The Global Dimension of Tomato Yellow Leaf Curl Disease: Current Status and Breeding Perspectives
Abstract
:1. Tomato Yellow Leaf Curl Disease Causing Agents: Tomato Yellow Leaf Curl Virus (TYLCV) and TYLCV-Like Viruses

2. Global Spreading of TYLCD: Efficient Transmission of Whitefly Vector and Dynamic Nature of the Virus
2.1. An Efficient Transmission Vector: Whiteflies
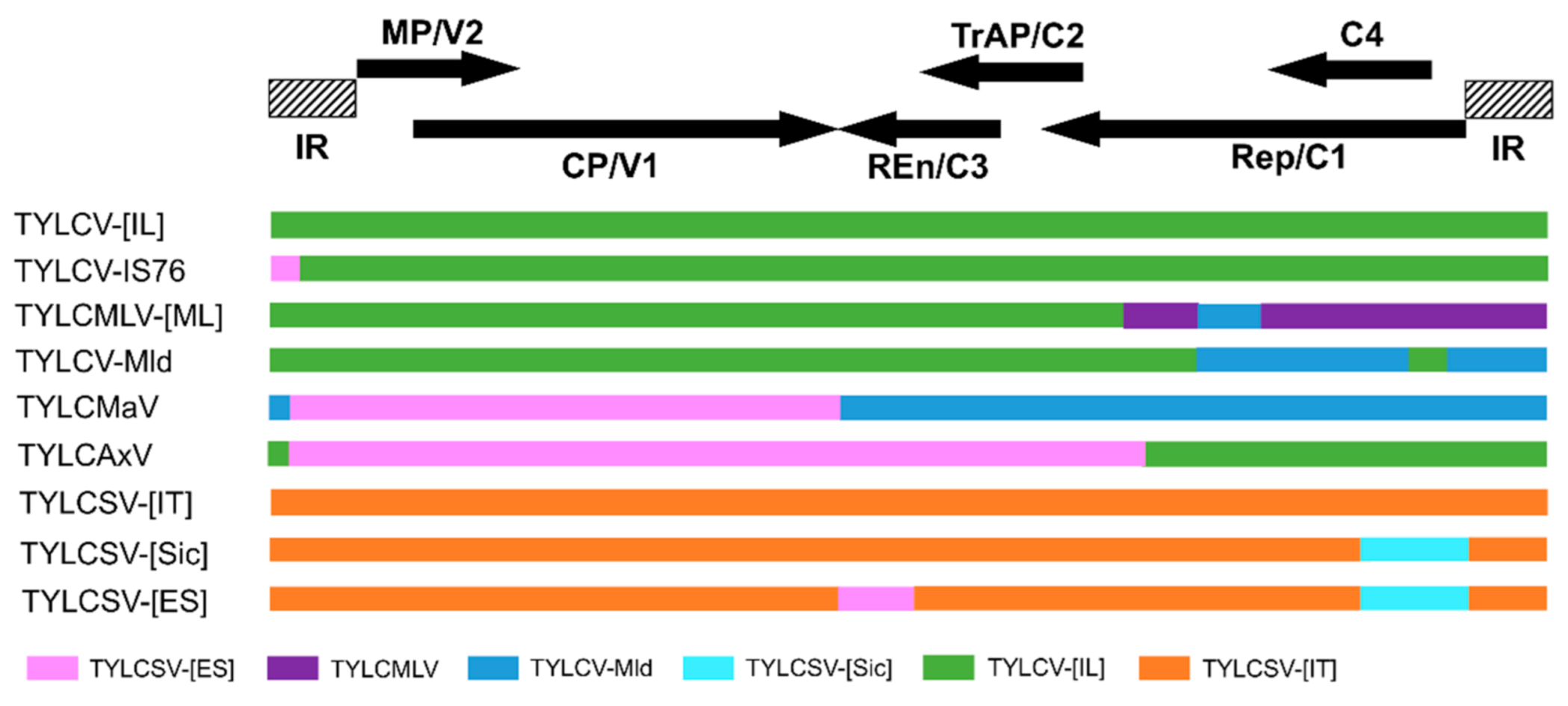
2.2. Driving Forces of Begomoviruses Evolution: Mutation and Recombination
3. Increasing Global Significance of TYLCD
3.1. Mixed Infection: An Incubator of New Recombinant Viruses
3.2. Alarming Scenario: Role of Betasatellites in TYLCV Epidemics
3.3. Emerging Problem: Plant Host Range Expansion
4. TYLCD Control: Mapped TYLCV Resistance Genes
5. Breeding Strategies: Mounting a Broad-Spectrum and Sustainable Begomovirus Resistance
5.1. Fishing in the Gene Pool: Natural Variation of Wild Tomato Relatives
5.2. A Challenging Task: Introgression Breeding for TYLCV Resistance
5.3. Pyramiding Resistance Genes: Towards Durable and Broad-Spectrum Resistance
5.4. Additional Source for Resistance: Dysfunctional Susceptibility Genes
5.5. Engineering Virus Resistance: Modification of Virus Genes
6. Conclusions and Prospects
- Screening of additional wild tomato accessions for natural resistance to TYLCV and related begomoviruses.
- Testing TYLCV-resistant tomato genotypes for resistance to TYLCV-betasatellite complexes.
- Testing the performance of tomato lines containing individual TYLCV-resistant loci (Ty-1 to Ty-6) and their combinations for resistance to other globally emerging begomoviruses, e.g., ToLCNDV.
- Combining the individual Ty-genes with whitefly-resistance genes in order to study whether such a combination will prolong the effectiveness of the virus resistance genes.
- Identification of the specific interactions between the proteins encoded by TYLCV (wild or mutants) and the proteins encoded by the Ty-gene alleles, as was done for the Ty-2 gene. This will allow us to forecast the effectiveness and durability of the Ty-genes in different tomato production areas by monitoring viral variants in the population.
- Screening for natural and/or induced mutations in S genes to get durable resistance to begomoviruses.
- Determining the role of mixed viral infections, more prevalent due to emergence of virus diseases and modifications of crops and vector geographical limits due to climate change, in modulating host resistance and durability.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P. Emerging viral diseases of tomato crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [Green Version]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef]
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 11 January 2021).
- Lapidot, M. Screening for TYLCV-resistance plants using whitefly-mediated inoculation. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 329–342. [Google Scholar] [CrossRef]
- Jeske, H. Barcoding of plant viruses with circular single-stranded DNA based on rolling circle amplification. Viruses 2018, 10, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abhary, M.; Patil, B.L.; Fauquet, C.M. Molecular biodiversity, taxonomy, and nomenclature of Tomato Yellow Leaf Curl-like Viruses. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 85–118. [Google Scholar] [CrossRef]
- Gronenborn, B. The tomato yellow leaf curl virus genome and function of its proteins. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 67–84. [Google Scholar] [CrossRef]
- Castillo, A.G.; Morilla, G.; Lozano, R.; Collinet, D.; Perez-Luna, A.; Kashoggi, A.; Bejarano, E. Identification of plant genes involved in TYLCV replication. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 207–221. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Li, T.; Fang, Q.; Zhang, Z.; Zhou, X. Tomato yellow leaf curl Yunnan virus, a new begomovirus species associated with tomato yellow leaf curl disease in China. J. Plant Pathol. 2016, 98, 337–340. [Google Scholar] [CrossRef]
- Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Evolution of geminiviruses and their satellites. FEBS Lett. 2009, 583, 1825–1832. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Wu, P.; Liu, P.; Gong, H.; Zhou, X. Characterization of alphasatellites associated with monopartite begomovirus/betasatellite complexes in Yunnan, China. Virol. J. 2010, 7, 178. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Tao, X.; Xie, Y.; Fauquet, C.M.; Zhou, X. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004, 78, 13966–13974. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Yang, X.; Xie, Y.; Cui, X.; Zhou, X. Tomato yellow leaf curl Thailand virus-[Y72] from Yunnan is a monopartite begomovirus associated with DNAβ. Virus Genes 2009, 38, 328–333. [Google Scholar] [CrossRef]
- Sivalingam, P.N.; Varma, A. Role of betasatellite in the pathogenesis of a bipartite begomovirus affecting tomato in India. Arch. Virol. 2012, 157, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, B.; Li, F.; Yang, Q.; Zhou, X. Research advances in geminiviruses. In Current Research Topics in Plant Virology; Wang, A., Zhou, X., Eds.; Springer: Cham, Switzerland, 2016; pp. 251–269. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization Global Database. 2021. Available online: https://gd.eppo.int (accessed on 23 February 2021).
- Mabvakure, B.; Martin, D.P.; Kraberger, S.; Cloete, L.; van Brunschot, S.; Geering, A.D.W.; Thomas, J.E.; Bananej, K.; Lett, J.-M.; Lefeuvre, P.; et al. Ongoing geographical spread of Tomato yellow leaf curl virus. Virology 2016, 498, 257–264. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varsani, A.; et al. The spread of Tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Barboza, N.; Blanco-Meneses, M.; Hallwass, M.; Moriones, E.; Inoue-Nagata, A.K. First report of Tomato yellow leaf curl virus in tomato in Costa Rica. Plant Dis. 2014, 98, 699. [Google Scholar] [CrossRef] [PubMed]
- Chinnaraja, C.; Ramkissoon, A.; Ramsubhag, A.; Jayaraj, J. First report of Tomato yellow leaf curl virus infecting tomatoes in Trinidad. Plant Dis. 2016, 100, 1958. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Pan, H.; Chu, D.; Yan, W.; Su, Q.; Liu, B.; Wang, S.; Wu, Q.; Xie, W.; Jiao, X.; Li, R.; et al. Rapid spread of Tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PLoS ONE 2012, 7, e34817. [Google Scholar] [CrossRef] [Green Version]
- Seal, S.; van den Bosch, F.; Jeger, M. Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit. Rev. Plant Sci. 2006, 25, 23–46. [Google Scholar] [CrossRef]
- He, Y.Z.; Wang, Y.M.; Yin, T.Y.; Fiallo-Olivé, E.; Liu, Y.Q.; Hanley-Bowdoin, L.; Wang, X.W. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar] [CrossRef]
- Ghanim, M. A review of the mechanisms and components that determine the transmission efficiency of Tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Res. 2014, 186, 47–54. [Google Scholar] [CrossRef]
- Rosen, R.; Kanakala, S.; Kliot, A.; Pakkianathan, B.C.; Farich, B.A.; Santana-Magal, N.; Elimelech, M.; Kontsedalov, S.; Lebedev, G.; Cilia, M.; et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 2015, 15, 1–8. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Ning, W.; Shi, X.; Liu, B.; Pan, H.; Wei, W.; Zeng, Y.; Sun, X.; Xie, W.; Wang, S.; Wu, Q.; et al. Transmission of Tomato yellow leaf curl virus by Bemisia tabaci as affected by whitefly sex and biotype. Sci. Rep. 2015, 5, 10744. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Kontsedalov, S.; Skaljac, M.; Brumin, M.; Sobol, I.; Czosnek, H.; Vavre, F.; Fleury, F.; et al. The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 2010, 84, 9310–9317. [Google Scholar] [CrossRef] [Green Version]
- Duffy, S.; Holmes, E.C. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus Tomato yellow leaf curl virus. J. Virol. 2008, 82, 957–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, L.; Zhang, J.; Zhou, X.; Li, H. Genetic structure and population variability of Tomato yellow leaf curl China virus. J. Virol. 2007, 81, 5902–5907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pooggin, M. How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int. J. Mol. Sci. 2013, 14, 15233–15259. [Google Scholar] [CrossRef] [Green Version]
- Moriones, E.; Navas-Castillo, J. Rapid evolution of the population of begomoviruses associated with the tomato yellow leaf curl disease after invasion of a new ecological niche. Span. J. Agric. Res. 2008, 6, 147–159. [Google Scholar] [CrossRef]
- Belabess, Z.; Dallot, S.; El-Montaser, S.; Granier, M.; Majde, M.; Tahiri, A.; Blenzar, A.; Urbino, C.; Peterschmitt, M. Monitoring the dynamics of emergence of a non-canonical recombinant of Tomato yellow leaf curl virus and displacement of its parental viruses in tomato. Virology 2015, 486, 291–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiallo-Olivé, E.; Trenado, H.P.; Louro, D.; Navas-Castillo, J. Recurrent speciation of a Tomato yellow leaf curl geminivirus in Portugal by recombination. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Castillo, A.G.; Collinet, D.; Deret, S.; Kashoggi, A.; Bejarano, E.R. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 2003, 312, 381–394. [Google Scholar] [CrossRef]
- Rodriguez-Negrete, E.; Lozano-Duran, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef] [Green Version]
- Moriones, E.; García-Andrés, S.; Navas-Castillo, J. Recombination in the TYLCV complex: A mechanism to increase genetic diversity. Implications for plant resistance development. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 119–138. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Sanchez-Campos, S.; Noris, E.; Louro, D.; Accotto, G.P.; Moriones, E. Natural recombination between Tomato yellow leaf curl virus-Is and Tomato leaf curl virus. J. Gen. Virol. 2000, 81, 2797–2801. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pendón, J.A.; Sánchez-Campos, S.; Fortes, I.M.; Moriones, E. Tomato yellow leaf curl sardinia virus, a begomovirus species evolving by mutation and recombination: A challenge for virus control. Viruses 2019, 11, 45. [Google Scholar] [CrossRef] [Green Version]
- Belabess, Z.; Peterschmitt, M.; Granier, M.; Tahiri, A.; Blenzar, A.; Urbino, C. The non-canonical Tomato yellow leaf curl virus recombinant that displaced its parental viruses in southern Morocco exhibits a high selective advantage in experimental conditions. J. Gen. Virol. 2016, 97, 3433–3445. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Caruso, A.G.; Davino, S. The nucleotide sequence of a recombinant Tomato yellow leaf curl virus strain frequently detected in Sicily isolated from tomato plants carrying the Ty-1 resistance gene. Arch. Virol. 2018, 163, 795–797. [Google Scholar] [CrossRef]
- Granier, M.; Tomassoli, L.; Manglli, A.; Nannini, M.; Peterschmitt, M.; Urbino, C. First report of TYLCV-IS141, a Tomato yellow leaf curl virus recombinant infecting tomato plants carrying the Ty-1 resistance gene in Sardinia (Italy). Plant Dis. 2019, 103, 1437. [Google Scholar] [CrossRef]
- Li, F.; Huang, C.; Li, Z.; Zhou, X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014, 10, e1003921. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, N.; Li, Z.; Xu, X.; Wang, Y.; Yang, X.; Liu, S.-S.; Wang, A.; Zhou, X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017, 13, e1006213. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.S.; van Loon, J.J.A.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.J.; Idris, A.M.; Al-Saady, N.A.; Al-Mahruki, M.S.; Al-Subhi, A.M.; Brown, J.K. A divergent isolate of Tomato yellow leaf curl virus from Oman with an associated DNAβ satellite: An evolutionary link between Asian and the Middle Eastern virus–satellite complexes. Virus Genes 2008, 36, 169–176. [Google Scholar] [CrossRef]
- Chen, L.F.; Rojas, M.; Kon, T.; Gamby, K.; Xoconostle-Cazares, B.; Gilbertson, R.L. A severe symptom phenotype in tomato in Mali is caused by a reassortant between a novel recombinant begomovirus (Tomato yellow leaf curl Mali virus) and a betasatellite. Mol. Plant Pathol. 2009, 10, 415–430. [Google Scholar] [CrossRef]
- Ito, T.; Kimbara, J.; Sharma, P.; Ikegami, M. Interaction of Tomato yellow leaf curl virus with diverse betasatellites enhances symptom severity. Arch. Virol. 2009, 154, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Ueda, S.; Onuki, M.; Yamashita, M.; Yamato, Y. Pathogenicity and insect transmission of a begomovirus complex between Tomato yellow leaf curl virus and Ageratum yellow vein betasatellite. Virus Genes 2012, 44, 338–344. [Google Scholar] [CrossRef]
- Voorburg, C.M.; Yan, Z.; Bergua-Vidal, M.; Wolters, A.M.A.; Bai, Y.; Kormelink, R. Ty-1, a universal resistance gene against geminiviruses that is compromised by co-replication of a betasatellite. Mol. Plant Pathol. 2020, 21, 160–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conflon, D.; Granier, M.; Tiendrébéogo, F.; Gentit, P.; Peterschmitt, M.; Urbino, C. Accumulation and transmission of alphasatellite, betasatellite and Tomato yellow leaf curl virus in susceptible and Ty-1-resistant tomato plants. Virus Res. 2018, 253, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelbart, D.; Chen, L.; Alon, T.; Dobrinin, S.; Levin, I.; Lapidot, M. The recent association of a DNA betasatellite with Tomato yellow leaf curl virus in Israel–A new threat to tomato production. Crop Prot. 2020, 128, 104995. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato yellow leaf curl virus: Impact, challenges, and management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Salati, R.; Nahkla, M.K.; Rojas, M.R.; Guzman, P.; Jaquez, J.; Maxwell, D.P.; Gilbertson, R.L. Tomato yellow leaf curl virus in the Dominican Republic: Characterization of an infectious clone, virus monitoring in whiteflies, and identification of reservoir hosts. Phytopathology 2002, 92, 487–496. [Google Scholar] [CrossRef] [Green Version]
- García-Arenal, F.; Zerbini, F.M. Life on the edge: Geminiviruses at the interface between crops and wild plant hosts. Annu. Rev. Virol. 2019, 6, 411–433. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, S.S.E.A.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato leaf curl New Delhi virus: A widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol. Plant Pathol. 2017, 18, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Moriones, E.; Praveen, S.; Chakraborty, S. Tomato leaf curl New Delhi virus: An emerging virus complex threatening vegetable and fiber crops. Viruses 2017, 9, 264. [Google Scholar] [CrossRef] [Green Version]
- Fortes, I.M.; Sánchez-Campos, S.; Fiallo-Olivé, E.; Díaz-Pendón, J.A.; Navas-Castillo, J.; Moriones, E. A novel strain of Tomato leaf curl New Delhi virus has spread to the Mediterranean basin. Viruses 2016, 8, 307. [Google Scholar] [CrossRef] [Green Version]
- Juárez, M.; Rabadán, M.P.; Martínez, L.D.; Tayahi, M.; Grande-Pérez, A.; Gómez, P. Natural hosts and genetic diversity of the emerging Tomato leaf curl New Delhi virus in Spain. Front. Microbiol. 2019, 10, 140. [Google Scholar] [CrossRef]
- Saunders, K.; Salim, N.; Mali, V.R.; Malathi, V.G.; Briddon, R.; Markham, P.G.; Stanley, J. Characterisation of Sri Lankan Cassava mosaic virus and Indian cassava mosaic virus: Evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 2002, 293, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Jones, C.M.; Devine, G.; Zhang, F.; Denholm, I.; Gorman, K. Insecticide resistance in Bemisia tabaci biotype Q (Hemiptera: Aleyrodidae) from China. Crop Prot. 2010, 29, 429–434. [Google Scholar] [CrossRef]
- Roditakis, E.; Grispou, M.; Morou, E.; Kristoffersen, J.B.; Roditakis, N.; Nauen, R.; Vontas, J.; Tsagkarakou, A. Current status of insecticide resistance in Q biotype Bemisia tabaci populations from Crete. Pest Manag. Sci. 2009, 65, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Hanson, P.M.; Green, S.K.; Kuo, G. Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop. Rep. 2006, 56, 17–18. [Google Scholar]
- Hutton, S.F.; Scott, J.W. Ty-6, a major begomovirus resistance gene located on chromosome 10. Rept. Tomato Genet. Coop. 2014, 64, 14–18. [Google Scholar]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular mapping of Ty-4, a new Tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soc. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; van-Oss, H.; et al. Mapping and introgression of a Tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Wolters, A.M.A. The NLR protein encoded by the resistance gene Ty-2 is triggered by the replication-associated protein Rep/C1 of Tomato yellow leaf curl virus. Front. Plant Sci. 2020, 11, 1384. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.; Bai, Y. The Tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Szinay, D.; Hutton, S.F.; de Jong, H.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Bai, Y. Chromosomal rearrangements between tomato and Solanum chilense hamper mapping and breeding of the TYLCV resistance gene Ty-1. Plant J. 2011, 68, 1093–1103. [Google Scholar] [CrossRef]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.F.; Bai, Y.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by Cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [Green Version]
- Hamera, S.; Song, X.; Su, L.; Chen, X.; Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 2012, 69, 104–115. [Google Scholar] [CrossRef]
- Belabess, Z.; Urbino, C.; Granier, M.; Tahiri, A.; Blenzar, A.; Peterschmitt, M. The typical RB76 recombination breakpoint of the invasive recombinant Tomato yellow leaf curl virus of Morocco can be generated experimentally but is not positively selected in tomato. Virus Res. 2018, 243, 44–51. [Google Scholar] [CrossRef]
- Hanson, P.M.; Bernacchi, D.; Green, S.; Tanksley, S.D.; Muniyappa, V.; Padmaja, A.S.; Chen, H.-M.; Kuo, G.; Fang, D.; Chen, J.-T. Mapping a wild tomato introgression associated with Tomato yellow leaf curl virus resistance in a cultivated tomato line. J. Am. Soc. Hortic. Sci. 2000, 15, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Wolters, A.M.A.; Caro, M.; Dong, S.; Finkers, R.; Gao, J.; Visser, R.G.F.; Wang, X.; Du, Y.; Bai, Y. Detection of an inversion in the Ty-2 region between S. lycopersicum and S. habrochaites by a combination of de novo genome assembly and BAC cloning. Theor. Appl. Genet. 2015, 128, 1987–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Cano, E.; Resende, R.O.; Boiteux, L.S.; Giordano, L.B.; Fernández-Muñoz, R.; Moriones, E. Phenotypic expression, stability, and inheritance of a recessive resistance to monopartite begomoviruses associated with tomato yellow leaf curl disease in tomato. Phytopathology 2008, 98, 618–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, L.B.; Silva-Lobo, V.L.; Santana, F.M.; Fonseca, M.E.N.; Boiteux, L.S. Inheritance of resistance to the bipartite Tomato chlorotic mottle begomovirus derived from Lycopersicon esculentum cv. ‘Tyking’. Euphytica 2005, 143, 27–33. [Google Scholar] [CrossRef]
- Wu, X.; He, W.T.; Tian, S.; Meng, D.; Li, Y.; Chen, W.; Li, L.; Tian, L.; Zhong, C.; Han, F.; et al. pelo is required for high efficiency viral replication. PLoS Pathog. 2014, 10, e1004034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, W.; Wu, J.; Ye, J.; Zheng, W.; Wang, S.; Zhu, X.; Zhou, J.; Pan, Z.; Zhang, B.; Zhu, S. A Pelota-like gene regulates root development and defence responses in rice. Ann. Bot. 2018, 122, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Feng, B.H.; Wang, H.M.; Xu, X.; Shi, Y.F.; He, Y.; Chen, Z.; Sathe, A.P.; Wu, J.L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.-C.; Smith, H.; Hutton, S. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.W.; Hutton, S.F.; Freeman, J.H. Fla. 8638B and Fla. 8624 tomato breeding lines with begomovirus resistance genes ty-5 plus Ty-6 and Ty-6, respectively. HortScience 2015, 50, 1405–1407. [Google Scholar] [CrossRef]
- Michelson, I.; Zamir, D.; Czosnek, H. Accumulation and translocation of Tomato yellow leaf curl virus (TYLCV) in a Lyco-persicon esculentum breeding line containing the L. chilense TYLCV tolerance gene Ty-1. Phytopathology 1994, 84, 928–933. [Google Scholar] [CrossRef]
- Barbieri, M.; Acciarri, N.; Sabatini, E.; Sardo, L.; Accotto, G.P.; Pecchioni, N. Introgression of resistance to two Mediterranean virus species causing tomato yellow leaf curl into a valuable traditional tomato variety. J. Plant Pathol. 2010, 92, 485–493. [Google Scholar] [CrossRef]
- Ohnishi, J.; Yamaguchi, H.; Saito, A. Analysis of the Mild strain of Tomato yellow leaf curl virus, which overcomes Ty-2. Arch. Virol. 2016, 161, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, M.S.; Jindal, S.K.; Sharma, A.; Prasanna, H.C. Tomato yellow leaf curl virus disease of tomato and its man-agement through resistance breeding: A review. J. Hortic. Sci. Biotechnol. 2020, 95, 425–444. [Google Scholar] [CrossRef]
- Yan, Z.; Pérez de Castro, A.; Díez, M.J.; Hutton, S.F.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y.; Li, J. Resistance to Tomato yellow leaf curl virus in tomato germplasm. Front. Plant Sci. 2018, 9, 1198. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.K.; Kalloo, M.K. Sources and inheritance of resistance to leaf curl virus in Lycopersicon. Theor. Appl. Genet. 1987, 73, 707–710. [Google Scholar] [CrossRef]
- Kasrani, M.A. Inheritance of resistance to Tomato yellow leaf curl virus (TYLCV) in Lycopersicon pimpinellifolium. Plant Dis. 1989, 73, 435–437. [Google Scholar] [CrossRef]
- Azizi, A.; Mozafari, J.; Shams-bakhsh, M. Phenotypic and molecular screening of tomato germplasm for resistance to Tomato yellow leaf curl virus. Iran. J. Biotechnol. 2008, 6, 199–206. [Google Scholar]
- De la Peña, R.; Kadirvel, P.; Venkatesan, S.; Kenyon, L.; Hughes, J. Integrated approaches to manage Tomato yellow leaf curl viruses. In Biocatalysis and Biomolecular Engineering; Hou, C.T., Shaw, J.-F., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 105–132. [Google Scholar] [CrossRef]
- El-Dougdoug, N.K.; Mahfouze, S.A.; Ahmed, S.A.; Othman, B.A.; Hazaa, M.M. Identification of biochemical and molecular markers in Tomato yellow leaf curl virus resistant tomato species. Sci. Agric. 2013, 2, 46–53. [Google Scholar]
- Ji, Y.; Scott, J.W.; Hanson, P.; Graham, E.; Maxwell, D.P. Sources of resistance, inheritance, and location of genetic loci conferring resistance to members of the tomato-infecting begomoviruses. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 343–362. [Google Scholar] [CrossRef]
- Jordá, C.; Picó, B.; Díez, M.J.; Nuez, F. Cribado de germoplasma resistente a TYLCV. Desarrollo de un método de diagnóstico adecuado. In Proceedings of the VIII Congreso Nacional de la Sociedad Española de Fitopatología, Córdoba, Spain, 23–27 September 1996; p. 218. [Google Scholar]
- Kasrawi, M.A.; Suwwan, M.A.; Mansour, A. Sources of resistance to Tomato yellow leaf curl virus (TYLCV) in Lycopersicon species. Euphytica 1988, 37, 61–64. [Google Scholar] [CrossRef]
- Pereira-Carvalho, R.C.; Boiteux, L.S.; Fonseca, M.E.N.; Díaz-Pendón, J.A.; Moriones, E.; Fernández-Muñoz, R.; Charchar, J.M.; Resende, R.O. Multiple resistance to Meloidogyne spp. and bipartite and monopartite Begomovirus spp. in wild Solanum (Lycopersicon) accessions. Plant Dis. 2010, 94, 179–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Identificación de nuevas fuentes de resistencia al virus del rizado amarillo del tomate (TYLCV). Actas Horticultura 2004, 41, 119–122. [Google Scholar]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Caracterización de entradas de Lycopersicon peruvianum y L. chilense por su resistencia al Tomato yellow leaf curl virus (TYLCV). Actas Portuguesas Horticultura 2005, 8, 48–54. [Google Scholar]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Exploiting partial resistance to Tomato yellow leaf curl virus derived from Solanum pimpinellifolium UPV16991. Plant Dis. 2008, 92, 1083–1090. [Google Scholar] [CrossRef] [Green Version]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Resistencia a la enfermedad del rizado amarillo del tomate en la especie silvestre Solanum lycopersicoides. Actas Horticultura 2010, 55, 169–170. [Google Scholar]
- Picó, B.; Díez, M.J. Screening Lycopersicon spp. for resistance to TYLCV. In Proceedings of the 2nd International Workshop on Bemisia and Geminiviral Disease, USDA-ARS, San Juan, Puerto Rico, 7–12 June 1998; p. 43. [Google Scholar]
- Picó, B.; Díez, M.J.; Nuez, F. Viral diseases causing the greatest economic losses to the tomato crop. II. The Tomato yellow leaf curl virus—A review. Sci. Hortic. 1996, 67, 151–196. [Google Scholar] [CrossRef]
- Picó, B.; Díez, M.J.; Nuez, F. Evaluation of whitefly-mediated inoculation techniques to screen Lycopersicon esculentum and wild relatives for resistance to Tomato yellow leaf curl virus. Euphytica 1998, 101, 259–271. [Google Scholar] [CrossRef]
- Picó, B.; Ferriol, M.; Diez, M.J.; Nuez, F. Developing tomato breeding lines resistant to Tomato yellow leaf curl virus. Plant Breed. 1999, 118, 537–542. [Google Scholar] [CrossRef]
- Picó, B.; Ferriol, M.; Diez, M.; Nuez, F. Cribado de fuentes de resistencia de Lycopersicon spp. al Tomato yellow leaf curl virus mediante agroinoculación en disco de hoja. Actas Horticultura 1999, 24, 105–112. [Google Scholar]
- Picó, B.; Sifres, A.; Elía, M.; Díez, M.J.; Nuez, F. Searching for new resistance sources to Tomato yellow leaf curl virus within a highly variable wild Lycopersicon genetic pool. Acta Physiol. Plant. 2000, 22, 344–350. [Google Scholar] [CrossRef]
- Picó, B.; Herraiz, J.; Ruiz, J.; Nuez, F. Widening the genetic basis of virus resistance in tomato. Sci. Hortic. 2002, 94, 73–89. [Google Scholar] [CrossRef]
- Pilowsky, M.; Cohen, S. Screening additional wild tomatoes for resistance to the whitefly borne Tomato yellow leaf curl virus. Acta Physiol. Plant 2000, 22, 351–353. [Google Scholar] [CrossRef]
- Soler, S.; Pico, B.; Sifres, A.; Diez, M.; De Frutos, R.; Nuez, F. Multiple virus resistance in a collection of Lycopersicon spp. In Proceedings of the Fifth Congress of the European Foundation for Plant Pathology, Taormina, Italy, 18–22 September 2000; European Foundation for Plant Pathology: Taormina, Italy, 2000; p. 17. [Google Scholar]
- Tomás, D.M.; Cañizares, M.C.; Abad, J.; Fernández-Muñoz, R.; Moriones, E. Resistance to Tomato yellow leaf curl virus accumulation in the tomato wild relative Solanum habrochaites associated with the C4 viral protein. Mol. Plant Microbe Interact. 2010, 24, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Vidavsky, F.; Czosnek, H. Tomato breeding lines resistant and tolerant to Tomato yellow leaf curl virus issued from Lycopersicon hirsutum. Phytopathology 1998, 88, 910–914. [Google Scholar] [CrossRef] [Green Version]
- Vidavski, F.; Czosnek, H.; Gazit, S.; Levy, D.; Lapidot, M. Pyramiding of genes conferring resistance to Tomato yellow leaf curl virus from different wild tomato species. Plant Breed. 2008, 127, 625–631. [Google Scholar] [CrossRef]
- Vidavsky, F.; Leviatov, S.; Milo, J.; Rabinowitch, H.; Kedar, N.; Czosnek, H. Response of tolerant breeding lines of tomato, Lycopersicon esculentum, originating from three different sources (L. peruvianum, L. pimpinellifolium and L. chilense) to early controlled inoculation by Tomato yellow leaf curl virus (TYLCV). Plant Breed. 1998, 117, 165–169. [Google Scholar] [CrossRef]
- Zakay, Y.; Navot, N.; Zeidan, M.; Kedar, N.; Rabinowitch, H.; Czosnek, H.; Zamir, D. Screening Lycopersicon accessions for resistance to Tomato yellow leaf curl virus: Presence of viral DNA and symptom development. Plant Dis. 1991, 75, 279–281. [Google Scholar] [CrossRef]
- Díez, M.J.; Nuez, F. Tomato. In Vegetables II. Handbook of Plant Breeding; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; pp. 249–323. [Google Scholar] [CrossRef]
- Maruthi, M.N.; Czosnek, H.; Vidavski, F.; Tarba, S.Y.; Milo, J.; Leviatov, S.; Venkatesh, H.M.; Padmaja, A.S.; Kulkarni, R.S.; Muniyappa, V. Comparison of resistance to Tomato leaf curl virus (India) and Tomato yellow leaf curl virus (Israel) among Lycopersicon wild species, breeding lines and hybrids. Eur. J. Plant Pathol. 2003, 109, 1–11. [Google Scholar] [CrossRef]
- Pereira-Carvalho, R.; Díaz-Pendón, J.; Fonseca, M.; Boiteux, L.; Fernández-Muñoz, R.; Moriones, E.; Resende, R.O. Recessive resistance derived from tomato cv. Tyking-limits drastically the spread of Tomato yellow leaf curl virus. Viruses 2015, 7, 2518–2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Evaluation of breeding tomato lines partially resistant to Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus derived from Lycopersicon chilense. Can. J. Plant Pathol. 2005, 27, 268–275. [Google Scholar] [CrossRef]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Inheritance of Tomato yellow leaf curl virus resistance derived from Solanum pimpinellifolium UPV16991. Plant Dis. 2007, 91, 879–885. [Google Scholar] [CrossRef] [Green Version]
- Pilowsky, M.; Cohen, S. Inheritance of resistance to Tomato yellow leaf curl virus in tomatoes. Phytopathology 1974, 64, 632–635. [Google Scholar] [CrossRef]
- Geneif, A.A. Breeding for resistance to Tomato leaf curl virus in tomatoes in the Sudan. In Proceedings of the VIII African Symposium on Horticultural Crops, Wad Medani, Sudan, 20–24 March 1983; Volume 143, pp. 469–484. [Google Scholar]
- Víquez-Zamora, M.; Caro, M.; Finkers, R.; Tikunov, Y.; Bovy, A.; Visser, R.G.F.; Bai, Y.; van Heusden, S. Mapping in the era of sequencing: High density genotyping and its application for mapping TYLCV resistance in Solanum pimpinellifolium. BMC Genom. 2014, 15, 1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilowsky, M.; Cohen, S. Tolerance to Tomato yellow leaf curl virus derived from Lycopersicon peruvianum. Plant Dis. 1990, 74, 248–250. [Google Scholar] [CrossRef]
- Julián, O.; Herráiz, J.; Corella, S.; di-Lolli, I.; Soler, S.; Díez, M.J.; Pérez de Castro, A. Initial development of a set of introgression lines from Solanum peruvianum PI 126944 into tomato: Exploitation of resistance to viruses. Euphytica 2013, 193, 183–196. [Google Scholar] [CrossRef]
- Hutton, S.F.; Scott, J.W. Fla. 7907C: A Fla. 7907 near-isogenic tomato inbred line containing the begomovirus resistance gene, Ty-1. HortScience 2017, 52, 658–660. [Google Scholar] [CrossRef] [Green Version]
- Hutton, S.F.; Ji, Y.; Scott, J.W. Fla. 8923: A tomato breeding line with begomovirus resistance gene Ty-3 in a 70-kb Solanum chilense introgression. HortScience 2015, 50, 1257–1259. [Google Scholar] [CrossRef] [Green Version]
- Pérez de Castro, A.; Julián, O.; Díez, M.J. Genetic control and mapping of Solanum chilense LA1932, LA1960 and LA1971-derived resistance to Tomato yellow leaf curl disease. Euphytica 2013, 190, 203–214. [Google Scholar] [CrossRef]
- Caro, M.; Verlaan, M.G.; Julián, O.; Finkers, R.; Wolters, A.M.A.; Hutton, S.F.; Scott, J.W.; Kormelink, R.; Visser, R.G.F.; Díez, M.J.; et al. Assessing the genetic variation of Ty-1 and Ty-3 alleles conferring resistance to Tomato yellow leaf curl virus in a broad tomato germplasm. Mol. Breed. 2015, 35, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, M. Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 2017, 26, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, H.C.; Sinha, D.P.; Rai, G.K.; Krishna, R.; Kashyap, S.P.; Singh, N.K.; Singh, M.; Malathi, V.G. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite Tomato leaf curl viruses of India. Plant Pathol. 2015, 64, 256–264. [Google Scholar] [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [Green Version]
- van Schie, C.C.; Takken, F.L. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Prins, M.W.; Van Enckevort, L.J.G.; Versluis, H.P. Geminivirus Resistant Plants. U.S. Patent No. 2020/0392530 A1, 27 June 2019. [Google Scholar]
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef] [Green Version]
- Bastet, A.; Robaglia, C.; Gallois, J.L. eIF4E resistance: Natural variation should guide gene editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.Y.; Thomas, M.R.; Rasheed, M.S.; Saeed, M.; Hanson, P.; De Barro, P.J.; Rezaian, M.A. A recessive allele (tgr-1) conditioning tomato resistance to geminivirus infection is associated with impaired viral movement. Phytopathology 2007, 97, 930–937. [Google Scholar] [CrossRef] [Green Version]
- Blair, M.W.; Rodriguez, L.M.; Pedraza, F.; Morales, F.; Beebe, S. Genetic mapping of the Bean golden yellow mosaic geminivirus resistance gene bgm-1 and linkage with potyvirus resistance in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2007, 114, 261–271. [Google Scholar] [CrossRef]
- Czosnek, H.; Eybishtz, A.; Sade, D.; Gorovits, R.; Sobol, I.; Bejarano, E.; Rosas-Díaz, T.; Lozano-Durán, R. Discovering host genes involved in the infection by the Tomato yellow leaf curl virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing. Viruses 2013, 5, 998–1022. [Google Scholar] [CrossRef] [Green Version]
- Kalinina, N.O.; Khromov, A.; Love, A.J.; Taliansky, M.E. CRISPR Applications in plant virology: Virus resistance and beyond. Phytopathology 2020, 110, 18–28. [Google Scholar] [CrossRef]
- Tomlinson, L.; Yang, Y.; Emenecker, R.; Smoker, M.; Taylor, J.; Perkins, S.; Smith, J.; MacLean, D.; Olszewski, N.E.; Jones, J.D.G. Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 2018, 17, 132–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Gady, A.L.; Hermans, F.W.; Van de Wal, M.H.; van Loo, E.N.; Visser, R.G.F.; Bachem, C.W. Implementation of two high through-put techniques in a novel application: Detecting point mutations in large EMS mutated plant populations. Plant Methods 2009, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissner, R.; Jacobson, Y.; Melamed, S.; Levyatuv, S.; Shalev, G.; Ashri, A.; Elkind, Y.; Levy, A. A new model system for tomato genetics. Plant J. 1997, 12, 1465–1472. [Google Scholar] [CrossRef]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef]
- Minoia, S.; Petrozza, A.; D’Onofrio, O.; Piron, F.; Mosca, G.; Sozio, G.; Cellini, F.; Bendahmane, A.; Carriero, F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, T.; Asamizu, E.; Mizoguchi, T.; Fukuda, N.; Matsukura, C.; Ezura, H. Mutant resources for the miniature tomato (Solanum lycopersicum L.) ‘Micro-Tom’. Hortic. J. 2009, 78, 6–13. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; Thomson, J.A. Transgenic strategies for developing crops resistant to geminiviruses. Plant Sci. 2009, 176, 1–11. [Google Scholar] [CrossRef]
- Ammara, U.E.; Mansoor, S.; Saeed, M.; Amin, I.; Briddon, R.W.; Al-Sadi, A.M. RNA interference-based resistance in transgenic tomato plants against Tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virol. J. 2015, 12, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antignus, Y.; Vunsh, R.; Lachman, O.; Pearlsman, M.; Maslenin, L.; Hananya, U.; Rosner, A. Truncated Rep gene originated from Tomato yellow leaf curl virus-Israel [Mild] confers strain-specific resistance in transgenic tomato. Ann. Appl. Biol. 2004, 144, 39–44. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [Green Version]
- Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Boosting plant immunity with CRISPR/Cas. Genome Biol. 2015, 16, 254. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Si, X.; Zhang, Y.; Zhang, H.; Zhang, F.; Gao, C. Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome Biol. 2018, 19, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, S.S.E.A.; Mansoor, S.; Ali, Z.; Tashkandi, M.; Mahfouz, M.M. Engineering plants for geminivirus resistance with CRISPR/Cas9 system. Trends Plant Sci. 2016, 21, 279–281. [Google Scholar] [CrossRef] [PubMed]


| Resistance Gene a | Genetic Source | Chromosome | Inheritance Pattern | Gene Identity c | Reference | |
|---|---|---|---|---|---|---|
| Accession/Line b | Species | |||||
| Ty-1 | LA1969 | S. chilense | 6 | Dominant | RDR | [75,78] |
| Ty-2 | B6013 | S. habrochaites | 11 | Dominant | NLR | [71,77,79] |
| Ty-3 | LA2779 | S. chilense | 6 | Dominant | RDR | [73,78] |
| Ty-4 | LA1932 | S. chilense | 3 | Incomplete dominant | [74] | |
| ty-5 | Tyking | S. lycopersicum | 4 | Recessive | Pelota | [76] |
| Ty-6 | LA2779 | S. chilense | 10 | Incomplete dominant | [72] | |
| Solanum spp. a | Number of Accessions b | ||
|---|---|---|---|
| Symptomless | Symptomatic | Segregating | |
| S. arcanum | 15 | 5 | 5 |
| S. cheesmaniae/S. galapagense | 0 | 9 | 0 |
| S. chilense | 54 | 6 | 4 |
| S. chmielewskii | 1 | 3 | 0 |
| S. corneliomulleri | 30 | 8 | 9 |
| S. habrochaites | 13 | 52 | 14 |
| S. huaylasense | 4 | 0 | 0 |
| S. lycopersicoides | 1 | 13 | 0 |
| S. neorickii | 2 | 5 | 2 |
| S. pennellii | 2 | 42 | 2 |
| S. peruvianum | 69 | 39 | 20 |
| S. pimpinellifolium | 9 | 455 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Wolters, A.-M.A.; Navas-Castillo, J.; Bai, Y. The Global Dimension of Tomato Yellow Leaf Curl Disease: Current Status and Breeding Perspectives. Microorganisms 2021, 9, 740. https://doi.org/10.3390/microorganisms9040740
Yan Z, Wolters A-MA, Navas-Castillo J, Bai Y. The Global Dimension of Tomato Yellow Leaf Curl Disease: Current Status and Breeding Perspectives. Microorganisms. 2021; 9(4):740. https://doi.org/10.3390/microorganisms9040740
Chicago/Turabian StyleYan, Zhe, Anne-Marie A. Wolters, Jesús Navas-Castillo, and Yuling Bai. 2021. "The Global Dimension of Tomato Yellow Leaf Curl Disease: Current Status and Breeding Perspectives" Microorganisms 9, no. 4: 740. https://doi.org/10.3390/microorganisms9040740
APA StyleYan, Z., Wolters, A.-M. A., Navas-Castillo, J., & Bai, Y. (2021). The Global Dimension of Tomato Yellow Leaf Curl Disease: Current Status and Breeding Perspectives. Microorganisms, 9(4), 740. https://doi.org/10.3390/microorganisms9040740







