COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control
Abstract
1. Introduction
2. Arbovirus Disease
2.1. Arbovirus
2.2. Arbovirus Disease Transmission
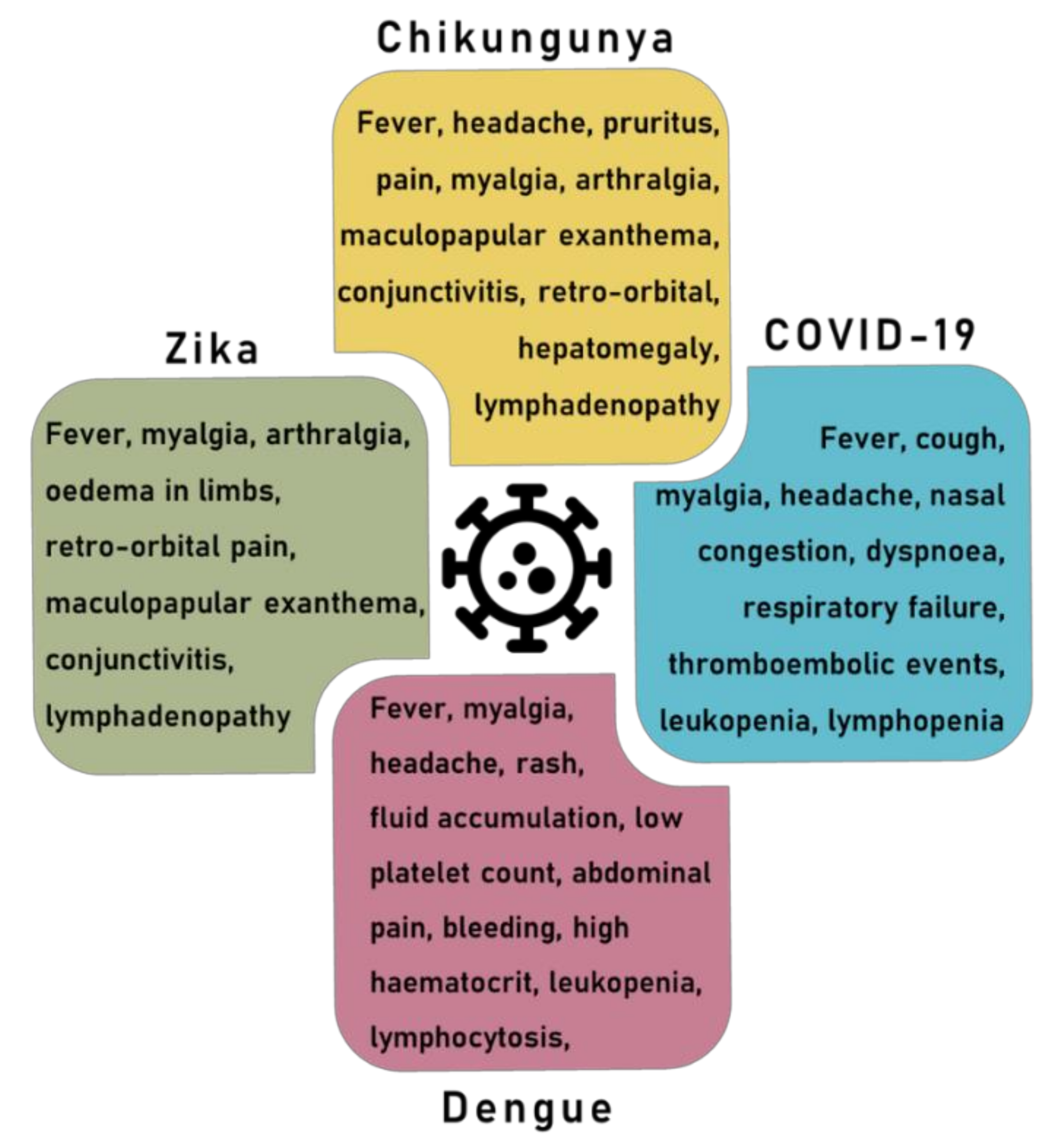
2.3. Arbovirus Disease Symptoms
2.4. Vaccine Development against Arbovirus
3. Coronavirus Disease
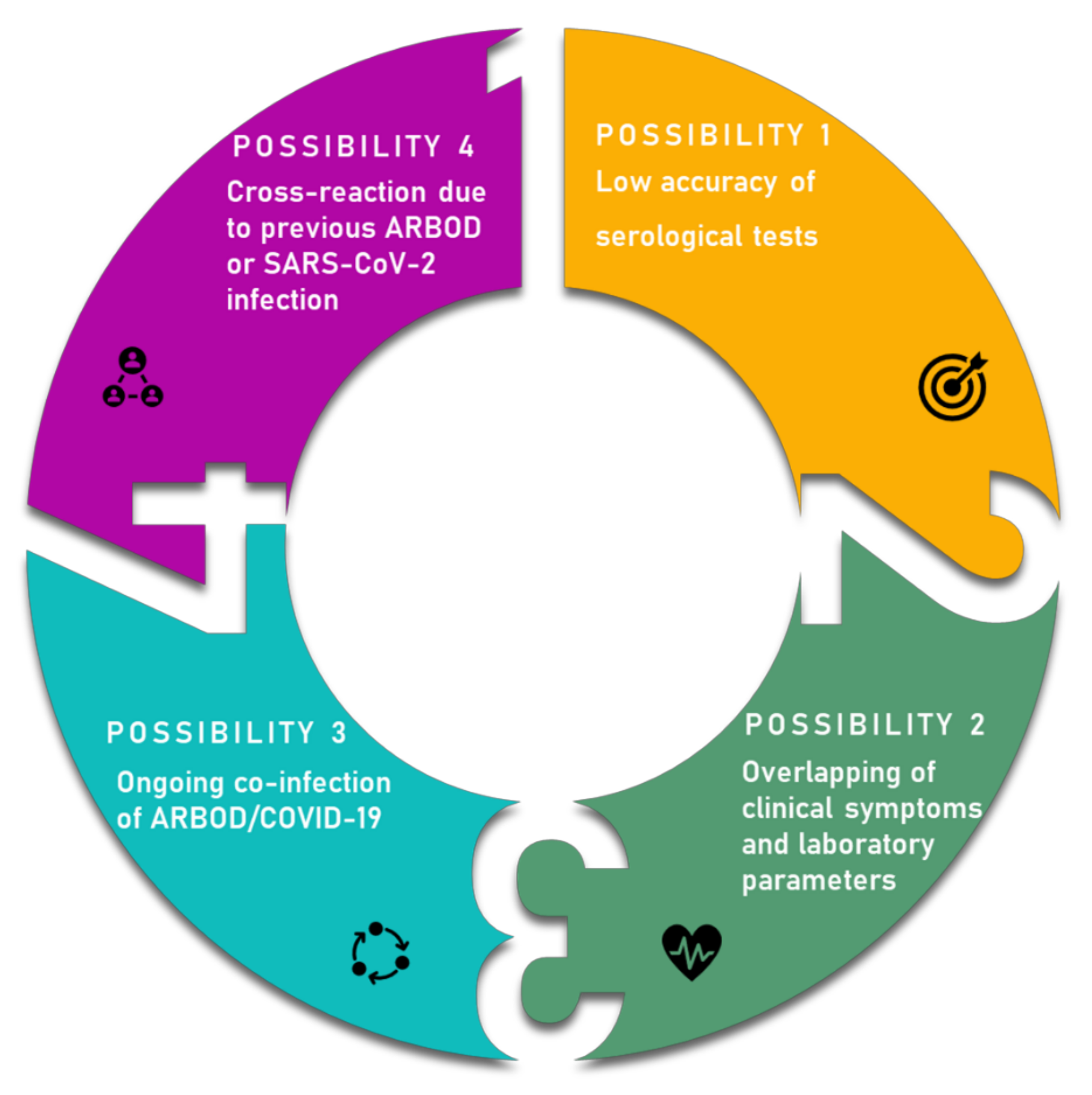
4. Overlapping Symptoms and Co-Infection
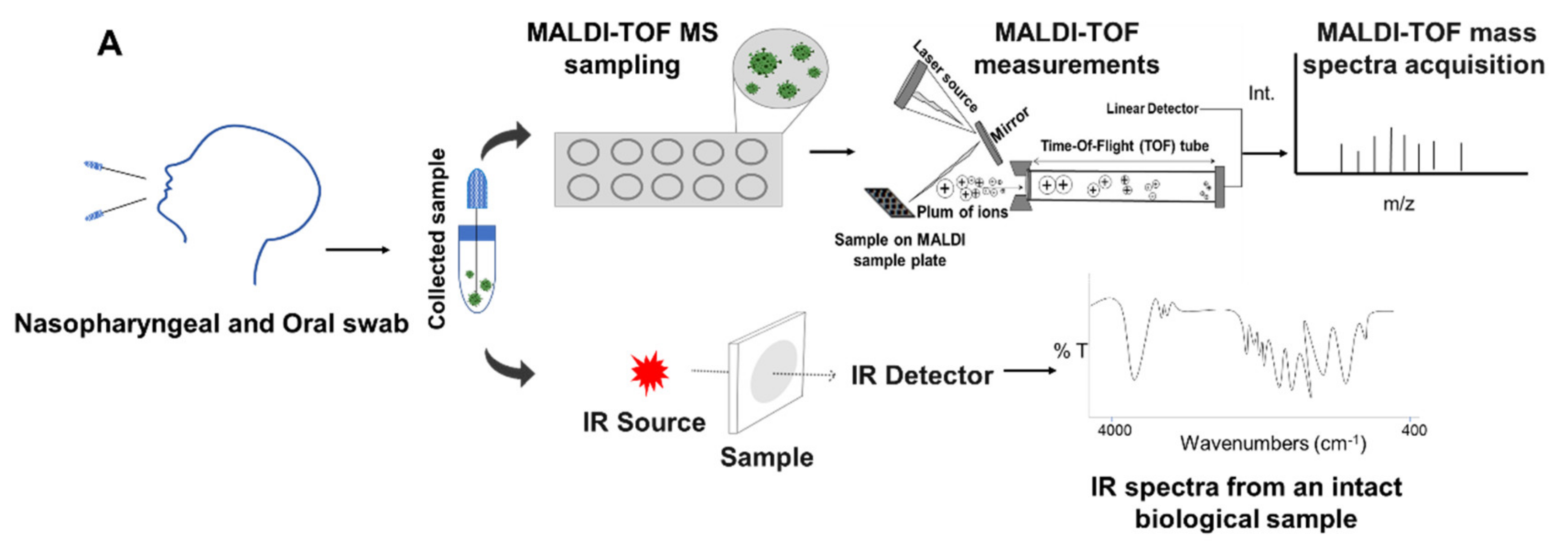
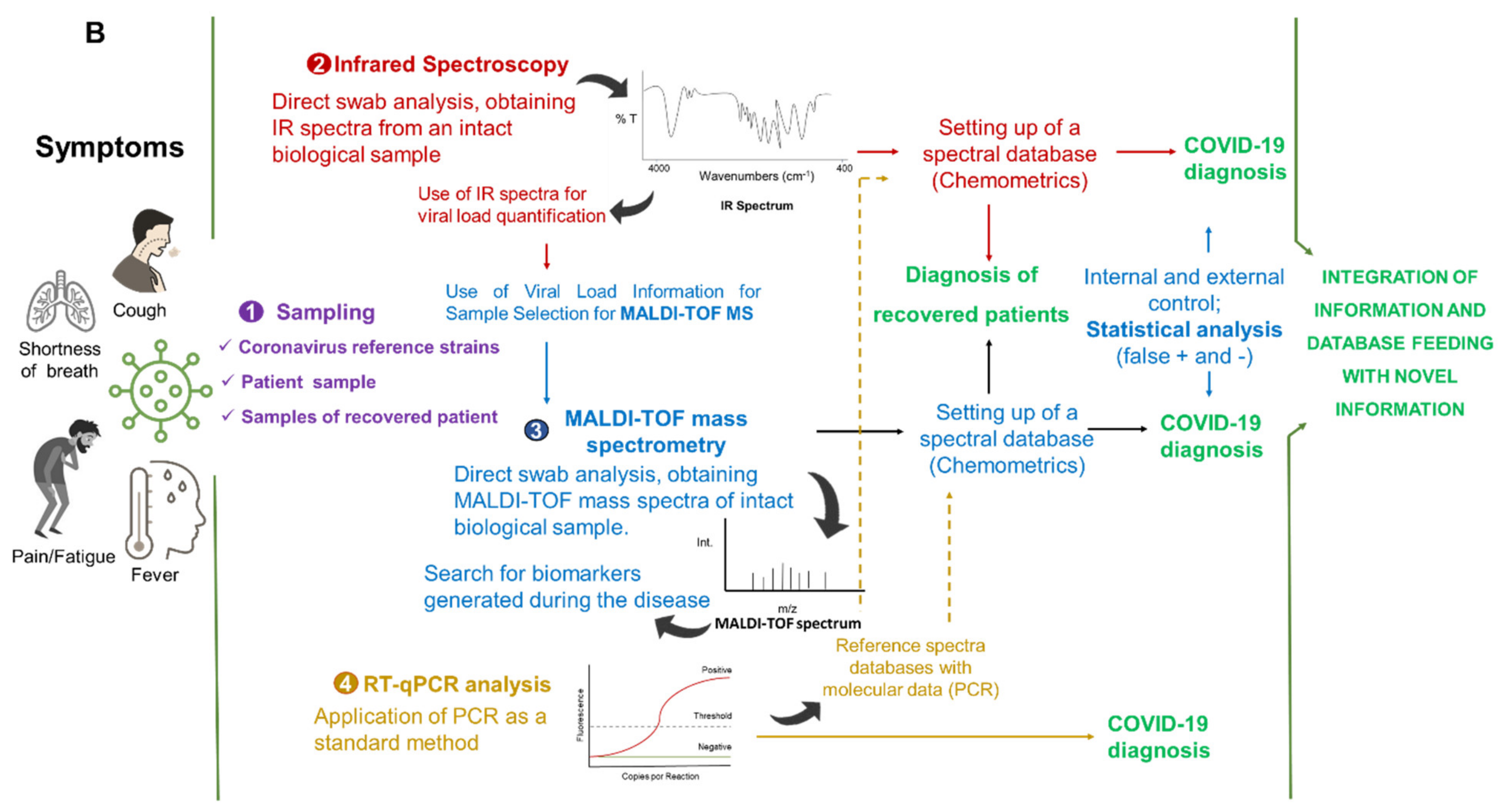
5. Integrated Analytical Platform for Fast and Cost-Effective of EID Diagnosis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–994. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Zoonoses. Available online: https://www.who.int/news-room/fact-sheets/detail/zoonoses (accessed on 30 January 2021).
- Haider, N.; Rothman-Ostrow, P.; Osman, A.Y.; Arruda, L.B.; Macfarlane-Berry, L.; Elton, L.; Thomason, M.J.; Yeboah-Manu, D.; Ansumana, R.; Kapata, N.; et al. COVID-19-Zoonosis or Emerging Infectious Disease? Front. Public Health 2020, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- El Amri, H.; Boukharta, M.; Zakham, F.; Ennaji, M.M. Emergence and Re-emergence of Viral Zoonotic Diseases: Concepts and Factors of Emerging and Re-emerging Globalization of Health Threats. In Emerging and Re-Emerging Viral Pathogens: Fundamental and Basic Virology Aspects of Human, Animal and Plant Pathogens; Ennaji, M.M., Ed.; Academic Press: Waltham, MA, USA, 2020; pp. 619–634. [Google Scholar] [CrossRef]
- El-Sayed, A.; Kamel, M. Climatic changes and their role in emergence and re-emergence of diseases. Environ. Sci. Pollut. Res. 2020, 27, 22336–22352. [Google Scholar] [CrossRef]
- Gibb, R.; Redding, D.W.; Chin, K.Q.; Donnelly, C.A.; Blackburn, T.M.; Newbold, T.; Jones, K.E. Zoonotic host diversity increases in human-dominated ecosystems. Nature 2020, 584, 398–402. [Google Scholar] [CrossRef]
- Faust, C.L.; McCallum, H.I.; Bloomfield, L.; Gottdenker, N.L.; Gillespie, T.R.; Torney, C.J.; Dobson, A.P.; Plowright, R.K. Pathogen spillover during land conversion. Ecol. Lett. 2018, 21, 471–483. [Google Scholar] [CrossRef]
- Baloch, S.; Baloch, M.A.; Zheng, T.; Pei, X. The Coronavirus Disease 2019 (COVID-19) Pandemic. Tohoku J. Exp. Med. 2020, 250, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Episode #21—COVID-19—Origins of the SARS-CoV-2 Virus. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/media-resources/science-in-5/episode-21---covid-19---origins-of-the-sars-cov-2-virus (accessed on 27 January 2021).
- United Nations Department of Economic and Social Affairs (UN DESA). World Economic Situation & Prospects: Report 12; United Nations: New York, NY, USA, 2021; ISBN 978-92-1-005498-0. [Google Scholar]
- International Labour Organization (ILO). Available online: https://www.ilo.org/wcmsp5/groups/public/---dgreports/---dcomm/documents/briefingnote/wcms_767028.pdf (accessed on 12 January 2021).
- Artsob, H.; Lindsay, R.; Drebot, M. Arboviruses. In International Encyclopedia of Public Health, 2nd ed.; Quah, S.R., Ed.; Academic Press: Waltham, MA, USA, 2017; pp. 154–160. [Google Scholar] [CrossRef]
- Freitas, L.P.; Gonçalves Cruz, O.; Lowe, R.; Sa Carvalho, M. Space-time clusters of dengue, chikungunya, and Zika cases in the city of Rio de Janeiro. Proc. R. Soc. B 2019, 286, 20191867. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control, an Agency of the European Union—ECDC. Dengue. Available online: https://www.ecdc.europa.eu/en/dengue (accessed on 27 January 2021).
- Spinicci, M.; Bartoloni, A.; Mantella, A.; Zammarchi, L.; Rossolini, G.M.; Antonelli, A. Low risk of serological cross-reactivity between dengue and COVID-19. Mem. Inst. Oswaldo Cruz 2020, 115, 1–2. [Google Scholar] [CrossRef]
- Malibari, A.A.; Al-Husayni, F.; Jabri, A.; Al-Amri, A.; Alharbi, M.A. Patient with Dengue Fever and COVID-19: Coinfection or Not? Cureus 2020, 12, 17–20. [Google Scholar] [CrossRef]
- Gijavanekar, C.; Drabek, R.; Soni, M.; Jackson, G.W.; Strych, U.; Fox, G.E.; Fofanov, Y. Detection and Typing of Viruses Using Broadly Sensitive Cocktail-PCR and Mass Spectrometric Cataloging Demonstration with Dengue Virus. J. Mol. Diagn. 2012, 14, 402–407. [Google Scholar] [CrossRef][Green Version]
- Fernandes, J.N.; Santos, L.M.B.; Chouin-carneiro, T.; Pavan, M.G.; Garcia, G.A.; David, M.R.; Beier, J.C.; Dowell, F.E. Rapid, non-invasive detection of Zika virus in Aedes aegypti mosquitoes by near-infrared spectroscopy. Sci. Adv. 2018, 4, eaat0496. [Google Scholar] [CrossRef]
- Khan, S.; Rehman, U. Spectroscopy as a tool for detection and monitoring of Coronavirus (COVID-19). Expert Rev. Mol. Diagn. 2020, 20, 1–3. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Disease Outbreaks. Available online: https://www.who.int/emergencies/diseases/en/ (accessed on 27 January 2021).
- McArthur, D.B. Emerging Infectious Diseases. Nurs. Clin. N. Am. 2019, 54, 297–311. [Google Scholar] [CrossRef]
- Weaver, S.C.; Chen, R.; Diallo, M. Chikungunya virus: Role of vectors in emergence from enzootic cycles. Annu. Rev. Entomol. 2020, 65, 313–332. [Google Scholar] [CrossRef]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Ross, R.W. The Newala epidemic. III. The virus: Isolation, pathogenic properties and relationship to the epidemic. Epidemiol. Infect. 1956, 54, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B.; Katzelnick, L.C.; Russell, P.K.; Markoff, L.; Aguiar, M.; Dans, L.R.; Dans, A.L. Ethics of a partially effective dengue vaccine: Lessons from the Philippines. Vaccine 2020, 38, 5572–5576. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Kraemer, M.U.G.; Brady, O.J.; Pigott, D.M.; Shearer, F.M.; Weiss, D.J.; Golding, N. Mapping global environmental suitability for Zika virus. eLife 2016, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cattarino, L.; Rodriguez-Barraquer, I.; Imai, N.; Cummings, D.A.T.; Ferguson, N.M. Mapping global variation in dengue transmission intensity. Sci. Transl. Med. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Pialoux, G.; Gaüzère, B.A.; Jauréguiberry, S.; Strobel, M. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis 2007, 7, 319–327. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 27 January 2021).
- Martín, J.L.; Brathwaite, O.; Zambrano, B.; Solórzano, J.O.; Bouckenooghe, A.; Dayan, G.H.; Guzmán, M.G. The epidemiology of dengue in the Americas over the last three decades: A worrisome reality. Am. J. Trop. Med. Hyg. 2010, 82, 128–135. [Google Scholar] [CrossRef]
- Vairo, F.; Aimè Coussoud-Mavoungou, M.P.; Ntoumi, F.; Castilletti, C.; Kitembo, L.; Haider, N.; Carletti, F. Chikungunya Outbreak in the Republic of the Congo, 2019-Epidemiological, Virological and Entomological Findings of a South-North Multidisciplinary Taskforce Investigation. Viruses 2020, 12, 1020. [Google Scholar] [CrossRef]
- Sardi, S.I.; Somasekar, S.; Naccache, S.N.; Bandeira, A.C.; Tauro, L.B.; Campos, G.S.; Chiub, C.Y. Coinfections of Zika and Chikungunya viruses in Bahia, Brazil, identified by metagenomic next-generation sequencing. J. Clin. Microbiol. 2016, 54, 2348–2353. [Google Scholar] [CrossRef]
- Metsky, H.C.; Matranga, C.B.; Wohl, S.; Schaffner, S.F.; Freije, C.A.; Winnicki, S.M.; Sabeti, P.C. Zika virus evolution and spread in the Americas. Nature 2017, 546, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bugallo, G.; Piedra, L.A.; Rodriguez, M.; Bisset, J.A.; Lourenço-de-Oliveira, R.; Weaver, S.C.; Vasilakis, N.; Vega-Rúa, A. Vector-borne transmission and evolution of Zika virus. Nat. Ecol. Evol. 2019, 3, 561–569. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Fact Sheets: Neglected Tropical Diseases. Available online: https://www.who.int/topics/tropical_diseases/factsheets/neglected/en/ (accessed on 27 January 2021).
- European Centre for Disease Prevention and Control. All Topics: A to Z. Available online: https://www.ecdc.europa.eu/en/all-topics#jump-Z (accessed on 27 January 2021).
- National Institute of Allergy and Infectious Diseases (NIAID’s). NIAID Emerging Infectious Diseases/Pathogens. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 27 January 2021).
- Communicable Disease Centre (CDC). Diseases: Neglected Tropical Diseases (NTDs). Available online: https://www.cdc.gov/globalhealth/ntd/diseases/index.html (accessed on 27 January 2021).
- Pan American Health Organization (PAHO). Zika. Available online: https://www.paho.org/es/temas/zika (accessed on 27 January 2021).
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Matusali, G.; Colavita, F.; Bordi, L.; Lalle, E.; Ippolito, G.; Capobianchi, M.R.; Castilletti, C. Tropism of the chikungunya virus. Viruses 2019, 11, 175. [Google Scholar] [CrossRef]
- Powers, A.M.; Brault, A.C.; Tesh, R.B.; Weaver, S.C. Re-emergence of chikungunya and o’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Naveca, F.G.; Claro, I.; Giovanetti, M.; de Jesus, J.G.; Xavier, J.; Iani, F.C.M.; Nascimento, V.A.D.; Souza, V.C.D.; Silveira, P.P.; Lourenço, J. Genomic, epidemiological and digital surveillance of Chikungunya virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 2018, 13, e0007065. [Google Scholar] [CrossRef] [PubMed]
- Sirinavin, S.; Nuntnarumit, P.; Supapannachart, S.; Boonkasidecha, S.; Techasaensiri, C.; Yoksarn, S. Vertical dengue infection: Case reports and review. Pediatr. Infect. Dis. J. 2004, 23, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Touret, Y.; Randrianaivo, H.; Michault, A.; Schuffenecker, I.; Kauffmann, E.; Lenglet, Y.; Barau, G.; Fourmaintraux, A. Early maternal-Fetal transmission of the Chikungunya virus. Presse Med. 2006, 35, 1656–1658. [Google Scholar] [CrossRef]
- Contopoulos-Ioannidis, D.; Newman-Lindsay, S.; Chow, C.; LaBeaud, A.D. Mother-to-child transmission of Chikungunya virus: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2018, 12, e0006510. [Google Scholar] [CrossRef]
- Peiter, P.C.; Pereira, R.D.S.; Moreira, M.C.N.; Nascimento, M.; Tavares, M.D.F.L.; Franco, V.D.C.; Cortes, J.J.C.; Campos, D.D.S.; Barcellos, C. Zika epidemic and microcephaly in Brazil: Challenges for access to health care and promotion in three epidemic areas. PLoS ONE 2020, 15, e0235010. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Teissier, A.; Rouault, E.; Teururai, S.; de Pina, J.J.; Nhan, T.X. Detection of chikungunya virus in saliva and urine. Virol. J. 2016, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I. The global burden of dengue: An analysis from the global burden of disease study. Lancet Infect. Dis. 2013, 16, 712–723. [Google Scholar] [CrossRef]
- Cardona-Ospina, J.A.; Arteaga-Livias, K.; Villamil-Gómez, W.E.; Pérez-Díaz, C.E.; Katterine Bonilla-Aldana, D.; Mondragon-Cardona, Á.; Solarte-Portilla, M. Dengue and COVID-19, overlapping epidemics? An analysis from Colombia. J. Med. Virol. 2021, 93, 522–527. [Google Scholar] [CrossRef]
- Akrami, K.M.; Nogueira, B.M.F.; Rosário, M.S.; Moraes, L.; Cordeiro, M.T.; Haddad, R.; Gomes, L.N. The re-emergence of Zika in Brazil in 2020: A case of Guillain-Barré Syndrome during the low season for arboviral infections. J. Travel Med. 2020, 27, 1–2. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastere, S.; Roche, C.; Vanhomwegen, J.; Dub, T. Guillain-Barré Syndrome outbreak caused by ZIKA virus infection in French Polynesia. Lancet 2017, 387, 1531–1539. [Google Scholar] [CrossRef]
- Schwartz, O.; Albert, M.L. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Hunsberger, S.; Ortega-Villa, A.M.; Powers, J.H.; Rincón León, H.A.; Caballero Sosa, S.; Ruiz Hernández, E.; Nájera Cancino, J.G.; Nason, M.; Lumbard, K.; Sepulveda, J.; et al. Patterns of signs, symptoms, and laboratory values associated with Zika, dengue, and undefined acute illnesses in a dengue endemic region: Secondary analysis of a prospective cohort study in southern Mexico. Int. J. Infect. Dis. 2020, 98, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Pessôa, R.; Patriota, J.V.; Lourdes de Souza, M.; Felix, A.C.; Mamede, N.; Sanabani, S.S. Investigation into an outbreak of dengue-like illness in Pernambuco, Brazil, revealed a cocirculation of Zika, Chikungunya, and dengue virus type 1. Medicine 2016, 95, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.M.O.; Tauro, L.B.; Kikuti, M.; Anjos, R.O.; Santos, V.C.; Gonçalves, T.S.F.; Paploski, I.A.D.; Moreira, P.S.S.; Nascimento, L.C.J.; Campos, G.S.; et al. Concomitant Transmission of Dengue, Chikungunya, and Zika Viruses in Brazil: Clinical and Epidemiological Findings from Surveillance for Acute Febrile Illness. Clin. Infect. Dis. 2019, 69, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Vellere, I.; Lagi, F.; Spinicci, M.; Mantella, A.; Mantengoli, E.; Corti, G.; Colao, M.G. Arbo-score: A rapid score for early identification of patients with imported arbovirosis caused by Dengue, Chikungunya and Zika virus. Microorganisms 2020, 8, 1731. [Google Scholar] [CrossRef]
- Diamond, M.S.; Ledgerwood, J.E.; Pierson, T.C. Zika virus vaccine development: Progress in the face of new challenges. Annu. Rev. Med. 2019, 70, 121–135. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Smith, P.G.; Luo, R.; Kelly-Cirino, C.; Curry, D.; Larson, H.; Durbin, A.; Chu, M.; Tharmaphornpilas, P.; Ng, L.C.; et al. Pre-vaccination screening strategies for the use of the CYD-TDV dengue vaccine: A meeting report. Vaccine 2019, 37, 5137–5146. [Google Scholar] [CrossRef]
- Prompetchara, E.; Ketloy, C.; Thomas, S.J.; Ruxrungtham, K. Dengue vaccine: Global development update. Asian Pac. J. Allergy Immunol. 2020, 38, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Villar, L.; Dayan, G.H.; Arredondo-García, J.L.; Rivera, D.M.; Cunha, R.; Deseda, C.; Reynales, H.; Costa, M.S.; Morales-Ramírez, J.O.; Carrasquilla, G.; et al. Efficacy of a Tetravalent Dengue Vaccine in Children in Latin America. N. Engl. J. Med. 2015, 372, 113–123. [Google Scholar] [CrossRef]
- Sridhar, S.; Luedtke, A.; Langevin, E.; Zhu, M.; Bonaparte, M.; Machabert, T.; Savarino, S.; Zambrano, B.; Moureau, A.; Khromava, A.; et al. Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N. Engl. J. Med. 2018, 379, 327–340. [Google Scholar] [CrossRef]
- Wilder-Smith, A. Dengue vaccine development: Status and future. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2020, 63, 40–44. [Google Scholar] [CrossRef]
- Biswal, S.; Borja-Tabora, C.; Martinez Vargas, L.; Velásquez, H.; Theresa Alera, M.; Sierra, V.; Johana Rodriguez-Arenales, E. Efficacy of a tetravalent dengue vaccine in healthy children aged 4-16 years: A randomised, placebo-controlled, phase 3 trial. Lancet 2020, 395, 1423–1433. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 27 January 2021).
- Meo, S.A.; Alhowikan, A.M.; Meo, I.M.; Halepoto, D.M. Novel coronavirus 2019-nCoV: Prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV 2012–2019. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2012–2019. [Google Scholar] [CrossRef]
- Zeng, Z.; Chen, D.; Tan, W.; Qiu, S.; Xu, D.; Liang, H. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: A study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 363–369. [Google Scholar] [CrossRef]
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Khalili, A.S.; Simonsen, L. Personal View Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Li, X.; Zai, J.; Zhao, Q.; Nie, Q.; Li, Y.; Foley, B.T.; Chaillon, A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020, 92, 602–611. [Google Scholar] [CrossRef]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Doxey, A.C.; Mossman, K.; Irving, A.T. Unraveling the Zoonotic Origin and Transmission of SARS-CoV-2. Trends Ecol. Evol. 2021, 36180–36184. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Komine-Aizawa, S.; Takada, K.; Hayakawa, S. Placental barrier against COVID-19. Placenta 2020, 15, 45–49. [Google Scholar] [CrossRef]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARSCoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef]
- Groß, R.; Conzelmann, C.; Müller, J.A.; Stenger, S.; Steinhart, K.; Kirchhoff, F.; Münch, J. Detection of SARS-CoV-2 in human breastmilk. Lancet 2020, 12, 1757–1758. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 5, 929–936. [Google Scholar] [CrossRef]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic features and clinical course of patients infected with SARS- CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, C.F.; Vora, N.; Byrne, T.; Lewer, D.; Kelly, G.; Heaney, J.; Gandhi, S. Pandemic peak SARS-CoV-2 infection and seroconversion rates in London frontline health-care workers. Lancet 2020, 396, e6–e7. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness—A living systematic review and meta-analysis. medRxiv 2020, 2, 13–22. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Li, Y.; Campbell, H.; Kulkarni, D.; Harpur, A.; Nundy, M.; Wang, X.; Nair, H. The temporal association of introducing and lifting non-pharmaceutical interventions with the time-varying reproduction number (R) of SARS-CoV-2: A modelling study across 131 countries. Lancet Infect. Dis. 2021, 21, 193–202. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Coronavirus Disease 2019 (COVID-19): Situation Report—24. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200213-sitrep-24-covid-19.pdf?sfvrsn=9a7406a4_4 (accessed on 27 January 2021).
- Najafimehr, H.; Mohamed Ali, K.; Safari, S.; Yousefifard, M.; Hosseini, M. Estimation of basic reproduction number for COVID-19 and the reasons for its differences. Int. J. Clin. Pract. 2020, 74, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020, 17, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.L.; Wang, Y.M.; Wu, Z.Q.; Xiang, Z.C.; Guo, L.; Xu, T.; Jiang, Y.Z.; Xiong, Y.; Li, Y.-J.; Li, X.W.; et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020, 5, 1015–1024. [Google Scholar] [CrossRef]
- Guijarro-Castro, C.; Rosón-González, M.; Abreu, A.; García-Arratibel, A.; Ochoa-Mulas, M. Síndrome de Guillain-Barré tras infección por SARS-CoV-2. Comentarios tras la publicación de 16 nuevos casos. Neurología 2020, 35, 412–415. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, D.; Zhou, H.; Liu, J.; Chen, S. Guillain-Barré syndrome associated with SARS-CoV-2 infection: Causality or coincidence? Lancet Neurol. 2020, 19, 383–384. [Google Scholar] [CrossRef]
- World Health Organization (WHO). SARS-CoV-2 Variants. Available online: https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ (accessed on 27 January 2021).
- Kirby, T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021, 5, e20–e21. [Google Scholar] [CrossRef]
- Wise, J. Patient with new strain of coronavirus is treated in intensive care at London hospital. BMJ 2012, 345, e6455. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 11 January 2021).
- Moraes, B.C.; Souza, E.B.; Sodré, G.R.C.; Ferreira, D.B.D.S.; Ribeiro, J.B.M. Seasonality of dengue reporting in state capitals in the Brazilian amazon and impacts of el niño/la niña. Cad. Saude Publ. 2019, 35, 1–7. [Google Scholar] [CrossRef]
- Lokida, D.; Lukman, N.; Salim, G.; Butar, D.P.B.; Kosasih, H.; Wulan, W.N.; Naysilla, A.M.; Djajady, Y.; Sari, R.A.; Arlinda, D.; et al. Diagnosis of COVID-19 in a Dengue-Endemic Area. Am. J. Trop. Med. Hyg. 2020, 103, 1220–1222. [Google Scholar] [CrossRef]
- Teotônio, I.M.S.N.; de Carvalho, J.L.; Castro, L.C.; Nitz, N.; Hagström, L.; Rios, G.G.; Oliveira, M.F.R.; Dallago, B.S.L.; Hecht, M. Clinical and biochemical parameters of COVID-19 patients with prior or active dengue fever. Acta Trop. 2021, 214, 105782. [Google Scholar] [CrossRef]
- Joob, B.; Wiwanitkit, V. COVID-19 can present with a rash and be mistaken for dengue. J. Am. Acad. Dermatol. 2020, 82. [Google Scholar] [CrossRef]
- Yan, G.; Lee, C.K.; Lam, L.T.M.; Yan, B.; Chua, Y.X.; Lim, A.Y.N.; Phang, K.F.; Kew, G.S.; Teng, H.; Ngai, C.H.; et al. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet Infect. Dis. 2020, 20, 536. [Google Scholar] [CrossRef]
- Prasitsirikul, W.; Pongpirul, K.; Pongpirul, W.A.; Panitantum, N.; Ratnarathon, A.C.; Hemachudha, T. Nurse infected with Covid-19 from a provisional dengue patient. Emerg. Microbes Infect. 2020, 9, 1354–1355. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Keler, S.; Kolodny, R.; Ben-Tal, N.; Atias-Varon, D.; Shlush, E.; Gerlic, M. Potential Antigenic Cross-reactivity Between Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Dengue Viruses. Clin. Infect. Dis. 2020, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kembuan, G.J. Dengue serology in Indonesian COVID-19 patients: Coinfection or serological overlap? IDCases 2020, 22, e00927. [Google Scholar] [CrossRef] [PubMed]
- Estofolete, C.F.; Machado, L.F.; Zini, N.; Luckemeyer, G.D.; Moraes, M.M.; dos Santos, T.M.I.L.; dos Santos, B.F.; Ruiz, L.G.P.; Vasilakis, N.; Lobo, S.M.A.; et al. Presentation of fatal stroke due to SARS-CoV-2 and dengue virus coinfection. J. Med. Virol. 2021, 93, 1770–1775. [Google Scholar] [CrossRef]
- Erukhimovitch, V.; Karpasasa, M.; Huleihel, M. Spectroscopic Detection and Identification of Infected cell with Herpes Viruses. Biopolymers 2009, 91, 61–67. [Google Scholar] [CrossRef]
- Chou, T.; Hsu, W.; Wang, C.; Chen, Y.; Fang, J. Rapid and specific influenza virus detection by functionalized magnetic nanoparticles and mass spectrometry. J. Nanobiotechnol. 2011, 9, 52. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Wang, X.; Yao, H.; Xu, X.; Zhang, P.; Zhang, M.; Shao, J.; Xiao, Y.; Wang, H. Limits of Detection of 6 Approved RT-PCR Kits for the Novel SARS-Coronavirus-2 (SARS-CoV-2). Clin. Chem 2020, 66, 977–979. [Google Scholar] [CrossRef]
- Pejcic, B.; Myers, M.; Ross, A. Mid-Infrared Sensing of Organic Pollutants in Aqueous Environments. Sensors 2009, 9, 6232–6253. [Google Scholar] [CrossRef]
- Sakudo, A.; Babac, K.; Ikutaa, K. Discrimination of influenza virus-infected nasal fluids by Vis-NIR spectroscopy. Clin. Chim. Acta 2012, 24, 130–134. [Google Scholar] [CrossRef]
- Nachtigall, F.M.; Pereira, A.; Trofymchuk, O.S.; Santos, L.S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020, 38, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Fraga, M.E.; Kozakiewicz, Z.; Lima, N. Fourier transform infrared as a powerful technique for the identification and characterization of filamentous fungi and yeasts. Res. Microbiol. 2010, 161, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Sousa, C.; Lopes, J.A.; Novais, A.; Peixe, L. A Front Line on Klebsiella pneumoniae Capsular Polysaccharide Knowledge: Fourier Transform Infrared Spectroscopy as an Accurate and Fast Typing Tool. mSystems 2020, 5, e00386-19. [Google Scholar] [CrossRef]
- Krokhin, O.; Li, Y.; Andonov, A.; Feldmann, H.; Flick, R.; Jones, S.; Stroeher, U.; Bastien, N.; Dasuri, K.V.N.; Cheng, K.; et al. Mass Spectrometric Characterization of Proteins from the SARS Virus. Mol. Cell Proteomics 2003, 2, 346–356. [Google Scholar] [CrossRef]
- Vitale, R.; Roine, E.; Bamford, D.H.; Corcell, A. Lipid fingerprints of intact viruses by MALDI-TOF/mass spectrometry. Biochim. Biophys. Acta 2013, 1831, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Hsu, I.H.; Sun, Y.C.; Wang, Y.K.; Wu, T.K. Immunocapture couples with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for rapid detection of type 1 dengue virus. J. Chromatogr. 2013, 1288, 21–27. [Google Scholar] [CrossRef]
- Magalhães, T.T.B.; Almeida, L.B.V. Scientific Report: Métodos Para Diagnóstico Molecular Em Pacientes Com Arboviroses Recorrentes Por Espectrometria de Massas: Íons Marcadores de Rápida Identificação Para Dengue (I, II E III), Zika E Chikungunya Vírus; Centro Universitário de Brasília- UNICEUB: Brasilia, Brazil, 2019. [Google Scholar]
- Luan, J.; Yuan, J.; Li, X.; Jin, S.; Yu, L.; Liao, M.; Zhang, H.; Xu, C. Multiplex Detection of 60 Hepatitis B Virus Variants by MALDI-TOF Mass Spectrometry. Clin. Chem. 2009, 1509, 1503–1509. [Google Scholar] [CrossRef]
- Tran, A.; Alby, K.; Kerr, A.; Jones, M.; Gilligan, P.H. Cost Savings Realized by Implementation of Routine Microbiological Identification by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2015, 53, 2473–2479. [Google Scholar] [CrossRef] [PubMed]
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 2017, 9, 1510. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. How the pandemic might play out in 2021 and beyond. Nature 2020, 584, 22–25. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, J.; Ferreira, E.C.; Santos, C. COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control. Microorganisms 2021, 9, 708. https://doi.org/10.3390/microorganisms9040708
Costa J, Ferreira EC, Santos C. COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control. Microorganisms. 2021; 9(4):708. https://doi.org/10.3390/microorganisms9040708
Chicago/Turabian StyleCosta, Jéssica, Eugénio C. Ferreira, and Cledir Santos. 2021. "COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control" Microorganisms 9, no. 4: 708. https://doi.org/10.3390/microorganisms9040708
APA StyleCosta, J., Ferreira, E. C., & Santos, C. (2021). COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control. Microorganisms, 9(4), 708. https://doi.org/10.3390/microorganisms9040708







