Human Endogenous Retroviruses in Glioblastoma Multiforme
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncology 2015, 17 (Suppl. 4), iv1–iv62. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H.; Kleihues, P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005, 109, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.; LaBombardi, V.; Green, S.; Pessin-Minsley, M.S.; Germano, I.M.; Rosenzweig, K.E. No circulating cytomegalovirus in five patients with glioblastoma multiforme. Anticancer Res. 2011, 31, 959–960. [Google Scholar] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 2018, 20 (Suppl. 4), iv1–iv86. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.L.; Schwartzbaum, J.A.; Wrensch, M.; Wiemels, J.L. Epidemiology of brain tumors. Neurol. Clin. 2007, 25, 867–890. [Google Scholar] [CrossRef] [PubMed]
- Barchana, M.; Margaliot, M.; Liphshitz, I. Changes in Brain Glioma Incidence and Laterality Correlates with Use of Mobile Phones–A Nationwide Population Based Study in Israel. Asian Pac. J. Cancer Prev. 2012, 13, 5857–5863. [Google Scholar] [CrossRef] [PubMed]
- Deltour, I.; Auvinen, A.; Feychting, M.; Johansen, C.; Klaeboe, L.; Sankila, R.; Schüz, J. Mobile Phone Use and Incidence of Glioma in the Nordic Countries 1979–2008: Consistency Check. Epidemiology 2012, 23, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Little, M.P.; Azizova, T.V.; Bazyka, D.; Bouffler, S.D.; Cardis, E.; Chekin, S.; Chumak, V.V.; Cucinotta, F.A.; de Vathaire, F.; Hall, P. Systematic review and meta-analysis of circulatory disease from exposure to low-level ionizing radiation and estimates of potential population mortality risks. Environ. Health Perspect. 2012, 120, 1503–1511. [Google Scholar] [CrossRef]
- Cobbs, C.S.; Harkins, L.; Samanta, M.; Gillespie, G.Y.; Bharara, S.; King, P.H.; Nabors, L.B.; Cobbs, C.G.; Britt, W.J. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002, 62, 3347–3350. [Google Scholar]
- Wrensch, M.; Minn, Y.; Chew, T.; Bondy, M.; Berger, M.S. Epidemiology of primary brain tumors: Current concepts and review of the literature. Neuro-Oncology 2002, 4, 278–299. [Google Scholar] [CrossRef]
- Saddawi-Konefka, R.; Crawford, J.R. Chronic Viral Infection and Primary Central Nervous System Malignancy. J. Neuroimmune Pharmacol. 2010, 5, 387–403. [Google Scholar] [CrossRef][Green Version]
- Alexander, B.M.; Cloughesy, T.F. Adult glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Hochhalter, C.B.; Carr, C.; O’Neill, B.E.; Ware, M.L.; Strong, M.J. The association between human cytomegalovirus and glioblastomas: A review. Neuroimmunol. Neuroinflammation 2017, 4, 96. [Google Scholar] [CrossRef]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef]
- Nelson, P.N.; Carnegie, P.; Martin, J.; Ejtehadi, H.D.; Hooley, P.; Roden, D.; Rowland-Jones, S.; Warren, P.; Astley, J.; Murray, P.G. Demystified… Human endogenous retroviruses. Mol. Pathol. 2003, 56, 11. [Google Scholar] [CrossRef]
- Kojima, K.K. Human transposable elements in Repbase: Genomic footprints from fish to humans. Mob. DNA 2018, 9, 2. [Google Scholar] [CrossRef]
- Lander, E.S. Initial sequencing and analysis of the human genome. International Human Genome Sequencing Consortium. Nature 2001, 409, 860–921. [Google Scholar]
- Cegolon, L.; Salata, C.; Weiderpass, E.; Vineis, P.; Palù, G.; Mastrangelo, G. Human endogenous retroviruses and cancer prevention: Evidence and prospects. BMC Cancer 2013, 13, 4. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. 2), 14572–14579. [Google Scholar] [CrossRef]
- Mager, D.; Stoye, J. Mammalian endogenous retroviruses. Microbiol. Spectr. 2015, 3, MDNA3-0009-2014. [Google Scholar] [CrossRef]
- Gilboa, E.; Mitra, S.W.; Goff, S.; Baltimore, D. A detailed model of reverse transcription and tests of crucial aspects. Cell 1979, 18, 93–100. [Google Scholar] [CrossRef]
- Swanstrom, R.; DeLorbe, W.J.; Bishop, J.M.; Varmus, H.E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: Viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc. Natl. Acad. Sci. USA 1981, 78, 124–128. [Google Scholar] [CrossRef]
- Yamamoto, T.; De Crombrugghe, B.; Pastan, I. Identification of a functional promoter in the long terminal repeat of Rous sarcoma virus. Cell 1980, 22, 787–797. [Google Scholar] [CrossRef]
- Jern, P.; Coffin, J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef]
- Leib-Mösch, C.; Seifarth, W.; Schön, U. Influence of human endogenous retroviruses on cellular gene expression. In Retroviruses and Primate Genome Evolution; Sverdlov, E., Ed.; Landes Bioscience: Georgetown, TX, USA, 2004; pp. 124–145. [Google Scholar]
- Medstrand, P.; Landry, J.-R.; Mager, D.L. Long Terminal Repeats Are Used as Alternative Promoters for the Endothelin B Receptor and Apolipoprotein C-I Genes in Humans. J. Biol. Chem. 2001, 276, 1896–1903. [Google Scholar] [CrossRef]
- Boller, K.; Schönfeld, K.; Lischer, S.; Fischer, N.; Hoffmann, A.; Kurth, R.; Tönjes, R.R. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 2008, 89, 567–572. [Google Scholar] [CrossRef]
- Faff, O.; Murray, A.B.; Schmidt, J.; Leib-Mösch, C.; Erfle, V.; Hehlmann, R. Retrovirus-like particles from the human T47D cell line are related to mouse mammary tumour virus and are of human endogenous origin. J. Gen. Virol. 1992, 73, 1087–1097. [Google Scholar] [CrossRef]
- Andersson, A.C.; Svensson, A.C.; Rolny, C.; Andersson, G.; Larsson, E. Expression of human endogenous retrovirus ERV3 (HERV-R) mRNA in normal and neoplastic tissues. Int. J. Oncol. 1998, 12, 309–322. [Google Scholar] [CrossRef]
- Löwer, R.; Löwer, J.; Tondera-Koch, C.; Kurth, R. A General Method for the Identification of Transcribed Retrovirus Sequences (R-U5 PCR) Reveals the Expression of the Human Endogenous Retrovirus Loci HERV-H and HERV-K in Teratocarcinoma Cells. Virology 1993, 192, 501–511. [Google Scholar] [CrossRef]
- Larsson, E.; Andersson, G. Beneficial Role of Human Endogenous Retroviruses: Facts and Hypotheses. Scand. J. Immunol. 1998, 48, 329–338. [Google Scholar] [CrossRef]
- Lindeskog, M.; Medstrand, P.; Blomberg, J. Sequence variation of human endogenous retrovirus ERV9-related elements in an env region corresponding to an immunosuppressive peptide: Transcription in normal and neoplastic cells. J. Virol. 1993, 67, 1122–1126. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Roemer, K.; Best, B.; Afting, M.; Schommer, S.; Seitz, G.; Hartmann, M.; Mueller-Lantzsch, N. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 1996, 56, 4362–4365. [Google Scholar] [PubMed]
- Sauter, M.; Schommer, S.; Kremmer, E.; Remberger, K.; Dölken, G.; Lemm, I.; Buck, M.; Best, B.; Neumann-Haefelin, D.; Mueller-Lantzsch, N. Human endogenous retrovirus K10: Expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 1995, 69, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Schiavetti, F.; Thonnard, J.; Colau, D.; Boon, T.; Coulie, P.G. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002, 62, 5510–5516. [Google Scholar] [PubMed]
- Contreras-Galindo, R.; Kaplan, M.H.; Leissner, P.; Verjat, T.; Ferlenghi, I.; Bagnoli, F.; Giusti, F.; Dosik, M.H.; Hayes, D.F.; Gitlin, S.D.; et al. Human Endogenous Retrovirus K (HML-2) Elements in the Plasma of People with Lymphoma and Breast Cancer. J. Virol. 2008, 82, 9329–9336. [Google Scholar] [CrossRef]
- Pichon, J.-P.; Bonnaud, B.; Cleuziat, P.; Mallet, F. Multiplex degenerate PCR coupled with an oligo sorbent array for human endogenous retrovirus expression profiling. Nucleic Acids Res. 2006, 34, e46. [Google Scholar] [CrossRef][Green Version]
- Wang-Johanning, F.; Liu, J.; Rycaj, K.; Huang, M.; Tsai, K.; Rosen, D.G.; Chen, D.-T.; Lu, D.W.; Barnhart, K.F.; Johanning, G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 2006, 120, 81–90. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Frost, A.R.; Jian, B.; Azerou, R.; Lu, D.W.; Chen, D.T.; Johanning, G.L. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer 2003, 98, 187–197. [Google Scholar] [CrossRef]
- Gimenez, J.; Montgiraud, C.; Pichon, J.-P.; Bonnaud, B.; Arsac, M.; Ruel, K.; Bouton, O.; Mallet, F. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010, 38, 2229–2246. [Google Scholar] [CrossRef]
- Pérot, P.; Mullins, C.S.; Naville, M.; Bressan, C.; Hühns, M.; Gock, M.; Kuhn, F.; Volff, J.-N.; Trillet-Lenoir, V.; Linnebacher, M.; et al. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget 2015, 6, 40095–40111. [Google Scholar] [CrossRef]
- Perron, H.; Geny, C.; Laurent, A.; Mouriquand, C.; Pellat, J.; Perret, J.; Seigneurin, J. Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particles. Res. Virol. 1989, 140, 551–561. [Google Scholar] [CrossRef]
- McCormick, A.L.; Brown, R.H.; Cudkowicz, M.E.; Al-Chalabi, A.; Garson, J.A. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology 2008, 70, 278–283. [Google Scholar] [CrossRef]
- Yolken, R.H.; Karlsson, H.; Yee, F.; Johnston-Wilson, N.; Torrey, E. Endogenous retroviruses and schizophrenia. Brain Res. Rev. 2000, 31, 193–199. [Google Scholar] [CrossRef]
- Singh, S.K. Endogenous retroviruses: Suspects in the disease world. Futur. Microbiol. 2007, 2, 269–275. [Google Scholar] [CrossRef]
- Gruchot, J.; Kremer, D.; Küry, P. Neural Cell Responses Upon Exposure to Human Endogenous Retroviruses. Front. Genet. 2019, 10, 655. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Klein, J.; Acikelli, A.H.; Wilk, C.; Saka, S.; Jastrow, H.; Wennemuth, G.; Dammann, P.; Giger-Pabst, U.; Khosrawipour, V.; et al. Cytotoxic stress induces transfer of mitochondria-associated human endogenous retroviral RNA and proteins between cancer cells. Oncotarget 2017, 8, 95945–95964. [Google Scholar] [CrossRef]
- Diem, O.; Schäffner, M.; Seifarth, W.; Leib-Mösch, C. Influence of antipsychotic drugs on human endogenous retrovirus (HERV) transcription in brain cells. PLoS ONE 2012, 7, e30054. [Google Scholar] [CrossRef]
- Flockerzi, A.; Ruggieri, A.; Frank, O.; Sauter, M.; Maldener, E.; Kopper, B.; Wullich, B.; Seifarth, W.; Müller-Lantzsch, N.; Leib-Mösch, C. Expression patterns of transcribed human endogenous retrovirus HERV-K (HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genom. 2008, 9, 354. [Google Scholar] [CrossRef]
- Kessler, A.F.; Wiesner, M.; Denner, J.; Kämmerer, U.; Vince, G.H.; Linsenmann, T.; Löhr, M.; Ernestus, R.I.; Hagemann, C. Expression-analysis of the human endogenous retrovirus HERV-K in human astrocytic tumors. BMC Res. Notes 2014, 7, 159. [Google Scholar] [CrossRef]
- Zhu, L.; Zheng, W.J. Informatics, data science, and artificial intelligence. JAMA 2018, 320, 1103–1104. [Google Scholar] [CrossRef]
- Criscione, S.W.; Zhang, Y.; Thompson, W.; Sedivy, J.M.; Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genom. 2014, 15, 583. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, G.J.; Forrest, A.R.; Chalk, A.M.; Schroder, K.; Hayashizaki, Y.; Carninci, P.; Hume, D.A.; Grimmond, S.M. A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics 2008, 91, 281–288. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Huda, A.; Lunyak, V.V.; Jordan, I.K. A Gibbs sampling strategy applied to the mapping of ambiguous short-sequence tags. Bioinformatics 2010, 26, 2501–2508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Law, C.W.; Alhamdoosh, M.; Su, S.; Dong, X.; Tian, L.; Smyth, G.K.; Ritchie, M.E. RNA-Seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Morpheus. Available online: https://software.broadinstitute.org/morpheus (accessed on 27 February 2021).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, A.D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Anand, L. chromoMap: An R package for Interactive Visualization and Annotation of Chromosomes. bioRxiv 2019, 605600. [Google Scholar] [CrossRef]
- Yang, D.; Jang, I.; Choi, J.; Kim, M.-S.; Lee, A.J.; Kim, H.; Eom, J.; Kim, D.; Jung, I.; Lee, B. 3DIV: A 3D-genome Interaction Viewer and database. Nucleic Acids Res. 2018, 46, D52–D57. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019, 48, D845–D855. [Google Scholar] [CrossRef]
- Lättekivi, F.; Kõks, S.; Keermann, M.; Reimann, E.; Prans, E.; Abram, K.; Silm, H.; Kõks, G.; Kingo, K. Transcriptional landscape of human endogenous retroviruses (HERVs) and other repetitive elements in psoriatic skin. Sci. Rep. 2018, 8, 4358. [Google Scholar] [CrossRef]
- Ho, X.D.; Nguyen, H.G.; Trinh, L.H.; Reimann, E.; Prans, E.; Kõks, G.; Maasalu, K.; Le, V.Q.; Nguyen, V.H.; Le, N.T.N.; et al. Analysis of the Expression of Repetitive DNA Elements in Osteosarcoma. Front. Genet. 2017, 8, 193. [Google Scholar] [CrossRef]
- Bralten, L.B.C.; Gravendeel, A.M.; Kloosterhof, N.K.; Sacchetti, A.; Vrijenhoek, T.; Veltman, J.A.; Bent, M.J.V.D.; Kros, J.M.; Hoogenraad, C.C.; Smitt, P.A.E.S.; et al. The CASPR2 cell adhesion molecule functions as a tumor suppressor gene in glioma. Oncogene 2010, 29, 6138–6148. [Google Scholar] [CrossRef]
- Sakthikumar, S.; Roy, A.; Haseeb, L.; Pettersson, M.E.; Sundström, E.; Marinescu, V.D.; Lindblad-Toh, K.; Forsberg-Nilsson, K. Whole-genome sequencing of glioblastoma reveals enrichment of non-coding constraint mutations in known and novel genes. Genome Biol. 2020, 21, 1–22. [Google Scholar] [CrossRef]
- Derks, J.; Reijneveld, J.C.; Douw, L. Neural network alterations underlie cognitive deficits in brain tumor patients. Curr. Opin. Oncol. 2014, 26, 627–633. [Google Scholar] [CrossRef]
- Heimans, J.J.; Reijneveld, J.C. Factors affecting the cerebral network in brain tumor patients. J. Neuro-Oncol. 2012, 108, 231–237. [Google Scholar] [CrossRef]
- Lee, Y.N.; Bieniasz, P.D. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007, 3, e10. [Google Scholar] [CrossRef]
- Guenette, S.Y.; Chen, J.; Jondro, P.D.; Tanzi, R.E. Association of a novel human FE65-like protein with the cytoplasmic domain of the beta-amyloid precursor protein. Proc. Natl. Acad. Sci. USA 1996, 93, 10832–10837. [Google Scholar] [CrossRef]
- Solans, A.; Estivill, X.; De La Luna, S. A new aspartyl protease on 21q22.3, BACE2, is highly similar to Alzheimer’s amyloid precursor protein beta-secretase. Cytogenet. Cell Genet. 2000, 89, 177–184. [Google Scholar] [CrossRef]
- Camporesi, E.; Lashley, T.; Gobom, J.; Lantero-Rodriguez, J.; Hansson, O.; Zetterberg, H.; Blennow, K.; Becker, B. Neuroligin-1 in brain and CSF of neurodegenerative disorders: Investigation for synaptic biomarkers. Acta Neuropathol. Commun. 2021, 9, 19. [Google Scholar] [CrossRef]
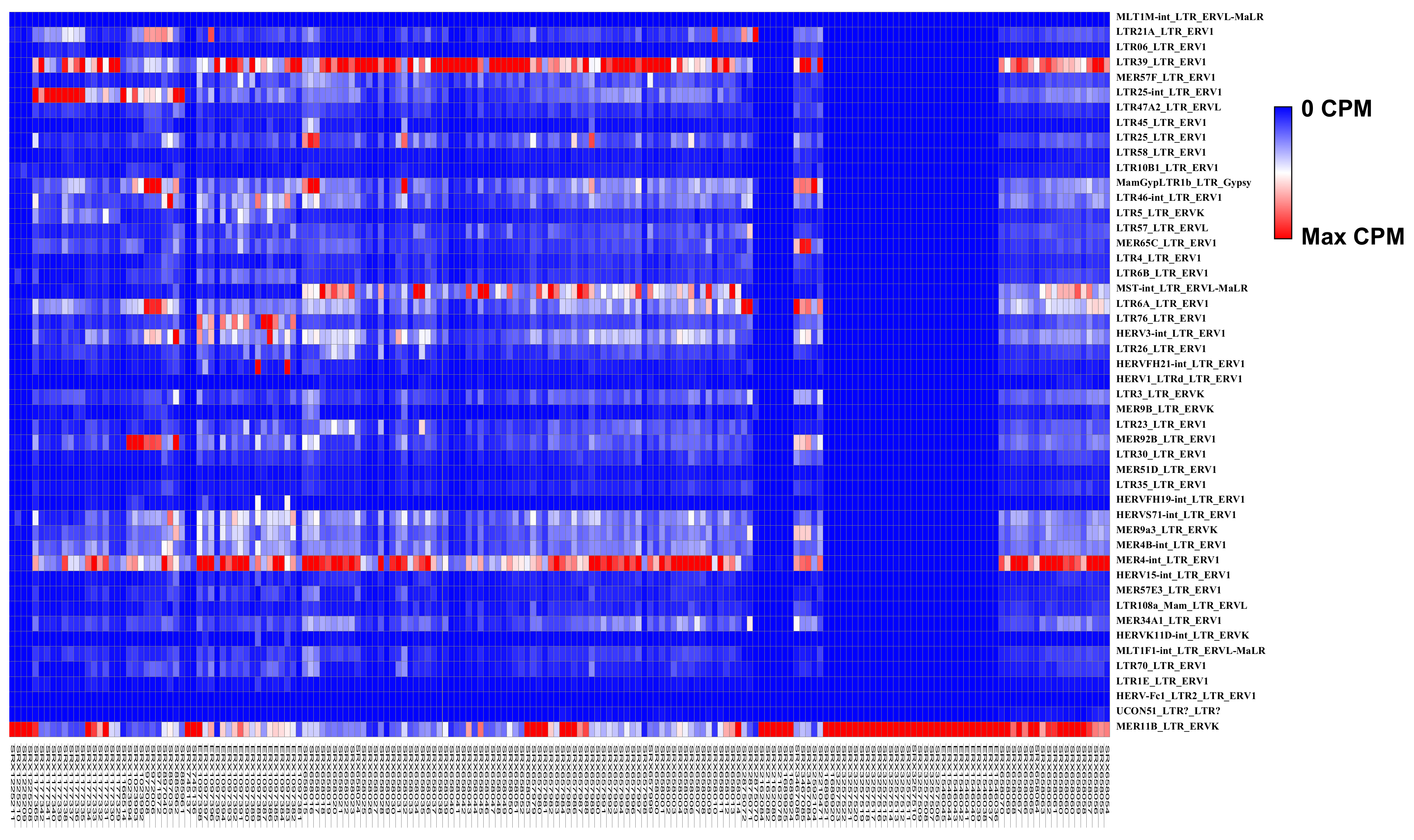




| LTR Elements | Superfamily | NB Mean CPM | GBM Mean CPM | Fold Changes GBM vs. NB |
|---|---|---|---|---|
| MLT1M-int_LTR_ERVL-MaLR | ERV3 | 0 | 0.03 | 13.85 |
| LTR21A_LTR_ERV1 | ERV1 | 48.57 | 249.19 | 5.13 |
| LTR06_LTR_ERV1 | ERV1 | 9.29 | 36.37 | 3.91 |
| LTR39_LTR_ERV1 | ERV1 | 281.42 | 947.75 | 3.37 |
| MER57F_LTR_ERV1 | ERV1 | 44.07 | 145.12 | 3.29 |
| LTR25-int_LTR_ERV1 | ERV1 | 98.72 | 324.11 | 3.28 |
| LTR47A2_LTR_ERVL | ERV3 | 26.76 | 87.46 | 3.27 |
| LTR45_LTR_ERV1 | ERV1 | 12.9 | 41.61 | 3.23 |
| LTR25_LTR_ERV1 | ERV1 | 53.83 | 161.48 | 3 |
| LTR58_LTR_ERV1 | ERV1 | 13.2 | 39.2 | 2.97 |
| LTR10B1_LTR_ERV1 | ERV1 | 31.4 | 92.43 | 2.94 |
| MamGypLTR1b_LTR_Gypsy | Gypsy | 106.56 | 305.69 | 2.87 |
| LTR46-int_LTR_ERV1 | ERV1 | 70.41 | 199.2 | 2.83 |
| LTR5_LTR_ERVK | ERV2 | 32.82 | 85.7 | 2.61 |
| LTR57_LTR_ERVL | ERV3 | 49.21 | 127.45 | 2.59 |
| MER65C_LTR_ERV1 | ERV1 | 60.88 | 156.72 | 2.57 |
| LTR4_LTR_ERV1 | ERV1 | 24.56 | 61.49 | 2.5 |
| LTR6B_LTR_ERV1 | ERV1 | 42.14 | 103.63 | 2.46 |
| MST-int LTR ERVL-MaLR | ERV3 | 132.56 | 320 | 2.41 |
| LTR6A_LTR_ERV1 | ERV1 | 137.45 | 325.28 | 2.37 |
| LTR76_LTR_ERV1 | ERV1 | 60.3 | 141.55 | 2.35 |
| HERV3-int_LTR_ERV1 | ERV1 | 118.4 | 275.59 | 2.33 |
| LTR26_LTR_ERV1 | ERV1 | 48.93 | 112.36 | 2.3 |
| HERVFH21-int_LTR_ERV1 | ERV1 | 22.23 | 50.74 | 2.28 |
| HERV1_LTRd_LTR_ERV1 | ERV1 | 6.46 | 14.7 | 2.28 |
| LTR3_LTR_ERVK | ERV2 | 65.17 | 145.43 | 2.23 |
| MER9B_LTR_ERVK | ERV2 | 16.12 | 35.17 | 2.18 |
| LTR23_LTR_ERV1 | ERV1 | 61.05 | 132.71 | 2.17 |
| MER92B_LTR_ERV1 | ERV1 | 96.38 | 209 | 2.17 |
| LTR30_LTR_ERV1 | ERV1 | 29.1 | 62.58 | 2.15 |
| MER51D_LTR_ERV1 | ERV1 | 11.86 | 25.37 | 2.14 |
| LTR35_LTR_ERV1 | ERV1 | 25.63 | 54.69 | 2.13 |
| HERVFH19-int_LTR_ERV1 | ERV1 | 10.32 | 21.85 | 2.12 |
| HERVS71-int_LTR_ERV1 | ERV1 | 122.04 | 256.2 | 2.1 |
| MER9a3_LTR_ERVK | ERV2 | 100.84 | 210.7 | 2.09 |
| MER4B-int_LTR_ERV1 | ERV1 | 105.81 | 221.07 | 2.09 |
| MER4-int_LTR_ERV1 | ERV1 | 317.48 | 656.31 | 2.07 |
| HERV15-int_LTR_ERV1 | ERV1 | 24.28 | 49.96 | 2.06 |
| MER57E3_LTR_ERV1 | ERV1 | 31.62 | 64.73 | 2.05 |
| LTR108a_Mam_LTR_ERVL | ERV3 | 25.92 | 52.95 | 2.04 |
| MER34A1_LTR_ERV1 | ERV1 | 77.33 | 157.29 | 2.03 |
| HERVK11D-int_LTR_ERVK | ERV2 | 6.01 | 12.2 | 2.03 |
| MLT1F1-int_LTR_ERVL-MaLR | ERV3 | 40.73 | 82.56 | 2.03 |
| LTR70_LTR_ERV1 | ERV1 | 37.51 | 75.05 | 2 |
| LTR1E_LTR_ERV1 | ERV1 | 45.06 | 21.22 | −2.12 |
| HERV-Fc1_LTR2_LTR_ERV1 | ERV1 | 0.34 | 0.16 | −2.16 |
| MER11B_LTR_ERVK | ERV2 | 25,765.36 | 3311.14 | −7.78 |
| Gene ID | Gene Name | DrugBank ID | Drug Name |
|---|---|---|---|
| SLC22A4 | solute carrier family 22 member 4 | DB00122 | Choline |
| DB00575 | Clonidine | ||
| DB01151 | Desipramine | ||
| DB00458 | Imipramine | ||
| DB01043 | Memantine | ||
| DB06691 | Mepyramine | ||
| DB00468 | Quinine | ||
| DB14754 | Solriamfetol | ||
| CA10 | carbonic anhydrase 10 | DB00909 | Zonisamide |
| GRIA2 | glutamate ionotropic receptor AMPA type subunit 2 | DB01351 | Amobarbital |
| DB00312 | Pentobarbital | ||
| DB00237 | Butabarbital | ||
| DB00241 | Butalbital | ||
| DB00306 | Talbutal | ||
| DB00418 | Secobarbital | ||
| DB00599 | Thiopental | ||
| DB00794 | Primidone | ||
| DB00849 | Methylphenobarbital | ||
| DB01174 | Phenobarbital | ||
| DB01353 | Butobarbital |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Yang, Y.; Zhang, N.; Soto, C.; Jiang, X.; An, Z.; Zheng, W.J. Human Endogenous Retroviruses in Glioblastoma Multiforme. Microorganisms 2021, 9, 764. https://doi.org/10.3390/microorganisms9040764
Yuan Z, Yang Y, Zhang N, Soto C, Jiang X, An Z, Zheng WJ. Human Endogenous Retroviruses in Glioblastoma Multiforme. Microorganisms. 2021; 9(4):764. https://doi.org/10.3390/microorganisms9040764
Chicago/Turabian StyleYuan, Zihao, Yuntao Yang, Ningyan Zhang, Claudio Soto, Xiaoqian Jiang, Zhiqiang An, and Wenjin Jim Zheng. 2021. "Human Endogenous Retroviruses in Glioblastoma Multiforme" Microorganisms 9, no. 4: 764. https://doi.org/10.3390/microorganisms9040764
APA StyleYuan, Z., Yang, Y., Zhang, N., Soto, C., Jiang, X., An, Z., & Zheng, W. J. (2021). Human Endogenous Retroviruses in Glioblastoma Multiforme. Microorganisms, 9(4), 764. https://doi.org/10.3390/microorganisms9040764








