Phylogenomic Insights into Distribution and Adaptation of Bdellovibrionota in Marine Waters
Abstract
1. Introduction
2. Methods
2.1. Genome Collection from Public Databases and Quality Control
2.2. Marine Water Samples Collection, DNA Extraction and Sequencing
2.3. Raw Data Processing, de Novo Assembly and Metagenomes Binning
2.4. Calculation of Bdellovibrionota Relative Abundance in Deep Sea
2.5. Genome Annotation and Metabolic Reconstruction
2.6. Phylogenetic Analysis of Conserved Proteins and 16S rRNA Genes
2.7. Availability of Data and Materials
3. Results
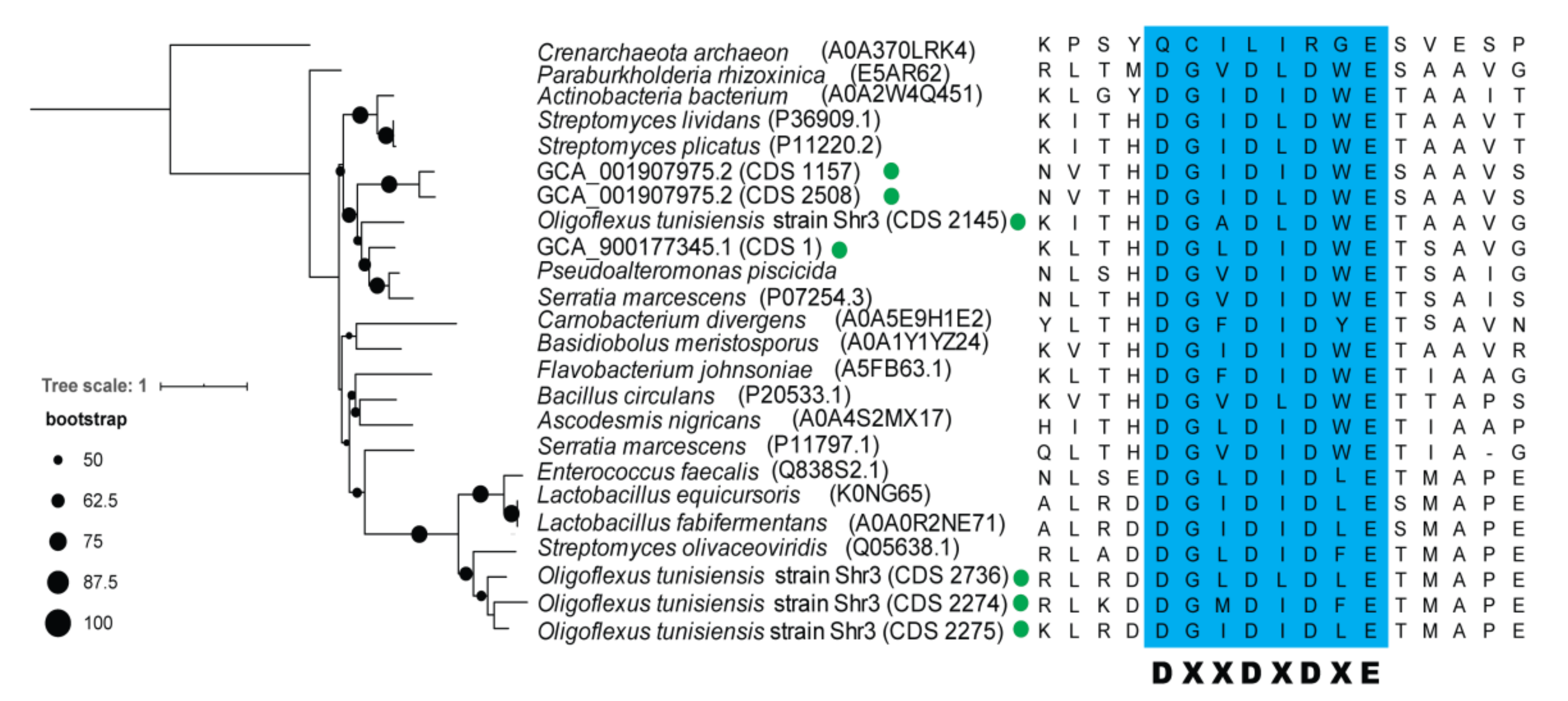
3.1. Phylogenomics of Bdellovibrionota
3.2. Relative Abundance of Bdellovibrionota in Marine Water Zones
3.3. Metabolic Potentials (Reconstruction) of Bdellovibrionota
3.4. Genes Involved in Survival of Bdellovibrionota in the Marine Environment
3.5. Oligoflexia as Potential Defender against Eukaryotic Pathogens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stolp, H.; Starr, M. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 1963, 29, 217–248. [Google Scholar] [CrossRef]
- Shilo, M.; Bruff, B. Lysis of Gram-Negative Bacteria by Host-Independent Ectoparasitic Bdellovibrio Bacteriovorus Isolates. J. Gen. Microbiol. 1965, 40, 317–318. [Google Scholar] [CrossRef]
- Thomashow, M.F.; Rittenberg, S.C. Intraperiplasmic Growth of Bdellovibrio-Bacteriovorus 109j - Solubilization of Escherichia-Coli Peptidoglycan. J. Bacteriol. 1978, 135, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Varon, M. Selection of Predation-Resistant Bacteria in Continuous Culture. Nature 1979, 277, 386–388. [Google Scholar] [CrossRef]
- Kadouri, D.E.; To, K.; Shanks, R.M.Q.; Doi, Y. Predatory Bacteria: A Potential Ally against Multidrug-Resistant Gram-Negative Pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef] [PubMed]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Onyile, O.; Rinaggio, J.; Connell, N.D.; et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio 2016, 7, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Cao, Q. New Bdellovibrio species mutant strain BDE-1F2 used for lysing pathogenic and/or potentially pathogenic bacteria associated with aquaculture, preferably Gram-negative and/or positive pathogenic bacteria e.g. Enterococcus. Univ. South China Technol. Patent number, CN111363689-A, 2020. [Google Scholar]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Jurkevitch, E.; Minz, D.; Ramati, B.; Barel, G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microb. 2000, 66, 2365–2371. [Google Scholar] [CrossRef]
- Sockett, R.E. Predatory Lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 2009, 63, 523–539. [Google Scholar] [CrossRef]
- Hobley, L.; Lerner, T.R.; Williams, L.E.; Lambert, C.; Till, R.; Milner, D.S.; Basford, S.M.; Capeness, M.J.; Fenton, A.K.; Atterbury, R.J.; et al. Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genom. 2012, 13, 670. [Google Scholar] [CrossRef] [PubMed]
- Paix, B.; Ezzedine, J.A.; Jacquet, S. Diversity, Dynamics, and Distribution of Bdellovibrio and Like Organisms in Perialpine Lakes. Appl. Environ. Microbio. 2019, 85, e02494-18. [Google Scholar] [CrossRef]
- Koval, S.F.; Hynes, S.H.; Flannagan, R.S.; Pasternak, Z.; Davidov, Y.; Jurkevitch, E. Bdellovibrio exovorus sp nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Micr. 2013, 63, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological Potential of Bdellovibrio and Like Organisms and Their Secreted Enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef]
- Williams, H.N.; Lymperopoulou, D.S.; Athar, R.; Chauhan, A.; Dickerson, T.L.; Chen, H.; Laws, E.; Berhane, T.K.; Flowers, A.R.; Bradley, N.; et al. Halobacteriovorax, an underestimated predator on bacteria: Potential impact relative to viruses on bacterial mortality. ISME J. 2016, 10, 491–499. [Google Scholar] [CrossRef]
- Nazarenko, E.L.; Crawford, R.J.; Ivanova, E.P. The Structural Diversity of Carbohydrate Antigens of Selected Gram-Negative Marine Bacteria. Mar. Drugs 2011, 9, 1914–1954. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, Z.M.; Li, J.; He, L.S.; Cui, G.J.; Li, W.L.; Chen, J.; Xin, Y.Z.; Cai, D.S.; Zhang, A.Q. Hadal water sampling by in situ microbial filtration and fixation (ISMIFF) apparatus. Deep Sea Res. Part I-Oceanogr. Res. Pap. 2019, 144, 132–137. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 January 2019).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. FastUniq: A fast de novo duplicates removal tool for paired short reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Gilna, P.; Li, W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 2009, 25, 1338–1340. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.M.; Huang, J.M.; Cui, G.J.; Li, W.L.; Li, J.; Wei, Z.F.; Chen, J.; Xin, Y.Z.; Cai, D.S.; Zhang, A.Q.; et al. In situ meta-omic insights into the community compositions and ecological roles of hadal microbes in the Mariana Trench. Environ. Microbiol. 2019, 21, 4092–4108. [Google Scholar] [CrossRef]
- Li, W.L.; Huang, J.M.; Zhang, P.W.; Cui, G.J.; Wei, Z.F.; Wu, Y.Z.; Gao, Z.M.; Han, Z.; Wang, Y. Periodic and Spatial Spreading of Alkanes and Alcanivorax Bacteria in Deep Waters of the Mariana Trench. Appl. Appl. Environ. Microbiol. 2019, 85, e02089-18. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Gish, W.; States, D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Multiple Alignment Using Hidden Markov Models. Third Int. Conf. Intel. Syst. Mol. Biol. 1995, 3, 114–120. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Alan, J.; Thomas, P.D.; Huang, X.D.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, W.; Li, Y.X.; Tam, C.; Bougouffa, S.; Wang, R.; Pei, B.; Chiang, H.; Leung, P.; Lu, Y.; et al. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun. 2019, 10, 517. [Google Scholar] [CrossRef]
- Tully, B.J. Metabolic diversity within the globally abundant Marine Group II Euryarchaea offers insight into ecological patterns. Nat. Commun. 2019, 10, 271. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Kwon, H.; Cho, G.; Kwon, J.; Choi, S.Y.; Mitchell, R.J. Viscosity has dichotomous effects on Bdellovibrio bacteriovorus HD100 predation. Environ. Microbiol. 2019, 21, 4675–4684. [Google Scholar] [CrossRef] [PubMed]
- Karunker, I.; Rotem, O.; Dori-Bachash, M.; Jurkevitch, E.; Sorek, R. A Global Transcriptional Switch between the Attack and Growth Forms of Bdellovibrio bacteriovorus. PLoS ONE 2013, 8, e61850. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Myrold, D.D.; Mueller, R.S. Distributions of Extracellular Peptidases Across Prokaryotic Genomes Reflect Phylogeny and Habitat. Front. Microbiol. 2019, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.; Heider, D.; Wendel, N.J.; Sperl, N.; Sieber, V. Bacterial Glycosyltransferases: Challenges and Opportunities of a Highly Diverse Enzyme Class Toward Tailoring Natural Products. Front. Microbiol. 2016, 7, 182. [Google Scholar] [CrossRef] [PubMed]
- Straley, S.C.; Lamarre, A.G.; Lawrence, L.J.; Conti, S.F. Chemotaxis of Bdellovibrio-Bacteriovorus toward Pure Compounds. J. Bacteriol. 1979, 140, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Morimoto, Y.V.; Oono, K.; Hayashi, F.; Oosawa, K.; Kudo, S.; Nakamura, S. Effect of the MotA(M206I) Mutation on Torque Generation and Stator Assembly in the Salmonella H+-Driven Flagellar Motor. J. Bacteriol. 2019, 201, e00727-18. [Google Scholar] [CrossRef]
- Lambert, C.; Smith, M.C.; Sockett, R.E. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 2003, 5, 127–132. [Google Scholar] [CrossRef]
- Kehry, M.R.; Bond, M.W.; Hunkapiller, M.W.; Dahlquist, F.W. Enzymatic damidation of methyl-accpting chem otaxis proteins in Escherichia coli catalyzed by the cheB gene prouct. Proc. Natl. Acad. Sci. USA 1983, 80, 3599–3603. [Google Scholar] [CrossRef] [PubMed]
- Batra, M.; Sharma, R.; Chandra, V.; Aggarwal, M.; Agarwal, U.; Gupta, P.; Singh, R.P.; Tomar, S. In silico and proteomic analysis of protein methyltransferase CheR from Bacillus subtilis. Int. J. Biol. Macromol. 2015, 77, 168–180. [Google Scholar] [CrossRef]
- Hamadeh, A.; Roberts, M.A.J.; August, E.; McSharry, P.E.; Maini, P.K.; Armitage, J.P.; Papachristodoulou, A. Feedback Control Architecture and the Bacterial Chemotaxis Network. PLoS Comput. Biol. 2011, 7, e1001130. [Google Scholar] [CrossRef] [PubMed]
- Ruby, E.G.; McCabe, J.B. Metabolism of periplasmic membrane-derived oligosaccharides by the predatory bacterium Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1988, 170, 646–652. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.M.; Zhou, Y.L.; Almeida, A.; Finn, R.D.; Danchin, A.; He, L.S. Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli. BMC Genom. 2020, 21, 408. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.J.; Cronan, J.E. The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Mol. Microbiol. 2011, 81, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G.J. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Nieder, J.B.; Hussels, M.; Bittl, R.; Brecht, M. Effect of TMAO and betaine on the energy landscape of photosystem I. Bba-Bioenergetics 2014, 1837, 849–856. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Booth, I.R.; Blount, P. The MscS and MscL Families of Mechanosensitive Channels Act as Microbial Emergency Release Valves. J. Bacteriol. 2012, 194, 4802–4809. [Google Scholar] [CrossRef] [PubMed]
- Bialecka-Fornal, M.; Lee, H.J.; Phillips, R. The Rate of Osmotic Downshock Determines the Survival Probability of Bacterial Mechanosensitive Channel Mutants. J. Bacteriol. 2015, 197, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Armoza-Zvuloni, R.; Schneider, A.; Shaked, Y. Rapid Hydrogen Peroxide Release during Coral-Bacteria Interactions. Front. Mar. Sci. 2016, 3, 124. [Google Scholar] [CrossRef]
- Gutierrez, C.; Devedjian, J.C. Osmotic Induction of Gene Osmc Expression in Escherichia-Coli K12. J. Mol. Biol. 1991, 220, 959–973. [Google Scholar] [CrossRef]
- Conter, A.; Gangneux, C.; Suzanne, M.; Gutierrez, C. Survival of Escherichia coli during long-term starvation: Effects of aeration, NaCl, and the rpoS and osmC gene products. Res. Microbiol. 2001, 152, 17–26. [Google Scholar] [CrossRef]
- Cho, Y.H.; Roe, J.H. Isolation and expression of the catA gene encoding the major vegetative catalase in Streptomyces coelicolor Muller. J. Bacteriol. 1997, 179, 4049–4052. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.S.; Oh, S.Y.; Chater, K.F.; Cho, Y.H.; Roe, J.H. H2O2-sensitive fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 2000, 275, 38254–38260. [Google Scholar] [CrossRef]
- Sun, D.; Crowell, S.A.; Harding, C.M.; De Silva, P.M.; Harrison, A.; Fernando, D.M.; Mason, K.M.; Santana, E.; Loewen, P.C.; Kumar, A.; et al. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci. 2016, 148, 31–40. [Google Scholar] [CrossRef]
- Rochat, T.; Miyoshi, A.; Gratadoux, J.J.; Duwat, P.; Sourice, S.; Azevedo, V.; Langella, P. High-level resistance to oxidative stress in Lactococcus lactis conferred by Bacillus subtilis catalase KatE. Microbiology 2005, 151, 3011–3018. [Google Scholar] [CrossRef]
- Lerner, T.R.; Lovering, A.L.; Bui, N.K.; Uchida, K.; Aizawa, S.; Vollmer, W.; Sockett, R.E. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 2012, 8, e1002524. [Google Scholar] [CrossRef]
- Lommatzsch, J.; Templin, M.F.; Kraft, A.R.; Vollmer, W.; Holtje, J.V. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J. Bacteriol. 1997, 179, 5465–5470. [Google Scholar] [CrossRef]
- Jennings, G.T.; Savino, S.; Marchetti, E.; Arico, B.; Kast, T.; Baldi, L.; Ursinus, A.; Holtje, J.V.; Nicholas, R.A.; Rappuoli, R.; et al. GNA33 from Neisseria meningitidis serogroup B encodes a membrane-bound lytic transglycosylase (MltA). Eur. J. Biochem. 2002, 269, 3722–3731. [Google Scholar] [CrossRef] [PubMed]
- White, C.L.; Kitich, A.; Gober, J.W. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 2010, 76, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Suefuji, K.; Valbuena, N.; Meehan, B.; Donegan, M.; Park, J.T. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 2005, 187, 3643–3649. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Didierjean, C.; Hecker, A.; Girardet, J.M.; Morel-Rouhier, M.; Gelhaye, E.; Favier, F. Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones. PLoS ONE 2016, 11, e0164678. [Google Scholar] [CrossRef][Green Version]
- Teixeira, M.C.; Fernandes, A.R.; Mira, N.P.; Becker, J.D.; Sa-Correia, I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006, 6, 230–248. [Google Scholar] [CrossRef]
- Luo, D.; Niu, X.; Yu, J.; Yan, J.; Gou, X.; Lu, B.R.; Liu, Y. Rice choline monooxygenase (OsCMO) protein functions in enhancing glycine betaine biosynthesis in transgenic tobacco but does not accumulate in rice (Oryza sativa L. ssp. japonica). Plant Cell Rep. 2012, 31, 1625–1635. [Google Scholar] [CrossRef]
- Diederichs, K.; Diez, J.; Greller, G.; Muller, C.; Breed, J.; Schnell, C.; Vonrhein, C.; Boos, W.; Welte, W. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 2000, 19, 5951–5961. [Google Scholar] [CrossRef]
- Ferreira, M.J.; de Sa-Nogueira, I. A Multitask ATPase Serving Different ABC-Type Sugar Importers in Bacillus subtilis. J. Bacteriol. 2010, 192, 5312–5318. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, C.J.; Faustoferri, R.C.; Quivey, R.G. RgpF Is Required for Maintenance of Stress Tolerance and Virulence in Streptococcus mutans. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Jang, C.H.; Piao, Y.L.; Huang, X.Q.; Yoon, E.J.; Park, S.H.; Lee, K.; Zhan, C.G.; Cho, H. Modeling and Re-Engineering of Azotobacter vinelandii Alginate Lyase to Enhance Its Catalytic Efficiency for Accelerating Biofilm Degradation. PLoS ONE 2016, 11, e0156197. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, R.F.; Paspaliari, D.K.; Larsen, T.; Storgaard, B.G.; Larsen, M.H.; Ingmer, H.; Palcic, M.M.; Leisner, J.J. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiol-Sgm 2013, 159, 833–847. [Google Scholar] [CrossRef]
- Tran, H.T.; Barnich, N.; Mizoguchi, E. Potential role of chitinases and chitin-binding proteins in host-microbial interactions during the development of intestinal inflammation. Histopathololgy 2011, 26, 1453–1464. [Google Scholar] [CrossRef]
- Tyler, L.; Bragg, J.N.; Wu, J.J.; Yang, X.H.; Tuskan, G.A.; Vogel, J.P. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genom. 2010, 11, 600. [Google Scholar] [CrossRef] [PubMed]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zayed, A.A.; Conceicao-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1109–1123. [Google Scholar] [CrossRef]
- Johnke, J.; Fraune, S.; Bosch, T.C.G.; Hentschel, U.; Schulenburg, H. Bdellovibrio and Like Organisms Are Predictors of Microbiome Diversity in Distinct Host Groups. Microbiol. Ecol. 2020, 79, 252–257. [Google Scholar] [CrossRef]
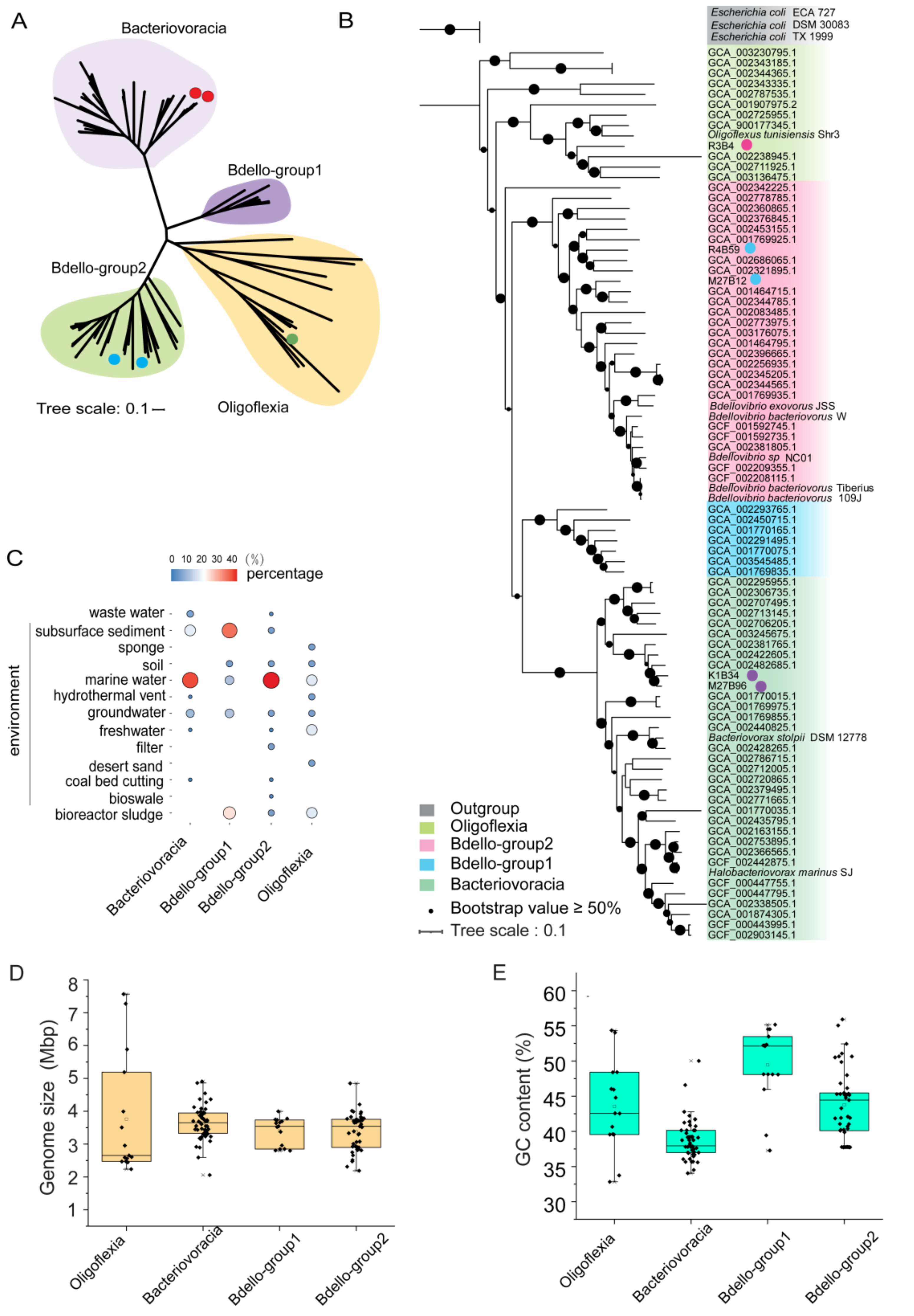
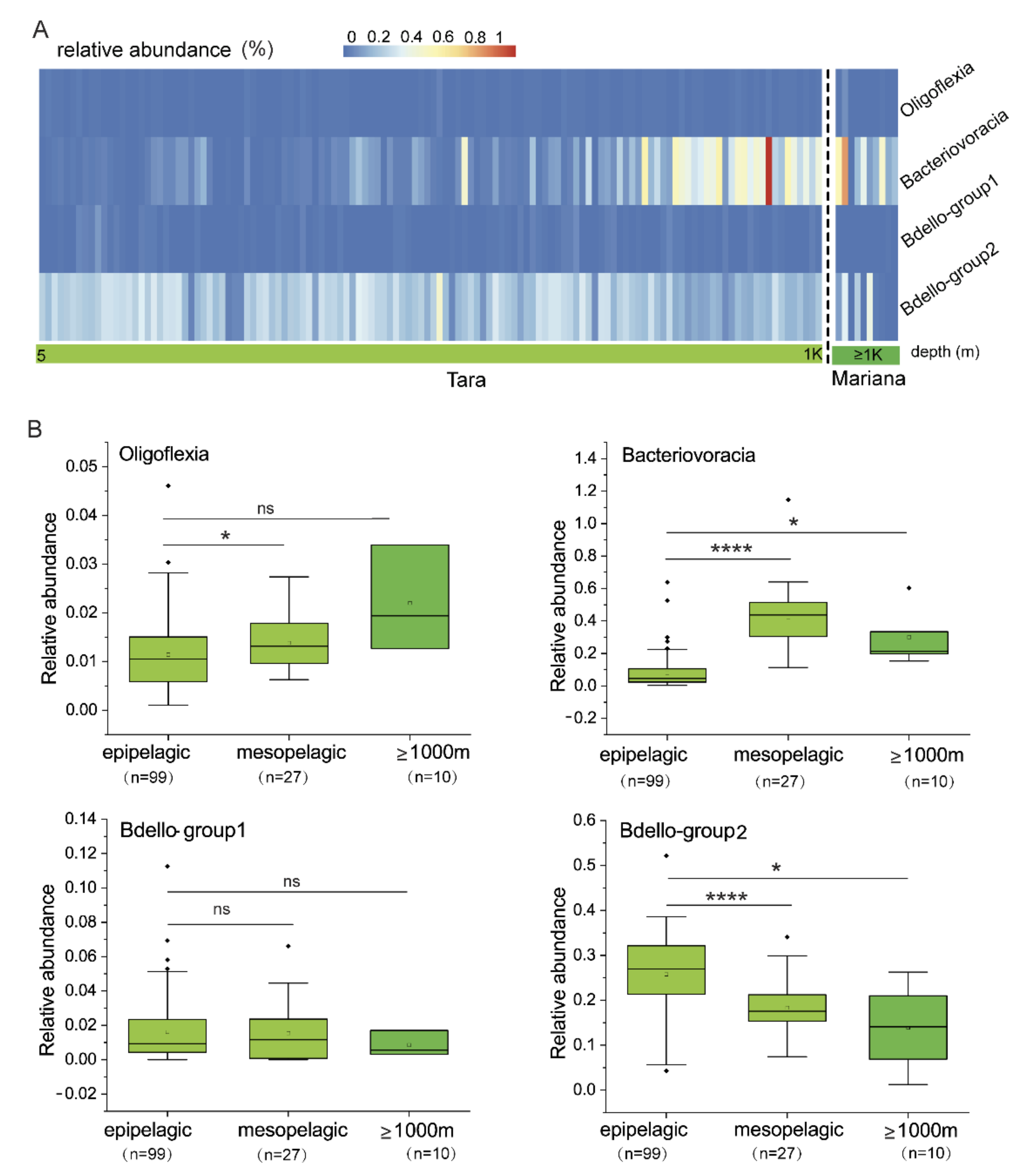
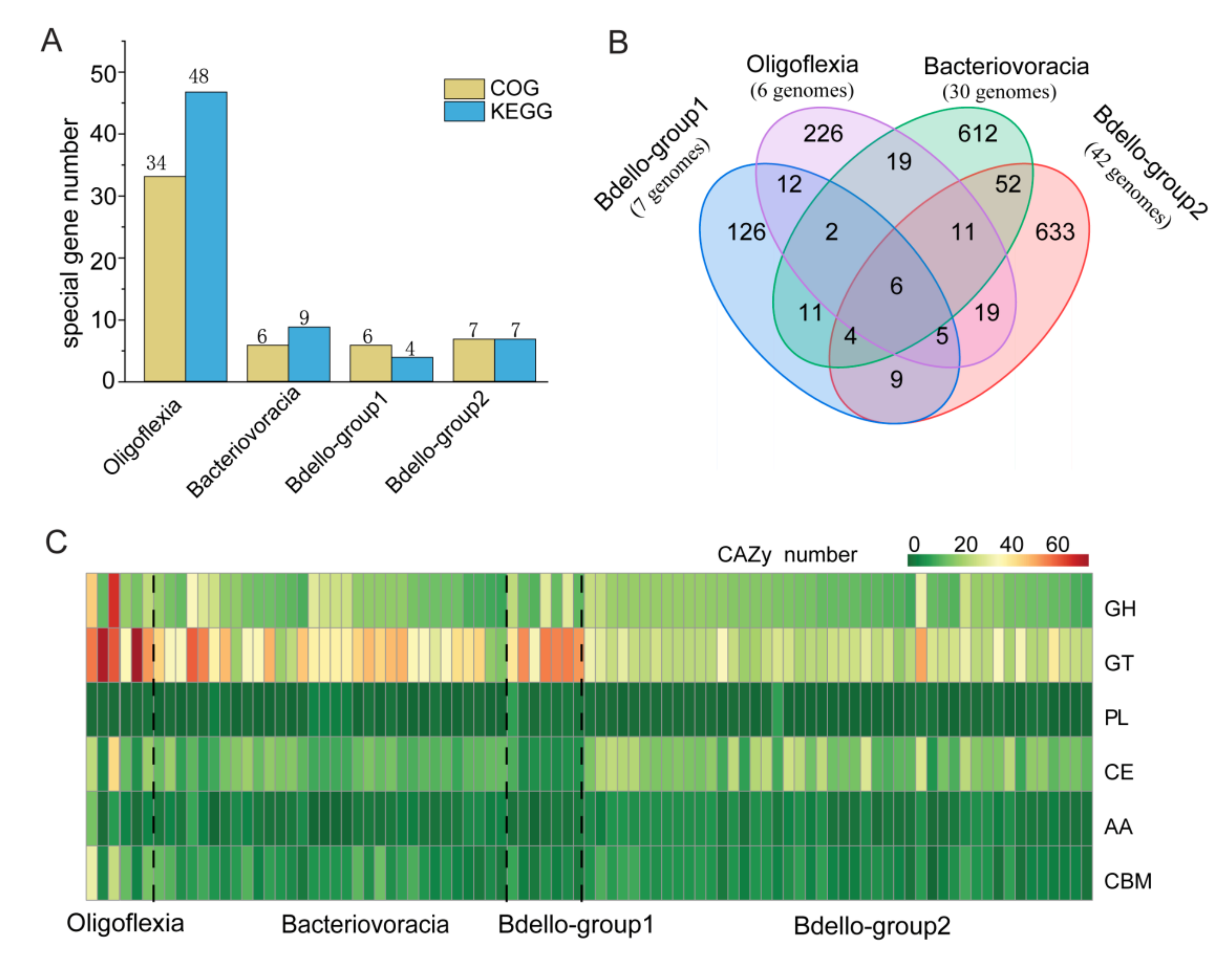
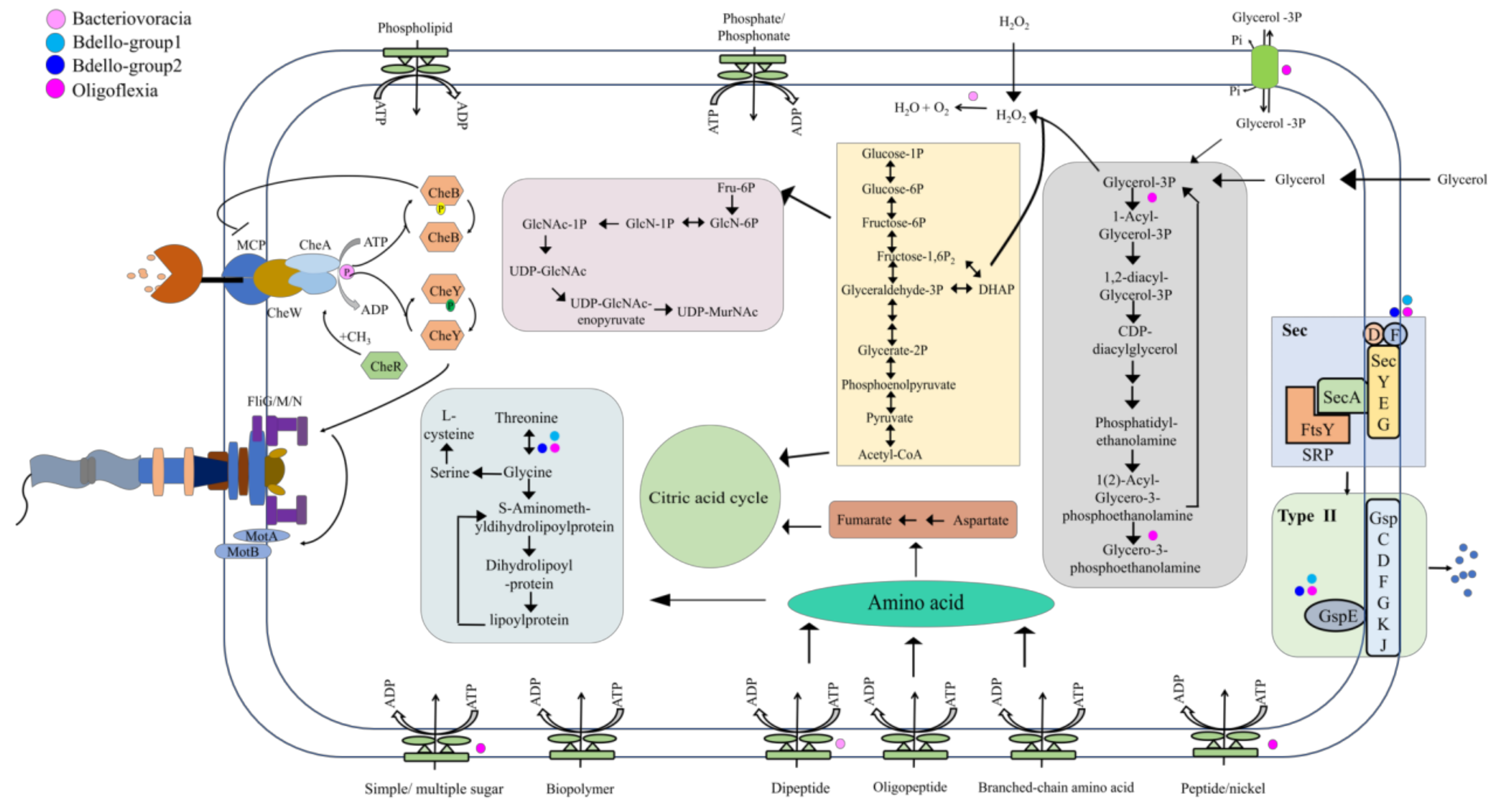
| Bin_id | K1B34 | M27B12 | M27B96 | R3B4 | R4B59 |
|---|---|---|---|---|---|
| Depth (m) | 1000 | 3000~5000 | 3000~5000 | 1646 | 5992 |
| Sampling site | 116.47° | 142.01° | 142.01° | 120.03° | 123.31° |
| (E; N) | 18.01° | 11.16° | 11.16° | 21.87° | 22.82° |
| Com. (%) | 75.16 | 98.21 | 80.15 | 81.83 | 71.94 |
| Con. (%) | 0.00 | 0.89 | 2.98 | 2.52 | 0.89 |
| N50 (bp) | 6396 | 295,316 | 5991 | 7831 | 3968 |
| No. contigs | 597 | 24 | 684 | 923 | 570 |
| Genome size (bp) | 3,229,602 | 4,849,992 | 3,554,382 | 5,883,831 | 2,188,444 |
| No. CDSs | 3741 | 4431 | 4172 | 5667 | 2566 |
| Genes Enriched in Marine Genomes of Bdellovibrionota.Group | KEGG | Non-Marine | Marine | Function |
|---|---|---|---|---|
| Bdello-group2 | K03442 | 11% | 89% | mscS; small conductance mechanosensitive channel |
| K08304 | 6% | 58% | mltA; membrane-bound lytic murein transglycosylase A [EC:4.2.2.-] | |
| K03571 | 17% | 58% | mreD; rod shape-determining protein MreD | |
| K02168 | 0% | 58% | betT, betS; choline/glycine/proline betaine transport protein | |
| K00147 | 11% | 53% | proA; glutamate-5-semialdehyde dehydrogenase [EC:1.2.1.41] | |
| K07393 | 0% | 53% | ECM4, yqjG; glutathionyl-hydroquinone reductase [EC:1.8.5.7] | |
| K09001 | 11% | 53% | anmK; anhydro-N-acetylmuramic acid kinase [EC:2.7.1.170] | |
| Bdello-group1 | K08641 | 0% | 100% | vanX; zinc D-Ala-D-Ala dipeptidase [EC:3.4.13.22] |
| K01273 | 0% | 100% | DPEP; membrane dipeptidase [EC:3.4.13.19] | |
| K05995 | 0% | 100% | pepE; dipeptidase E [EC:3.4.13.21] | |
| K15773 | 0% | 100% | hipB; HTH-type transcriptional regulator/antitoxin HipB | |
| K03442 | 0% | 100% | mscS; small conductance mechanosensitive channel | |
| K02168 | 0% | 100% | betT, betS; choline/glycine/proline betaine transport protein | |
| K06218 | 0% | 100% | relE, stbE; mRNA interferase RelE/StbE | |
| K07339 | 17% | 100% | hicA; mRNA interferase HicA [EC:3.1.-.-] | |
| K18843 | 0% | 100% | hicB; antitoxin HicB | |
| K19092 | 0% | 100% | parE1_3_4; toxin ParE1/3/4 | |
| Bacteriovoracia | K02168 | 21% | 78% | betT, betS; choline/glycine/proline betaine transport protein |
| K04063 | 11% | 78% | osmC; lipoyl-dependent peroxidase | |
| K19271 | 5% | 56% | catA; chloramphenicol O-acetyltransferase type A [EC:2.3.1.28] | |
| K03781 | 0% | 56% | katE, CAT, catB, srpA; catalase [EC:1.11.1.6] | |
| Oligoflexia | K01177 | 0% | 100% | beta-amylase [EC:3.2.1.2] |
| K10111 | 0% | 100% | malK; multiple sugar transport system ATP-binding protein [EC:3.6.3.-] | |
| K10112 | 0% | 100% | msmX; multiple sugar transport system ATP-binding protein | |
| K07272 | 0% | 100% | rgpF; rhamnosyltransferase [EC:2.4.1.-] | |
| K05841 | 0% | 100% | sterol 3beta-glucosyltransferase [EC:2.4.1.173] | |
| K03313 | 0% | 100% | nhaA; Na+:H+ antiporter, NhaA family | |
| K19294 | 0% | 100% | algI; alginate O-acetyltransferase complex protein AlgI | |
| K00499 | 0% | 100% | CMO; choline monooxygenase [EC:1.14.15.7] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.-M.; Zhou, Y.-L.; Wei, Z.-F.; Wang, Y. Phylogenomic Insights into Distribution and Adaptation of Bdellovibrionota in Marine Waters. Microorganisms 2021, 9, 757. https://doi.org/10.3390/microorganisms9040757
Li Q-M, Zhou Y-L, Wei Z-F, Wang Y. Phylogenomic Insights into Distribution and Adaptation of Bdellovibrionota in Marine Waters. Microorganisms. 2021; 9(4):757. https://doi.org/10.3390/microorganisms9040757
Chicago/Turabian StyleLi, Qing-Mei, Ying-Li Zhou, Zhan-Fei Wei, and Yong Wang. 2021. "Phylogenomic Insights into Distribution and Adaptation of Bdellovibrionota in Marine Waters" Microorganisms 9, no. 4: 757. https://doi.org/10.3390/microorganisms9040757
APA StyleLi, Q.-M., Zhou, Y.-L., Wei, Z.-F., & Wang, Y. (2021). Phylogenomic Insights into Distribution and Adaptation of Bdellovibrionota in Marine Waters. Microorganisms, 9(4), 757. https://doi.org/10.3390/microorganisms9040757






