culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef]
- Dantas, R.C.C.; Silva, R.T.E.; Ferreira, M.L.; Goncalves, I.R.; Araujo, B.F.; Campos, P.A.; Royer, S.; Batistao, D.; Gontijo-Filho, P.P.; Ribas, R.M. Molecular epidemiological survey of bacteremia by multidrug resistant Pseudomonas aeruginosa: The relevance of intrinsic resistance mechanisms. PLOS ONE 2017, 12, e0176774. [Google Scholar] [CrossRef] [PubMed]
- Atkin, S.D.; Abid, S.; Foster, M.; Bose, M.; Keller, A.; Hollaway, R.; Sader, H.S.; Greenberg, D.E.; Finklea, J.D.; Castanheira, M.; et al. Multidrug-resistant Pseudomonas aeruginosa from sputum of patients with cystic fibrosis demonstrates a high rate of susceptibility to ceftazidime-avibactam. Infect. Drug Resist 2018, 11, 1499–1510. [Google Scholar] [CrossRef]
- Rafiee, R.; Eftekhar, F.; Tabatabaei, S.A.; Minaee Tehrani, D. Prevalence of Extended-Spectrum and Metallo beta-Lactamase Production in AmpC beta-Lactamase Producing Pseudomonas aeruginosa Isolates From Burns. Jundishapur J. Microbiol. 2014, 7, e16436. [Google Scholar] [CrossRef] [PubMed]
- Berrazeg, M.; Jeannot, K.; Ntsogo Enguene, V.Y.; Broutin, I.; Loeffert, S.; Fournier, D.; Plesiat, P. Mutations in beta-Lactamase AmpC Increase Resistance of Pseudomonas aeruginosa Isolates to Antipseudomonal Cephalosporins. Antimicrob. Agents Chemother. 2015, 59, 6248–6255. [Google Scholar] [CrossRef] [PubMed]
- Treepong, P.; Kos, V.N.; Guyeux, C.; Blanc, D.S.; Bertrand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Z.; Chu, H.Q.; Han, L.Z.; Zhang, Z.M.; Li, B.; Zhao, L.; Xu, L. Resistant mechanisms and molecular epidemiology of imipenem-resistant Acinetobacter baumannii. Mol. Med. Rep. 2016, 14, 2483–2488. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uwingabiye, J.; Lemnouer, A.; Roca, I.; Alouane, T.; Frikh, M.; Belefquih, B.; Bssaibis, F.; Maleb, A.; Benlahlou, Y.; Kassouati, J.; et al. Clonal diversity and detection of carbapenem resistance encoding genes among multidrug-resistant Acinetobacter baumannii isolates recovered from patients and environment in two intensive care units in a Moroccan hospital. Antimicrob. Resist. Infect. Control. 2017, 6, 99. [Google Scholar] [CrossRef]
- Castilho, S.R.A.; Godoy, C.S.M.; Guilarde, A.O.; Cardoso, J.L.; Andre, M.C.P.; Junqueira-Kipnis, A.P.; Kipnis, A. Acinetobacter baumannii strains isolated from patients in intensive care units in Goiania, Brazil: Molecular and drug susceptibility profiles. PLOS ONE 2017, 12, e0176790. [Google Scholar] [CrossRef]
- Teo, J.W.; Chew, K.L.; Lin, R.T. Transmissible colistin resistance encoded by mcr-1 detected in clinical Enterobacteriaceae isolates in Singapore. Emerg. Microbes Infect. 2016, 5, e87. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.J.; Doi, Y.; Patil, S.; Huang, X.; Tian, G.B. Emergence of the Plasmid-Mediated mcr-1 Gene in Colistin-Resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother 2016, 60, 3862–3863. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Bardet, L.; Dubourg, G.; Fichaux, M.; Rolain, J.M. mcr-1 plasmid-mediated colistin resistance gene detection in an Enterobacter cloacae clinical isolate in France. J. Glob. Antimicrob. Resist. 2017, 10, 35–36. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Mazzacane, S. Spread of mcr-1-Driven Colistin Resistance on Hospital Surfaces, Italy. Emerg Infect Dis 2018, 24, 1752–1753. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Wu, S.J.; Chang, T.W.; Wang, C.F.; Suen, C.S.; Hwang, M.J.; Chang, M.D.; Chen, Y.T.; Liao, Y.D. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J. Biol. Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef] [PubMed]
- Khattab, M.; Nour, M.; ElSheshtawy, N. Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. J. Microb. Biochem. Technol. 2015, 7, 274–277. [Google Scholar]
- Heidary, Z.; Bandani, E.; Eftekhary, M.; Jafari, A.A. Virulence genes profile of multidrug resistant Pseudomonas aeruginosa isolated from Iranian children with UTIs. Acta Med. Iran. 2016, 54, 201–210. [Google Scholar] [PubMed]
- Al-Dahmoshi, H.O.; Al-Khafaji, N.S.; Jeyad, A.A.; Shareef, H.K.; Al-Jebori, R.F. Molecular detection of some virulence traits among Pseudomonas aeruginosa isolates, Hilla-Iraq. Biomed. Pharmacol. J. 2018, 11, 835–842. [Google Scholar] [CrossRef]
- Galdino, A.C.M.; Branquinha, M.H.; Santos, A.L.; Viganor, L. Pseudomonas aeruginosa and its arsenal of proteases: Weapons to battle the host. In Pathophysiological Aspects of Proteases; Chakraborti, S., Dhalla, N., Eds.; Springer: Singapore, 2017; pp. 381–397. [Google Scholar]
- Yousefi-Avarvand, A.; Khashei, R.; Ebrahim-Saraie, H.S.; Emami, A.; Zomorodian, K.; Motamedifar, M. The frequency of exotoxin A and exoenzymes S and U genes among clinical isolates of Pseudomonas aeruginosa in Shiraz, Iran. Int. J. Mol. Cell. Med. 2015, 4, 167–173. [Google Scholar]
- Adwan, G. Detection of Type III secretion toxins encoding-genes of Pseudomonas aeruginosa isolates in the West Bank-Palestine. J. Adv. Biol. Biotechnol. 2017, 11, 1–10. [Google Scholar] [CrossRef][Green Version]
- Mahdavi, M.; Zahraei Salehi, T.; Amini, K.; Mobasseri, P. Frequency of exoY, exoS, exoT and exoU genes among Pseudomonas aeruginosa Isolated from patients in Tehran hospitals by Multiplex PCR. Iran. J. Med. Microbiol. 2017, 11, 9–17. [Google Scholar]
- Dong, D.; Zou, D.; Liu, H.; Yang, Z.; Huang, S.; Liu, N.; He, X.; Liu, W.; Huang, L. Rapid detection of Pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front. Microbiol. 2015, 6, 1100. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.R.; Linker, A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [CrossRef] [PubMed]
- May, T.B.; Shinabarger, D.; Maharaj, R.; Kato, J.; Chu, L.; DeVault, J.D.; Roychoudhury, S.; Zielinski, N.A.; Berry, A.; Rothmel, R.K.; et al. Alginate synthesis by Pseudomonas aeruginosa: A key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 1991, 4, 191–206. [Google Scholar] [CrossRef]
- Mitov, I.; Strateva, T.; Markova, B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz. J. Microbiol. 2010, 41, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Rashno Taee, S.; Khansarinejad, B.; Abtahi, H.; Najafimosleh, M.; Ghaznavi-Rad, E. Detection of algD, oprL and exoA Genes by New Specific Primers as an Efficient, Rapid and Accurate Procedure for Direct Diagnosis of Pseudomonas aeruginosa Strains in Clinical Samples. Jundishapur J. Microbiol. 2014, 7, e13583. [Google Scholar] [CrossRef] [PubMed]
- Habibi, A.; Honarmand, R. Profile of Virulence Factors in the Multi-Drug Resistant Pseudomonas aeruginosa Strains of Human Urinary Tract Infections (UTI). Iran. Red Crescent Med. J. 2015, 17, e26095. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elmaraghy, N.; Abbadi, S.; Elhadidi, G.; Hashem, A.; Yousef, A. Virulence genes in Pseudomonas aeruginosa strains isolated at Suez Canal University Hospitals with respect to the site of infection and antimicrobial resistance. Int. J. Clin. Microbiol. Biochem. Technol. 2019, 2, 8–19. [Google Scholar]
- Cotar, A.I.; Chifiriuc, M.C.; Dinu, S.; Bucur, M.; Iordache, C.; Banu, O.; Dracea, O.; Larion, C.; Lazar, V. Screening of molecular virulence markers in Staphylococcus aureus and Pseudomonas aeruginosa strains isolated from clinical infections. Int. J. Mol. Sci. 2010, 11, 5273–5291. [Google Scholar] [CrossRef]
- Ertugrul, B.M.; Oryasin, E.; Lipsky, B.A.; Willke, A.; Bozdogan, B. Virulence genes fliC, toxA and phzS are common among Pseudomonas aeruginosa isolates from diabetic foot infections. Infect. Dis. 2017, 50, 273–279. [Google Scholar] [CrossRef]
- Fadhil, L.; Al-Marzoqi, A.H.; Al Taee, Z.M.; Shalan, A.A. Molecular and phenotypic study of virulence genes in a pathogenic strain of Pseudomonas aeruginosa isolated from various clinical origins by PCR: Profiles of genes and toxins. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 590–598. [Google Scholar]
- Ra’oof, W.A.M. Distribution of algD, lasB, pilB and nan1 genes among MDR clinical isolates of Pseudomonas aeruginosa in respect to site of infection. Tikrit Med. J. 2011, 17, 148–160. [Google Scholar]
- Nahar, N.; Asad, S.; Ahmed, T.; Setu, N.I.; Kayser, M.S.; Islam, M.S.; Kamrul, M.; Islam, M.M.R.; Al Aman, D.A.; Rashid, R.B. In silico assessment of the genotypic distribution of virulence and antibiotic resistance genes in Pseudomonas aeruginosa. J. Appl. Pharm. Sci. 2017, 7, 55–61. [Google Scholar]
- Ahmed, M.U.; Dunn, L.; Ivanova, E.P. Evaluation of current molecular approaches for genotyping of Campylobacter jejuni strains. Foodborne Pathog. Dis. 2012, 9, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Tabatabaee, A.; Behzadi, P.; Kheiri, R. Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of Escherichia coli strains isolated from different animal stool specimens. Iran. J. Pathol. 2017, 12, 25–34. [Google Scholar] [CrossRef] [PubMed]
| Groups (for mPCR) | Target Gene | Primers Sequence (5′-3′) | Amplicon Size (pb) | Virulence Fator (Gene Product) | References |
|---|---|---|---|---|---|
| Group I | exoS | CTTGAAGGGACTCGACAAGG (F) TTCAGGTCCGCGTAGTGAAT (R) | 504 | Exotoxin S | [19] |
| exoT | CAATCATCTCAGCAGAACCC (F) TGTCGTAGAGGATCTCCTG | 1159 | Exotoxin T | [20] | |
| phzI | CATCAGCTTAGCAATCCC (F) CGGAGAAACTTTTCCCTC (R) | 392 | Phenazine prodution | [20] | |
| phzM | ATGGAGAGCGGGATCGACAG (F) ATGCGGGTTTCCATCGGCAG (R) | 875 | Phenazine prodution | [20] | |
| apr | TGTCCAGCAATTCTCTTGC (F) CGTTTTCCACGGTGACC (R) | 1017 | Protease | [20] | |
| pilA | ACAGCATCCAACTGAGCG (F) TTGACTTCCTCCAGGCTG (R) | 1675 | Type IV fimbria | [20] | |
| Group II | oprI | ATGAACAACGTTCTGAAATTCTCTGCT (F) CTTGCGGCTGGCTTTTTCCAG (R) | 248 | Outer membrane lipoprotein I | [21] |
| oprL | ATGGAAATGCTGAAATTCGGC (F) CTTCTTCAGCTCGACGCGACG (R) | 504 | Peptidoglycan-associated lipoprotein | [21] | |
| pilB | TCGAACTGATGATCGTGG (F) CTTTCGGAGTGAACATCG (R) | 408 | Fimbrial protein | [20] | |
| nan | AGGATGAATACTTATTTTGAT (F) TCACTAAATCCATCTCTGACCCGATA (R) | 1316 | Putative neuraminidase | [19] | |
| algD | AAGGCGGAAATGCCATCTCC (F) AGGGAAGTTCCGGGCGTTTG (R) | 300 | GDP-mannose 6-dehydrogenase (alginate production) | [22] | |
| plcH | GCACGTGGTCATCCTGATGC (F) TCCGTAGGCGTCGACGTAC (R) | 608 | Hemolytic phospholipase C | [23] | |
| exoY | TATCGACGGTCATCGTCAGGT (F) TTGATGCACTCGACCAGCAAG (R) | 1035 | Adenylate cyclase | [1] | |
| Group III | toxA | GACAACGCCCTCAGCATCACCAGC (F) CGCTGGCCCATTCGCTCCAGCGCT (R) | 396 | Exotoxin A | [24] |
| lasB | GAATGAACGAAGCGTTCTCCGAC (F) TGGCGTCGACGAACACCTCG (R) | 284 | Protease | [1] | |
| aprA | GTCGACCAGGCGGCGGAGCAGATA (F) GCCGAGGCCGCCGTAGAGGATGTC (R) | 993 | Alkaline protease | [23] | |
| Prot IV | TATTTCGCCGACTCCCTGTA (F) GAATAGACGCCGCTGAAATC (R) | 752 | Protease type IV | [25] | |
| plcN | TCCGTTATCGCAACCAGCCCTACG (F) TCGCTGTCGAGCAGGTCGAAC (R) | 481 | Non-hemolytic phospholipase C | [26] |
| Groups (for mPCR) | Target Gene | Primer Sequence (5′-3) | Amplicon Size (pb) | Gene Product | Reference |
|---|---|---|---|---|---|
| Group IV | blaOxa23 | GATCGGATTGGAGAACCAGA (F) ATTTCTGACCGCATTTCCAT (R) | 501 | Oxacilinase | [27] |
| blaOxa24 | GGTTAGTTGGCCCCCTTAAA (F) AGTTGAGCGAAAAGGGGATT (R) | 246 | Oxacilinase | [27] | |
| blaOxa51 | TAATGCTTTGATCGGCCTTG (F) TGGATTGCACTTCATCTTGG (R) | 353 | Oxacilinase | [27] | |
| blaOxa58 | AAGTATTGGGGCTTGTGCTG (F) CCCCTCTGCGCTCTACATAC (R) | 599 | Oxacilinase | [27] | |
| Group V | ampC | CGGCTCGGTGAGCAAGACCTTC (F) AGTCGCGGATCTGTGCCTGGTC (R) | 218 | Cephalosporinase | [28] |
| mcr-1 | CAGTATAATTGCCGTAATTATCCCACCGT (F) GTCTCGGCTTGGTCGGTCTGTAG (R) | 400 | LPS-modifying enzyme | This study |
| Gender | Tissue Fragment (n = 11)/% | Blood Culture (n = 40)/% | Tracheal Secretion (n = 37)/% | Urine (n = 11)/% | p Value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Male | 4 | 36.4 | 18 | 45 | 21 | 56.8 | 8 | 72.7 | ns |
| Female | 7 | 63.6 | 22 | 55 | 16 | 43.2 | 3 | 27.3 | ns |
| Age group (years) | p < 0.05 | ||||||||
| up to 11 | 0 | - | 7 | 17.5 | - | - | - | - | |
| 12 to 19 | 0 | - | 1 | 2.5 | 2 | 5.4 | 1 | 9.1 | |
| 20 to 60 | 3 | 27.3 | 9 | 22.5 | 11 | 29.7 | 3 | 27.3 | |
| over 60 (years) | 8 | 72.7 | 23 | 57.5 | 24 | 64.9 | 7 | 63.6 | |
| Antimicrobial | MIC (mg/mL) | Number of Isolates | % Resistance |
|---|---|---|---|
| Cefoxitin | ≥64 | 99 | 100.0 |
| Ceftazidime | ≥64 | 70 | 70.7 |
| Cefepime | ≥64 | 61 | 61.6 |
| Ampicilin/sulbactam | ≥32/16 | 99 | 100.0 |
| Piperacilin/tazobactam | ≥128/4 | 74 | 74.7 |
| Aztreonam | ≥32 | 39 | 39.4 |
| Ciprofloxacin | ≥4 | 66 | 66.7 |
| Gentamicin | ≥16 | 58 | 58.6 |
| Amikacin | ≥64 | 44 | 44.4 |
| Imipenem | ≥16 | 57 | 57.6 |
| Meropenem | ≥16 | 56 | 56.6 |
| Colistin | ≥8 | 1 | 1.0 |
| Polymyxin B | ≤0.5 | 0 | 0.0 |
| Tigecycline | ≥16 | 99 | 100.0 |
| Fragment of Tissue | Blood Culture | Tracheal Secretion | Urine | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | No | % | ||
| Amikacin | 5 | 45.5 | 16 | 40.0 | 17 | 45.9 | 6 | 54.5 | ns |
| Gentamycin | 6 | 54.5 | 17 | 42.5 | 25 | 67.6 | 10 | 90.9 | <0.05 |
| Cefepime | 10 | 90.9 | 19 | 47.5 | 25 | 67.6 | 7 | 63.6 | <0.05 |
| Cefotaxime | 11 | 100.0 | 40 | 100 | 37 | 100.0 | 11 | 100.0 | <0.05 |
| Cefoxitin | 11 | 100.0 | 40 | 100 | 37 | 100.0 | 11 | 100.0 | <0.05 |
| Ceftazidime | 11 | 100.0 | 19 | 47.5 | 31 | 83.8 | 9 | 81.8 | <0.05 |
| Imipenem | 10 | 90.9 | 9 | 22.5 | 30 | 81.1 | 8 | 72.7 | <0.05 |
| Meropenem | 10 | 90.9 | 9 | 22.5 | 29 | 78.4 | 8 | 72.7 | <0.05 |
| Ampicillin/Sulbactam | 11 | 100.0 | 40 | 100 | 37 | 100.0 | 11 | 100.0 | ns |
| Piperacillin/Tazobactam | 9 | 81.8 | 25 | 62.5 | 30 | 81.1 | 10 | 90.9 | ns |
| Aztreonam | 9 | 81.8 | 7 | 17.5 | 17 | 45.9 | 6 | 54.5 | <0.05 |
| Ciprofloxacin | 11 | 100.0 | 22 | 55.0 | 25 | 67.6 | 8 | 72.7 | <0.05 |
| Polymyxin B | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | ns |
| Polymyxin E | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 1.0 | |
| Tigecycline | 11 | 100.0 | 40 | 100.0 | 37 | 100.0 | 11 | 100.0 | ns |
| Clinical Specimen | Genes Detected in Clinical Isolates of Pseudomonas aeruginosa | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oprl | oprL | lasB | exoY | toxA | exoT | algD | plcN | phzl | plcH | pilA | ExoS | phzM | pilB | apr | Prot IV | nan-1 | aprA | |
| Blood culture | 40 | 40 | 34 | 32 | 31 | 30 | 23 | 32 | 27 | 11 | 1 | 15 | 13 | 5 | 7 | 5 | 6 | 5 |
| Tracheal secretion | 37 | 37 | 35 | 35 | 34 | 33 | 32 | 26 | 29 | 25 | 6 | 19 | 13 | 13 | 10 | 10 | 5 | 0 |
| Urine | 11 | 11 | 9 | 9 | 6 | 8 | 9 | 8 | 6 | 7 | 2 | 5 | 2 | 3 | 1 | 2 | 3 | 1 |
| Fragment of tissue | 11 | 11 | 11 | 11 | 10 | 11 | 11 | 9 | 8 | 8 | 5 | 6 | 6 | 6 | 4 | 3 | 0 | 0 |
| Total genes | 99 | 99 | 89 | 87 | 81 | 82 | 75 | 75 | 70 | 51 | 14 | 45 | 34 | 27 | 22 | 20 | 14 | 6 |
| Clades | Base Pair Fragments (bp) | Number of Isolates in Each Group |
|---|---|---|
| G01 | 350–500–1000 | 8 |
| G02 | 350–650–1000 | 5 |
| G03 | 150–350-650–1000 | 7 |
| G04 | 150–350-400 | 13 |
| G05 | 250–350–750 | 2 |
| G06 | 250–350–750–1000 | 7 |
| G07 | 250–350–750–1000–2500 | 5 |
| G08 | 250–350–750–1000–1300–2500 | 3 |
| G09 | 150–250–350–400 | 5 |
| G10 | 350–650–850–2000 | 2 |
| G11 | 350–400–550–700–800–1100–2000 | 2 |
| G12 | 350–400–700–800–1100–2000 | 2 |
| G13 | 350–550–800–1100–2000 | 5 |
| Isolated in brackets | Isolates with heterogeneous profiles with different base pair sizes | 33 |
| Total | 99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nitz, F.; de Melo, B.O.; da Silva, L.C.N.; de Souza Monteiro, A.; Marques, S.G.; Monteiro-Neto, V.; de Jesus Gomes Turri, R.; Junior, A.D.S.; Conceição, P.C.R.; Magalhães, H.J.C.; et al. Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa. Microorganisms 2021, 9, 786. https://doi.org/10.3390/microorganisms9040786
Nitz F, de Melo BO, da Silva LCN, de Souza Monteiro A, Marques SG, Monteiro-Neto V, de Jesus Gomes Turri R, Junior ADS, Conceição PCR, Magalhães HJC, et al. Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa. Microorganisms. 2021; 9(4):786. https://doi.org/10.3390/microorganisms9040786
Chicago/Turabian StyleNitz, Fabiana, Bruna Oliveira de Melo, Luís Cláudio Nascimento da Silva, Andrea de Souza Monteiro, Sirlei Garcia Marques, Valério Monteiro-Neto, Rosimary de Jesus Gomes Turri, Antonio Dantas Silva Junior, Patrícia Cristina Ribeiro Conceição, Hilário José Cardoso Magalhães, and et al. 2021. "Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa" Microorganisms 9, no. 4: 786. https://doi.org/10.3390/microorganisms9040786
APA StyleNitz, F., de Melo, B. O., da Silva, L. C. N., de Souza Monteiro, A., Marques, S. G., Monteiro-Neto, V., de Jesus Gomes Turri, R., Junior, A. D. S., Conceição, P. C. R., Magalhães, H. J. C., Zagmignan, A., Ferro, T. A. F., & Bomfim, M. R. Q. (2021). Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa. Microorganisms, 9(4), 786. https://doi.org/10.3390/microorganisms9040786









