Investigation of the Initial Host Response of Naïve Atlantic Salmon (Salmo salar) Inoculated with Paramoeba perurans
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Husbandry
2.2. Paramoeba perurans Challenge and Sample Collection
2.3. Histology
2.4. Proteomics FASP
2.5. Mass Spectrometry
2.6. Data Processing and Bioinformatics
2.7. Data Analysis
3. Results
3.1. In Vivo Challenge, RT-PCR and Histopathology
3.2. Gill Proteomics
3.3. Serum Proteomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munday, B.L. Diseases of salmonids. In Proceedings of the Workshop on Diseases of Australian Fish and Shellfish; Australian Fish Health Reference Laboratory: Regional Veterinary Laboratory: Benalla, Australia, 1986; pp. 127–141. [Google Scholar]
- Rodger, H.D. Amoebic gill disease (AGD) in farmed salmon (Salmo salar) in Europe. Fish Vet. J. 2014, 14, 16–27. [Google Scholar]
- Taylor, R.S.; Muller, W.J.; Cook, M.T.; Kube, P.D.; Elliott, N.G. Gill observations in Atlantic salmon (Salmo salar, L.) during repeated amoebic gill disease (AGD) field exposure and survival challenge. Aquaculture 2009, 290, 1–8. [Google Scholar] [CrossRef]
- Oldham, T.; Rodger, H.; Nowak, B.F. Incidence and distribution of amoebic gill disease (AGD)—An epidemiological review. Aquaculture 2016, 457, 35–42. [Google Scholar] [CrossRef]
- Adams, M.B.; Nowak, B.F. Amoebic gill disease: Sequential pathology in cultured Atlantic salmon, Salmo salar L. J. Fish Dis. 2003, 26, 601–614. [Google Scholar] [CrossRef]
- Roberts, S.D.; Powell, M.D. Freshwater bathing alters the mucous layer of marine Atlantic salmon Salmo salar L. J. Fish Biol. 2008, 72, 1864–1870. [Google Scholar] [CrossRef]
- Martinsen, K.H.; Thorisdottir, A.; Lillehammer, M. Effect of hydrogen peroxide as treatment for amoebic gill disease in Atlantic salmon (Salmo salar L.) in different temperatures. Aquac. Res. 2018, 49, 1733–1739. [Google Scholar] [CrossRef]
- Kube, P.D.; Taylor, R.S.; Elliott, N.G. Genetic variation in parasite resistance of Atlantic salmon to amoebic gill disease over multiple infections. Aquaculture 2012, 364–365, 165–172. [Google Scholar] [CrossRef]
- Young, N.D.; Cooper, G.A.; Nowak, B.F.; Koop, B.F.; Morrison, R.N. Coordinated down-regulation of the antigen processing machinery in the gills of amoebic gill disease-affected Atlantic salmon (Salmo salar L.). Mol. Immunol. 2008, 45, 2581–2597. [Google Scholar] [CrossRef]
- Loo, G.H.; Sutton, D.L.; Schuller, K.A. Cloning and functional characterisation of a peroxiredoxin 1 (NKEF A) cDNA from Atlantic salmon (Salmo salar) and its expression in fish infected with Neoparamoeba perurans. Fish Shellfish Immunol. 2012, 32, 1074–1082. [Google Scholar] [CrossRef]
- Bridle, A.R.; Morrison, R.N.; Cupit Cunningham, P.M.; Nowak, B.F. Quantitation of immune response gene expression and cellular localisation of interleukin-1beta mRNA in Atlantic salmon, Salmo salar L., affected by amoebic gill disease (AGD). Vet. Immunol. Immunopathol. 2006, 114, 121–134. [Google Scholar] [CrossRef]
- Marcos-López, M.; Calduch-Giner, J.A.; Mirimin, L.; MacCarthy, E.; Rodger, H.D.; O’Connor, I.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J.; Piazzon, M.C. Gene expression analysis of Atlantic salmon gills reveals mucin 5 and interleukin 4/13 as key molecules during amoebic gill disease. Sci. Rep. 2018, 8, 13689. [Google Scholar] [CrossRef]
- Benedicenti, O.; Collins, C.; Wang, T.; McCarthy, U.; Secombes, C.J. Which Th pathway is involved during late stage amoebic gill disease? Fish Shellfish Immunol. 2015, 46, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Pennacchi, Y.; Leef, M.J.; Crosbie, P.B.B.; Nowak, B.F.; Bridle, A.R. Evidence of immune and inflammatory processes in the gills of AGD-affected Atlantic salmon, Salmo salar L. Fish Shellfish Immunol. 2014, 36, 563–570. [Google Scholar] [CrossRef]
- Nowak, B.; Cadoret, K.; Feist, S.W.; Bean, T.P. Laser-capture dissection and immunohistochemistry reveals chloride and mucous-cell specific gene expression in gills of seawater acclimated Atlantic salmon Salmo salar. J. Fish Biol. 2013, 83, 1459–1467. [Google Scholar] [CrossRef]
- Gross, K.A.; Powell, M.D.; Butler, R.; Morrison, R.N.; Nowak, B.F. Changes in the innate immune response of Atlantic salmon, Salmo salar L., exposed to experimental infection with Neoparamoeba sp. J. Fish Dis. 2005, 28, 293–299. [Google Scholar] [CrossRef]
- Valdenegro-Vega, V.A.; Polinski, M.; Bridle, A.; Crosbie, P.; Leef, M.; Nowak, B.F. Effects of single and repeated infections with Neoparamoeba perurans on antibody levels and immune gene expression in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 2015, 42, 522–529. [Google Scholar] [CrossRef]
- Marcos-López, M.; Espinosa Ruiz, C.; Rodger, H.D.; O’Connor, I.; MacCarthy, E.; Esteban, M.Á. Local and systemic humoral immune response in farmed Atlantic salmon (Salmo salar L.) under a natural amoebic gill disease outbreak. Fish Shellfish Immunol. 2017, 66, 207–216. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. PROTEOMICS in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345. [Google Scholar] [CrossRef]
- Valdenegro-Vega, V.A.; Crosbie, P.; Bridle, A.; Leef, M.; Wilson, R.; Nowak, B.F. Differentially expressed proteins in gill and skin mucus of Atlantic salmon (Salmo salar) affected by amoebic gill disease. Fish Shellfish Immunol. 2014, 40, 69–77. [Google Scholar] [CrossRef]
- Magdeldin, S.; Enany, S.; Yoshida, Y.; Xu, B.; Zhang, Y.; Zureena, Z.; Lokamani, I.; Yaoita, E.; Yamamoto, T. Basics and recent advances of two dimensional- polyacrylamide gel electrophoresis. Clin. Proteom. 2014, 11, 16. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Aasebø, E.; Mjaavatten, O.; Vaudel, M.; Bruserud, Ø.; Berven, F.; Selheim, F. Reliable FASP-based procedures for optimal quantitative proteomic and phosphoproteomic analysis on samples from acute myeloid leukemia patients. Biol. Proced. Online 2016, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Jensen, O.N.; Møller, I.M.; Hebelstrup, K.H.; Rogowska-Wrzesinska, A. Evaluation of sample preparation methods for mass spectrometry-based proteomic analysis of barley leaves. Plant Methods 2018, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, P.B.B.; Bridle, A.R.; Cadoret, K.; Nowak, B.F. In vitro cultured Neoparamoeba perurans causes amoebic gill disease in Atlantic salmon and fulfils Koch’s postulates. Int. J. Parasitol. 2012, 42, 511–515. [Google Scholar] [CrossRef]
- Downes, J.K.; Henshilwood, K.; Collins, E.M.; Ryan, A.; O’Connor, I.; Rodger, H.D.; MacCarthy, E.; Ruane, N.M. A longitudinal study of amoebic gill disease on a marine Atlantic salmon farm utilising a real-time PCR assay for the detection of Neoparamoeba perurans. Aquac. Environ. Interact. 2015, 7, 239–251. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2013, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef]
- Løvoll, M.; Johnsen, H.; Boshra, H.; Bøgwald, J.; Sunyer, J.O.; Dalmo, R.A. The ontogeny and extrahepatic expression of complement factor C3 in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 2007, 23, 542–552. [Google Scholar] [CrossRef]
- Noris, M.; Remuzzi, G. Overview of Complement Activation and Regulation. Semin. Nephrol. 2013, 33, 479–492. [Google Scholar] [CrossRef]
- Yoshida, Y.; Kato, H.; Ikeda, Y.; Nangaku, M. Pathogenesis of Atypical Hemolytic Uremic Syndrome. J. Atheroscler. Thromb. 2019, 26, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Meri, T.; Amdahl, H.; Lehtinen, M.J.; Hyvärinen, S.; McDowell, J.V.; Bhattacharjee, A.; Meri, S.; Marconi, R.; Goldman, A.; Jokiranta, T.S. Correction: Microbes Bind Complement Inhibitor Factor H via a Common Site. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Józsi, M. Factor H Family Proteins in Complement Evasion of Microorganisms. Front. Immunol. 2017, 8, 571. [Google Scholar] [CrossRef]
- Hourcade, D.E. The Role of Properdin in the Assembly of the Alternative Pathway C3 Convertases of Complement. J. Biol. Chem. 2006, 281, 2128–2132. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.K.; Moore, T.L. Presence of plasma complement regulatory proteins clusterin (Apo J) and vitronectin (S40) on circulating immune complexes (CIC). Clin. Exp. Immunol. 2006, 145, 398–406. [Google Scholar] [CrossRef]
- Morgan, P. Chapter 34-CD59. In Factsbook, 2nd ed.; Barnum, S., Schein, T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 361–367. ISBN 978-0-12-810420-0. [Google Scholar]
- Powell, M.D.; Leef, M.J.; Roberts, S.D.; Jones, M.A. Neoparamoebic gill infections: Host response and physiology in salmonids. J. Fish Biol. 2008, 73, 2161–2183. [Google Scholar] [CrossRef]
- Chalmers, L.; Taylor, J.F.; Roy, W.; Preston, A.C.; Migaud, H.; Adams, A. A comparison of disease susceptibility and innate immune response between diploid and triploid Atlantic salmon (Salmo salar) siblings following experimental infection with Neoparamoeba perurans, causative agent of amoebic gill disease. Parasitology 2017, 144, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.L.; Boison, S.A.; Lillehammer, M.; Norris, A.; Gjerde, B. Genome-wide association mapping and accuracy of predictions for amoebic gill disease in Atlantic salmon (Salmo salar). Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Robledo, D.; Hamilton, A.; Gutiérrez, A.P.; Bron, J.E.; Houston, R.D. Characterising the mechanisms underlying genetic resistance to amoebic gill disease in Atlantic salmon using RNA sequencing. BMC Genom. 2020, 21, 271. [Google Scholar] [CrossRef]
- Dietrich, M.A.; Adamek, M.; Bilinska, B.; Hejmej, A.; Steinhagen, D.; Ciereszko, A. Characterization, expression and antibacterial properties of apolipoproteins A from carp (Cyprinus carpio L.) seminal plasma. Fish Shellfish Immunol. 2014, 41, 389–401. [Google Scholar] [CrossRef]
- Figueroa, D.M.; Gordon, E.M.; Yao, X.; Levine, S.J. Chapter 13—Apolipoproteins as context-dependent regulators of lung inflammation. In Mechanisms and Manifestations of Obesity in Lung Disease; Johnston, R.A., Suratt, B.T., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 301–326. ISBN 978-0-12-813553-2. [Google Scholar]
- Westerterp, M.; Berbeée, J.F.; Pires, N.M.M.; Van Mierlo, G.J.D.; Kleemann, R.; Romijn, J.A.; Havekes, L.M.; Rensen, P.C.N. Apolipoprotein C-I Is Crucially Involved in Lipopolysaccharide-Induced Atherosclerosis Development in Apolipoprotein E–Knockout Mice. Circulation 2007, 116, 2173–2181. [Google Scholar] [CrossRef]
- Berbée, J.F.P.; van der Hoogt, C.C.; de Haas, C.J.C.; van Kessel, K.P.M.; Dallinga-Thie, G.M.; Romijn, J.A.; Havekes, L.M.; van Leeuwen, H.J.; Rensen, P.C.N. Plasma apolipoprotein CI correlates with increased survival in patients with severe sepsis. Intensive Care Med. 2008, 34, 907–911. [Google Scholar] [CrossRef]
- Xu, X.R.; Wang, Y.; Adili, R.; Ju, L.; Spring, C.M.; Jin, J.W.; Yang, H.; Neves, M.A.D.; Chen, P.; Yang, Y.; et al. Apolipoprotein A-IV binds αIIbβ3 integrin and inhibits thrombosis. Nat. Commun. 2018, 9, 3608. [Google Scholar] [CrossRef] [PubMed]
- Vowinkel, T.; Mori, M.; Krieglstein, C.F.; Russell, J.; Saijo, F.; Bharwani, S.; Turnage, R.H.; Davidson, W.S.; Tso, P.; Granger, D.N.; et al. Apolipoprotein A-IV inhibits experimental colitis. J. Clin. Investig. 2004, 114, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Roula, D.; Theiler, A.; Luschnig, P.; Sturm, G.J.; Tomazic, P.V.; Marsche, G.; Heinemann, A.; Sturm, E.M. Apolipoprotein A-IV acts as an endogenous anti-inflammatory protein and is reduced in treatment-naïve allergic patients and allergen-challenged mice. Allergy 2020, 75, 392–402. [Google Scholar] [CrossRef]
- Yang, Y.; Fu, Q.; Zhou, T.; Li, Y.; Liu, S.; Zeng, Q.; Wang, X.; Jin, Y.; Tian, C.; Qin, Z.; et al. Analysis of apolipoprotein genes and their involvement in disease response of channel catfish after bacterial infection. Dev. Comp. Immunol. 2017, 67, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, M.; Liu, M.; Ji, Y.; Li, Z. TNF-alpha and IL-6 inhibit apolipoprotein A-IV production induced by linoleic acid in human intestinal Caco2 cells. J. Inflamm. 2015, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.N.; Zou, J.; Secombes, C.J.; Scapigliati, G.; Adams, M.B.; Nowak, B.F. Molecular cloning and expression analysis of tumour necrosis factor-alpha in amoebic gill disease (AGD)-affected Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 2007, 23, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Sathyan, N.; Philip, R.; Chaithanya, E.R.; Kumar, P.R.A.; Sanjeevan, V.N.; Singh, I.S.B. Characterization of Histone H2A Derived Antimicrobial Peptides, Harriottins, from Sicklefin Chimaera Neoharriotta pinnata (Schnakenbeck, 1931) and Its Evolutionary Divergence with respect to CO1 and Histone H2A. ISRN Mol. Biol. 2013, 2013, 930216. [Google Scholar] [CrossRef]
- Borelli, V.; Vita, F.; Shankar, S.; Soranzo, M.R.; Banfi, E.; Scialino, G.; Brochetta, C.; Zabucchi, G. Human Eosinophil Peroxidase Induces Surface Alteration, Killing, and Lysis of Mycobacterium tuberculosis. Infect. Immun. 2003, 71, 605–613. [Google Scholar] [CrossRef]
- Allen, R.C.; Henery, M.L.; Allen, J.C.; Hawks, R.J.; Stephens, J.T. Myeloperoxidase and Eosinophil Peroxidase Inhibit Endotoxin Activity and Increase Mouse Survival in a Lipopolysaccharide Lethal Dose 90% Model. J. Immunol. Res. 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Lefkowitz, D.L.; Lincoln, J.A.; Howard, K.R.; Stuart, R.; Lefkowitz, S.S.; Allen, R.C. Macrophage-Mediated Candidacidal Activity is Augmented by Exposure to Eosinophil Peroxidase: A Paradigm for Eosinophil-Macrophage Interaction. Inflammation 1997, 21, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Kim, S.; Choi, Y.; Nielsen, T.B.; Yan, J.; Lu, A.; Ruan, J.; Lee, H.-R.; Wu, H.; Spellberg, B.; et al. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 2019, 20, 276–287. [Google Scholar] [CrossRef]
- Burgener, S.S.; Baumann, M.; Basilico, P.; Remold-O’Donnell, E.; Touw, I.P.; Benarafa, C. Myeloid conditional deletion and transgenic models reveal a threshold for the neutrophil survival factor Serpinb1. Biol. Chem. 2016, 397, 897–905. [Google Scholar] [CrossRef]
- Park, B.; Buti, L.; Lee, S.; Matsuwaki, T.; Spooner, E.; Brinkmann, M.M.; Nishihara, M.; Ploegh, H.L. Granulin Is a Soluble Cofactor for Toll-like Receptor 9 Signaling. Immunity 2011, 34, 505–513. [Google Scholar] [CrossRef]
- Tatematsu, M.; Funami, K.; Ishii, N.; Seya, T.; Obuse, C.; Matsumoto, M. LRRC59 Regulates Trafficking of Nucleic Acid–Sensing TLRs from the Endoplasmic Reticulum via Association with UNC93B1. J. Immunol. 2015, 195, 4933–4942. [Google Scholar] [CrossRef]
- Singh, B.K.; Kambayashi, T. The Immunomodulatory Functions of Diacylglycerol Kinase ζ. Front. Cell Dev. Biol. 2016, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Iliev, D.B.; Skjæveland, I.; Jørgensen, J.B. CpG oligonucleotides bind TLR9 and RRM-Containing proteins in Atlantic Salmon (Salmo salar). BMC Immunol. 2013, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Wang, Z.-G.; Segev, N.; Hu, S.; Minshall, R.D.; Dull, R.O.; Zhang, M.; Malik, A.B.; Hu, G. Rab11a Mediates Vascular Endothelial-Cadherin Recycling and Controls Endothelial Barrier Function. Arter. Thromb. Vasc. Biol. 2016, 36, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015, 458, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Hertle, E.; Van Der Kallen, C.J.H.; Schalkwijk, C.G.; Stehouwer, C.D.A.; Van Greevenbroek, M.M.J. Associations of dicarbonyl stress with complement activation: The CODAM study. Diabetologia 2020, 63, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, C.; Perraud, A.-L. Chapter 26-Magnesium and the Immune Response. In Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals; Collins, J.F., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 319–331. ISBN 978-0-12-802168-2. [Google Scholar]
- Matsuda-Lennikov, M.; Biancalana, M.; Zou, J.; Ravell, J.C.; Zheng, L.; Kanellopoulou, C.; Jiang, P.; Notarangelo, G.; Jing, H.; Masutani, E.; et al. Magnesium transporter 1 (MAGT1) deficiency causes selective defects in N-linked glycosylation and expression of immune-response genes. J. Biol. Chem. 2019, 294, 13638–13656. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Huang, X.; Guosong, L.; Yang, C.; Liu, G.; Chengying, Y.; Wu, Y. MAGT1-mediated disturbance of Mg2+ homeostasis lead to exhausted of HBV-infected NK and CD8+ T cells. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, T.; Rüther, U.; Metzger, R.; Copeland, N.G.; Jenkins, N.A.; Lobel, P.; Hoflack, B. Gene and pseudogene of the mouse cation-dependent mannose 6-phosphate receptor. Genomic organization, expression, and chromosomal localization. J. Biol. Chem. 1992, 267, 12211–12219. [Google Scholar] [CrossRef]
- Bergström, J.H.; Birchenough, G.M.H.; Katona, G.; Schroeder, B.O.; Schütte, A.; Ermund, A.; Johansson, M.E.V.; Hansson, G.C. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. USA 2016, 113, 13833–13838. [Google Scholar] [CrossRef]
- Hou, P.; Chen, F.; Yong, H.; Lin, T.; Li, J.; Pan, Y.; Jiang, T.; Li, M.; Chen, Y.; Song, J.; et al. PTBP3 contributes to colorectal cancer growth and metastasis via translational activation of HIF-1α. J. Exp. Clin. Cancer Res. 2019, 38, 301. [Google Scholar] [CrossRef]
- Babic, I.; Sharma, S.; Black, D.L. A Role for Polypyrimidine Tract Binding Protein in the Establishment of Focal Adhesions. Mol. Cell. Biol. 2009, 29, 5564–5577. [Google Scholar] [CrossRef]
- Momeni, H.R.; Kanje, M. The calpain inhibitor VI prevents apoptosis of adult motor neurons. Neuroreport 2005, 16, 1065–1068. [Google Scholar] [CrossRef]
- Kumar, V.; Everingham, S.; Hall, C.; Greer, P.A.; Craig, A.W.B. Calpains promote neutrophil recruitment and bacterial clearance in an acute bacterial peritonitis model. Eur. J. Immunol. 2013, 44, 831–841. [Google Scholar] [CrossRef]
- Nassar, D.; Letavernier, E.; Baud, L.; Aractingi, S.; Khosrotehrani, K. Calpain Activity Is Essential in Skin Wound Healing and Contributes to Scar Formation. PLoS ONE 2012, 7, e37084. [Google Scholar] [CrossRef]
- Park, S.G.; Shin, H.; Shin, Y.K.; Lee, Y.; Choi, E.-C.; Park, B.-J.; Kim, S. The Novel Cytokine p43 Stimulates Dermal Fibroblast Proliferation and Wound Repair. Am. J. Pathol. 2005, 166, 387–398. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, X.; Xu, Y.; Liu, Q.; Jiang, X.; Wang, S.; Guo, W.; Zhou, Y. Thymosin β4 is involved in the antimicrobial immune response of Golden pompano, Trachinotus ovatus. Fish Shellfish Immunol. 2017, 69, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Kang, M.; Jo, M.J.; Seo, Y.B.; Park, N.G.; Kim, G.-D. Anti-Inflammatory Activity of β-thymosin Peptide Derived from Pacific Oyster (Crassostrea gigas) on NO and PGE2 Production by Down-Regulating NF-κB in LPS-Induced RAW264.7 Macrophage Cells. Mar. Drugs 2019, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Diotallevi, M.; Checconi, P.; Palamara, A.T.; Celestino, I.; Coppo, L.; Holmgren, A.; Abbas, K.; Peyrot, F.; Mengozzi, M.; Ghezzi, P. Glutathione Fine-Tunes the Innate Immune Response toward Antiviral Pathways in a Macrophage Cell Line Independently of Its Antioxidant Properties. Front. Immunol. 2017, 8, 1239. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Minetti, C.A.; Nakhasi, H.L.; Chen, S.W.; Barbehenn, E.; Nunes, P.H.; Nguyen, N.Y. Isolation, cDNA cloning, and characterization of an 18-kDa hemagglutinin and amebocyte aggregation factor from Limulus polyphemus. J. Biol. Chem. 1992, 267, 22452–22459. [Google Scholar] [CrossRef]
- Bruhn, K.W.; Spellberg, B. Transferrin-mediated iron sequestration as a novel therapy for bacterial and fungal infections. Curr. Opin. Microbiol. 2015, 27, 57–61. [Google Scholar] [CrossRef]
- Liu, W.; Yan, M.; Liu, Y.; Wang, R.; Li, C.; Deng, C.; Singh, A.; Coleman, W.G., Jr.; Rodgers, G.P. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 2010, 107, 11056–11061. [Google Scholar] [CrossRef]
- Maizels, R.M.; McSorley, H.J. Regulation of the host immune system by helminth parasites. J. Allergy Clin. Immunol. 2016, 138, 666–675. [Google Scholar] [CrossRef]
- Wynne, J.W.; O’Sullivan, M.G.; Cook, M.T.; Stone, G.; Nowak, B.F.; Lovell, D.R.; Elliott, N.G. Transcriptome Analyses of Amoebic Gill Disease-affected Atlantic Salmon (Salmo salar) Tissues Reveal Localized Host Gene Suppression. Mar. Biotechnol. 2008, 10, 388–403. [Google Scholar] [CrossRef]

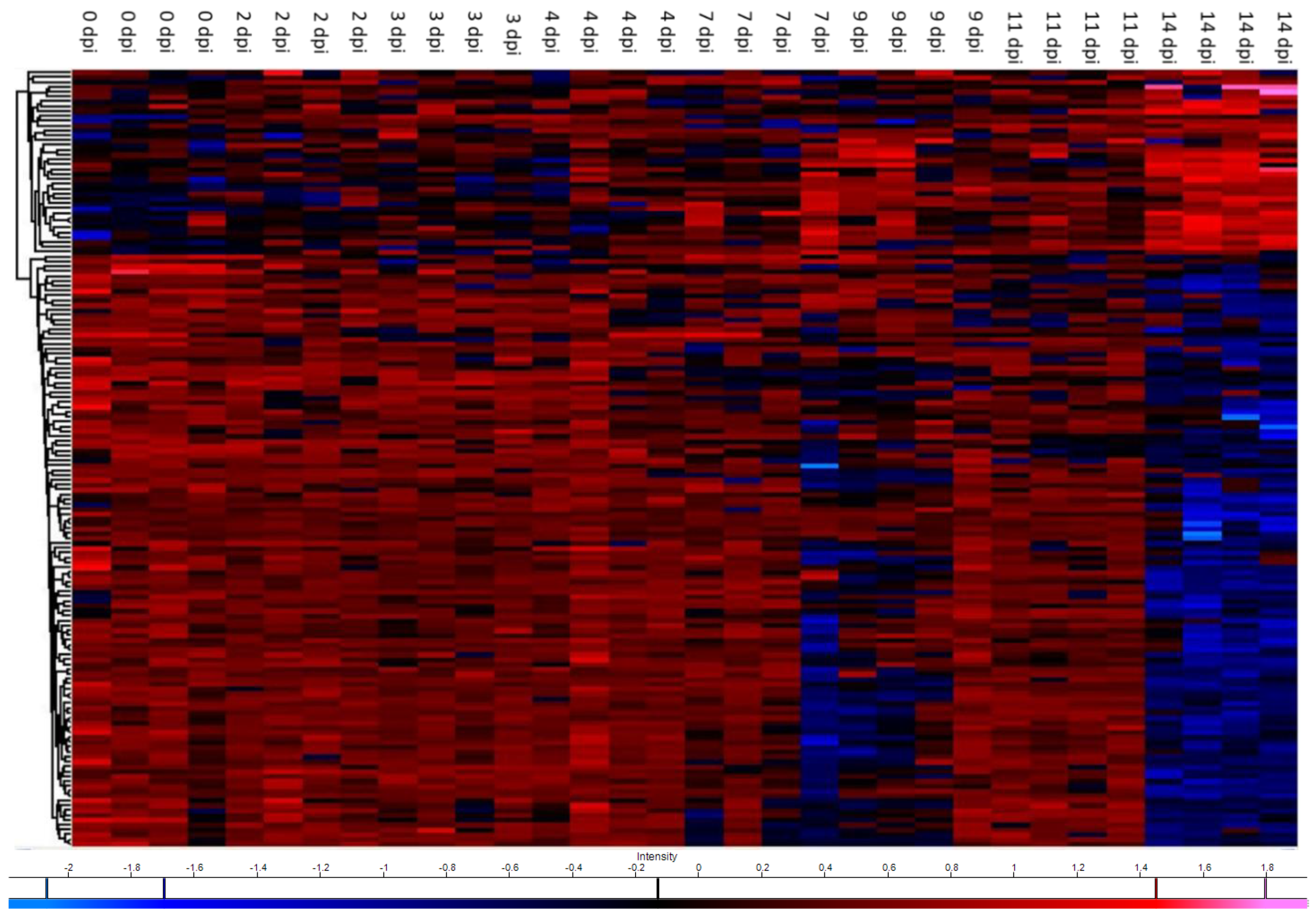
| Name | Gene | 2 dpi | 3 dpi | 4 dpi | 7 dpi | 9 dpi | 11 dpi | 14 dpi | Peptide Matches | % Sequence Coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | A0A1S3QBL3 | −1.76 | −1.61 | −1.35 | 6 | 22.00 | ||||
| Aminoacyl tRNA synthase complex-interacting multifunctional protein 2-like | A0A1S3S257 | −1.82 | 2 | 26.6 | ||||||
| Apolipoprotein A-I isoform X1 | A0A1S3NQ06 | −0.91 | 33 | 78.2 | ||||||
| Apolipoprotein A-I precursor | B5XBH3 | −1.33 | 31 | 78.6 | ||||||
| Apolipoprotein A-I-like | A0A1S3N6U9 | 2.08 | 2.72 | 7 | 34.50 | |||||
| Apolipoprotein A-IV | B5XCQ3 | 1.73 | 1.73 | 2.19 | 7 | 36.10 | ||||
| Apolipoprotein A-IV | B5X8U6 | 1.45 | 1.30 | 6 | 29.00 | |||||
| Apolipoprotein C-I isoform X1 | A0A1S3RYY8 | −0.62 | 8 | 55.2 | ||||||
| Apolipoprotein C-I-like | A0A1S3N6L4 | −1.51 | 8 | 60.90 | ||||||
| Apolipoprotein Eb-like | A0A1S3RXT3 | −2.40 | 17 | 59.80 | ||||||
| Apolipoprotein Eb-like | A0A1S3N6Y0 | −0.83 | 18 | 69.1 | ||||||
| Calcium uniporter protein, mitochondrial | A0A1S3QKG0 | 1.82 | 3 | 11.50 | ||||||
| Calpain-2 catalytic subunit-like | A0A1S3Q7N2 | −2.05 | 22 | 43.80 | ||||||
| Calpain-9-like isoform X1 | A0A1S3L6Q3 | −1.85 | 17 | 32.50 | ||||||
| Cation-dependent mannose—6-phosphate receptor | B5X109 | 2.01 | 3 | 10.40 | ||||||
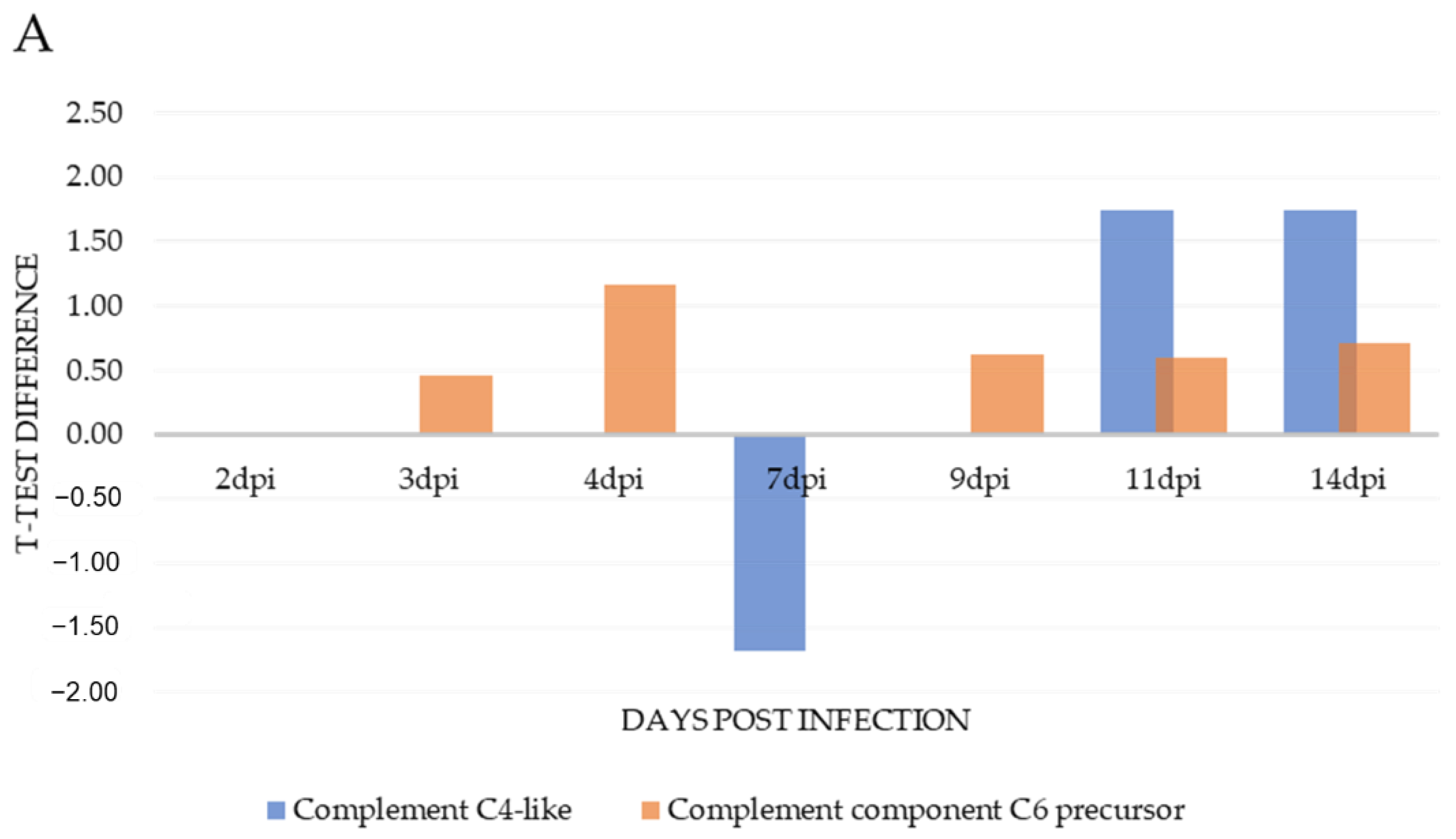
| Complement C4-like(LOC106605920) | A0A1S3RYT8 | −1.68 | 1.75 | 1.75 | 22 | 18.20 | ||||
| Complement component C6 precursor | C0H9G0 | 0.45 | 1.16 | 0.63 | 0.59 | 0.71 | 6 | 7.8 | ||
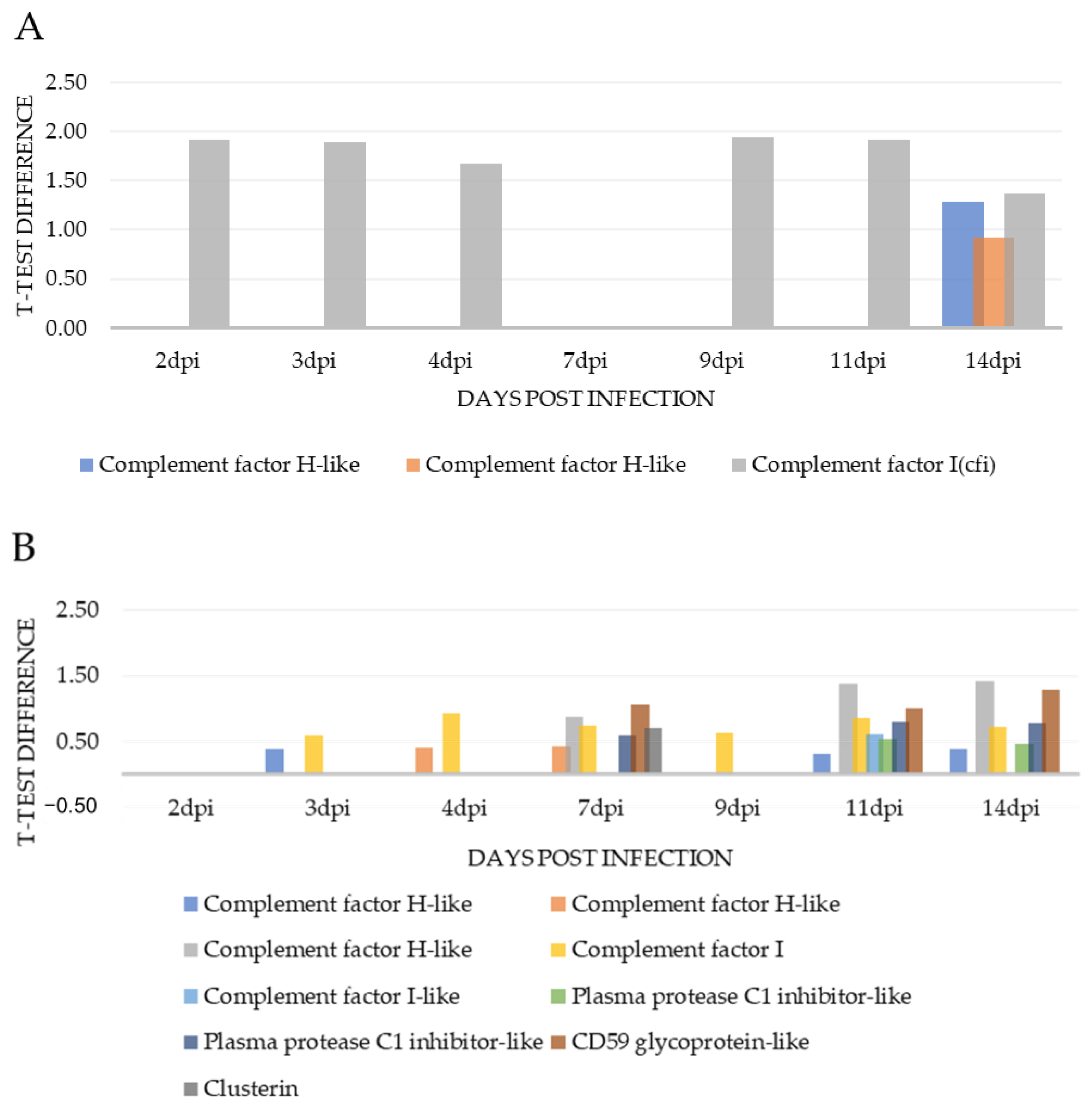
| Complement factor H-like | A0A1S3QR20 | 1.28 | 37 | 50.1 | ||||||
| Complement factor H-like | A0A1S3KK78 | 0.93 | 12 | 36.9 | ||||||
| Complement factor I(cfi) | A0A1S3QGX2 | 1.92 | 1.89 | 1.68 | 1.94 | 1.92 | 1.37 | 6 | 13.10 | |
| Deoxynucleoside triphosphate triphosphohydrolase SAMHD1-like(LOC106593439) | A0A1S3QJS3 | −0.62 | −1.59 | −1.39 | 5 | 37.80 | ||||
| Dicarbonyl and L-xylulose reductase(dcxr) | B5X5V8 | −2.10 | −2.22 | −2.96 | 3 | 13.10 | ||||
| DnaJ homolog subfamily A member 2 | C0HBK7 | 6.13 | 4.73 | 11 | 24.80 | |||||
| DnaJ homolog subfamily C member 3-like | A0A1S3SDE5 | −1.96 | 8 | 15.30 | ||||||
| Eosinophil peroxidase-like(LOC106572735) | A0A1S3MI56 | −2.11 | −1.74 | −1.47 | −1.15 | −2.05 | 34 | 50.50 | ||
| Granulins-like isoform X2 | A0A1S3NEN1 | −2.12 | −2.40 | −2.09 | −1.77 | 2 | 3.10 | |||
| Heterogeneous nuclear ribonucleoprotein A0 | B5X0T7 | −2.12 | −1.52 | 9 | 28.70 | |||||
| Histone H2A | B5X851 | −1.80 | 7 | 41.40 | ||||||
| Inositol-1-monophosphatase | N0GT48 | −1.66 | 3 | 14.60 | ||||||
| Intestinal mucin-like protein(LOC106595068) | 106595068 | 1.05 | 0.78 | 1.29 | 26 | 39.10 | ||||
| Leucine-rich repeat-containing protein 59-like | A0A1S3Q6W9 | −2.09 | 4 | 13.30 | ||||||
| Leukocyte elastase inhibitor | B5X4J0 | −1.06 | −1.06 | −1.59 | −1.62 | 8 | 28.50 | |||
| Magnesium transporter protein 1-like | A0A1S3QGC3 | −2.23 | −2.42 | 5 | 17.10 | |||||
| Mucin-2-like | A0A1S3QHF9 | 0.93 | 1.10 | 19 | 22.30 | |||||
| Mucin-5AC-like | A0A1S3R574 | 0.63 | 17 | 54.80 | ||||||
| Mucin-5B-like | A0A1S3QG78 | 0.81 | 36 | 49.60 | ||||||
| Myosin, light polypeptide 3-3 | B5DGT3 | −2.45 | 5 | 34.80 | ||||||
| Nuclear Receptor Coactivator 5 | B5X1C7 | −2.71 | 10 | 16.00 | ||||||
| Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha isoform X3 | A0A1S3NAV0 | 0.30 | −1.53 | 4 | 11.40 | |||||
| Pollen-specific leucine-rich repeat extensin-like protein 1 | A0A1S3QST0 | 1.73 | 4 | 23.40 | ||||||
| Polypyrimidine tract-binding protein 2 | B5X232 | 1.54 | 13 | 34.20 | ||||||
| Polypyrimidine tract-binding protein 3-like | A0A1S3QGP4 | −1.58 | 1.02 | 8 | 23.30 | |||||
| Ras-related protein Rab-11A(rb11a) | B5X4W2 | −3.09 | −3.01 | −3.51 | −2.73 | 9 | 39.40 | |||
| Sodium/potassium-transporting ATPase subunit alpha-1-like(LOC106610479) | A0A1S3SLV8 | −1.26 | −0.96 | −2.63 | −2.81 | −3.32 | 39 | 38.60 | ||
| Trafficking protein particle complex subunit | C0H7U8 | −1.76 | 4 | 24.40 | ||||||
| Transgelin | B9ENC8 | −1.52 | 19 | 77.70 | ||||||
| Zymogen granule membrane protein 16-like | A0A1S3KWW8 | 0.81 | 0.94 | 1.84 | 5 | 38.50 |
| Name | Gene | 2 dpi | 3 dpi | 4 dpi | 7 dpi | 9 dpi | 11 dpi | 14 dpi | Peptide Matches | % Sequence Coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| Apolipoprotein A-I isoform X1 | A0A1S3NQ06 | −0.32 | −0.31 | 48 | 85.5 | |||||
| Apolipoprotein A-I precursor | B5XBH3 | −0.92 | 32 | 84.7 | ||||||
| Apolipoprotein A-I-like | A0A1S3N6U9 | 1.65 | 1.66 | 27 | 74.1 | |||||
| Apolipoprotein A-IV | B5XCQ3 | 3.76 | 4.63 | 6.67 | 4.72 | 6.43 | 6.13 | 11 | 38.4 | |
| Apolipoprotein A-IV | B5X8U6 | 2.06 | 1.76 | 1.52 | 1.73 | 17 | 62.7 | |||
| Apolipoprotein A-IV-like | A0A1S3N6W3 | 1.49 | 1.21 | 1.58 | 1.42 | 1.36 | 21 | 61.6 | ||
| Apolipoprotein C-I isoform X1 | A0A1S3RYY8 | −1.29 | 9 | 60.9 | ||||||
| Apolipoprotein Eb-like | A0A1S3RXT3 | −0.99 | −1.60 | −1.06 | 21 | 54.1 | ||||
| Apolipoprotein Eb-like | A0A1S3M355 | 1.91 | 2.87 | 2.77 | 11 | 35.2 | ||||
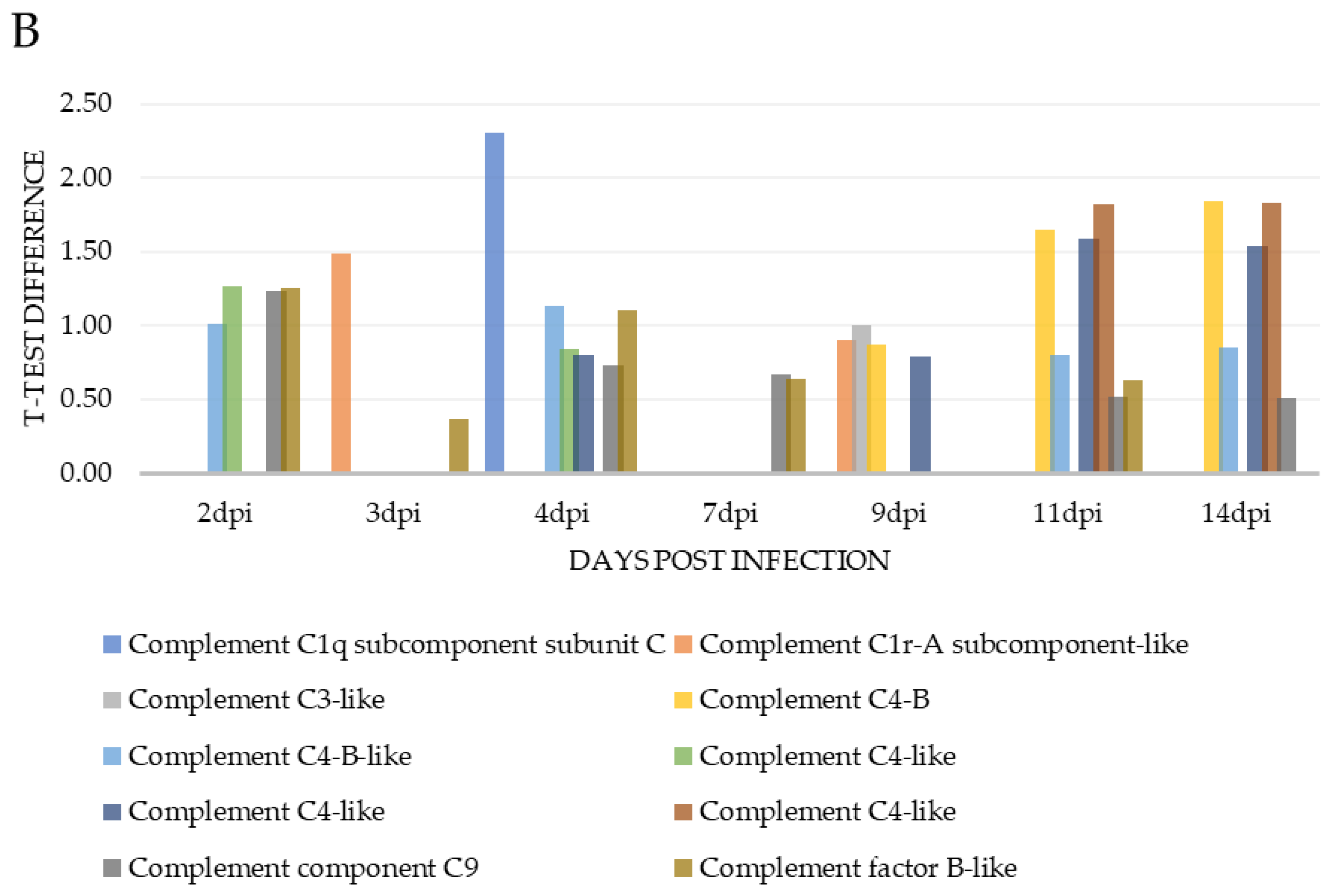
| CD59 glycoprotein-like | A0A1S3QTX5 | 1.07 | 1.01 | 1.28 | 4 | 48.9 | ||||
| Clusterin | C0H9Y2 | 0.70 | 8 | 15.8 | ||||||
| Complement C1q subcomponent subunit C | C0HB93 | 2.30 | 4 | 14.5 | ||||||
| Complement C1q-like protein 2 | A0A1S3LRM2 | 0.85 | 7 | 28.1 | ||||||
| Complement C1q-like protein 4 precursor | B9EPU5 | 0.82 | 10 | 42.9 | ||||||
| Complement C1r-A subcomponent-like | A0A1S3RZ46 | 1.48 | 0.90 | 7 | 16.1 | |||||
| Complement C1r-A subcomponent-like isoform X2 | A0A1S3MRC6 | 0.93 | 14 | 22.5 | ||||||
| Complement C2-like | A0A1S3NRD5 | 0.43 | 0.75 | 0.80 | 41 | 52.6 | ||||
| Complement C3-like | A0A1S3L7F1 | 0.34 | 0.39 | 0.25 | 67 | 30.4 | ||||
| Complement C3-like | A0A1S3ML57 | 1.00 | 9 | 33.3 | ||||||
| Complement C4-B | A0A1S3ML18 | 0.87 | 1.65 | 1.84 | 13 | 49 | ||||
| Complement C4-B-like | A0A1S3NRS7 | 1.01 | 1.13 | 0.80 | 0.84 | 28 | 13.6 | |||
| Complement C4-like | A0A1S3T028 | 1.26 | 0.83 | 23 | 12.4 | |||||
| Complement C4-like | A0A1S3RYT8 | 0.80 | 0.78 | 1.58 | 1.53 | 54 | 32.6 | |||
| Complement C4-like | A0A1S3QML7 | 1.82 | 1.83 | 35 | 50.1 | |||||
| Complement component C9 | A0A1S3LT49 | 1.24 | 0.73 | 0.67 | 0.51 | 0.50 | 23 | 45.3 | ||
| Complement factor B-like | A0A1S3LZP4 | 1.26 | 0.37 | 1.10 | 0.63 | 0.63 | 15 | 22.4 | ||
| Complement factor B-like | A0A1S3L2E5 | 0.75 | 0.93 | 0.59 | 0.63 | 0.85 | 33 | 35.7 | ||
| Complement factor H-like | A0A1S3KK78 | 0.39 | 0.31 | 0.40 | 20 | 71.1 | ||||
| Complement factor H-like | A0A1S3QR20 | 0.40 | 0.42 | 48 | 50.3 | |||||
| Complement factor H-like | A0A1S3KK91 | 0.87 | 1.37 | 1.42 | 6 | 32.4 | ||||
| Complement factor I | A0A1S3QGX2 | 0.59 | 0.93 | 0.75 | 0.64 | 0.85 | 0.72 | 26 | 44.1 | |
| Complement factor I-like | A0A1S3QGL8 | 0.61 | 13 | 47.8 | ||||||
| Glutathione peroxidase | A0A1S3MM95 | 2.78 | 1.72 | 6 | 38.2 | |||||
| Hemagglutinin/amebocyte aggregation factor-like isoform X1 | A0A1S3MZH7 | −1.06 | −1.07 | −1.01 | 6 | 38.1 | ||||
| Olfactomedin-4-like | A0A1S3MMF4 | 0.96 | 1.21 | 0.88 | 7 | 12.4 | ||||
| Olfactomedin-4-like | A0A1S3KM81 | 0.93 | 1.20 | 0.99 | 9 | 18.3 | ||||
| Pentaxin | B5X672 | −0.51 | −0.65 | −0.69 | −0.77 | 13 | 55 | |||
| Plasma protease C1 inhibitor-like | A0A1S3RGH0 | 0.53 | 0.47 | 25 | 39.8 | |||||
| Plasma protease C1 inhibitor-like | A0A1S3SJI8 | 0.59 | 0.81 | 0.79 | 14 | 26.2 | ||||
| Properdin-like | A0A1S3LII4 | 0.42 | 0.31 | 0.39 | 13 | 39.8 | ||||
| Serotransferrin | A0A1S2WYW0 | 1.50 | 82 | 78 | ||||||
| Serotransferrin | A0A1S3R1S1 | −0.35 | −0.27 | 84 | 78.2 | |||||
| Serotransferrin-1-like | A0A1S3R1U4 | 0.62 | −0.92 | −0.85 | 25 | 43 | ||||
| Thymosin beta | B5XAM0 | 2.53 | 3.00 | 2.79 | 2.46 | 4 | 76.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCormack, M.; Dillon, E.; O’Connor, I.; MacCarthy, E. Investigation of the Initial Host Response of Naïve Atlantic Salmon (Salmo salar) Inoculated with Paramoeba perurans. Microorganisms 2021, 9, 746. https://doi.org/10.3390/microorganisms9040746
McCormack M, Dillon E, O’Connor I, MacCarthy E. Investigation of the Initial Host Response of Naïve Atlantic Salmon (Salmo salar) Inoculated with Paramoeba perurans. Microorganisms. 2021; 9(4):746. https://doi.org/10.3390/microorganisms9040746
Chicago/Turabian StyleMcCormack, Michelle, Eugene Dillon, Ian O’Connor, and Eugene MacCarthy. 2021. "Investigation of the Initial Host Response of Naïve Atlantic Salmon (Salmo salar) Inoculated with Paramoeba perurans" Microorganisms 9, no. 4: 746. https://doi.org/10.3390/microorganisms9040746
APA StyleMcCormack, M., Dillon, E., O’Connor, I., & MacCarthy, E. (2021). Investigation of the Initial Host Response of Naïve Atlantic Salmon (Salmo salar) Inoculated with Paramoeba perurans. Microorganisms, 9(4), 746. https://doi.org/10.3390/microorganisms9040746






