Network Analysis Based on Unique Spectral Features Enables an Efficient Selection of Genomically Diverse Operational Isolation Units
Abstract
1. Introduction
2. Materials and Methods
2.1. Lactobacillus Brevis and Benchmark MALDI Datasets
2.2. Isolation, MALDI-TOF MS Sample Preparation and Data Acquisition
2.3. SPeDE Dereplication, Hierarchical Clustering and Network Analysis
2.4. Genome Sequencing, Assembly and Analysis of 14 Case Study Isolates
3. Results
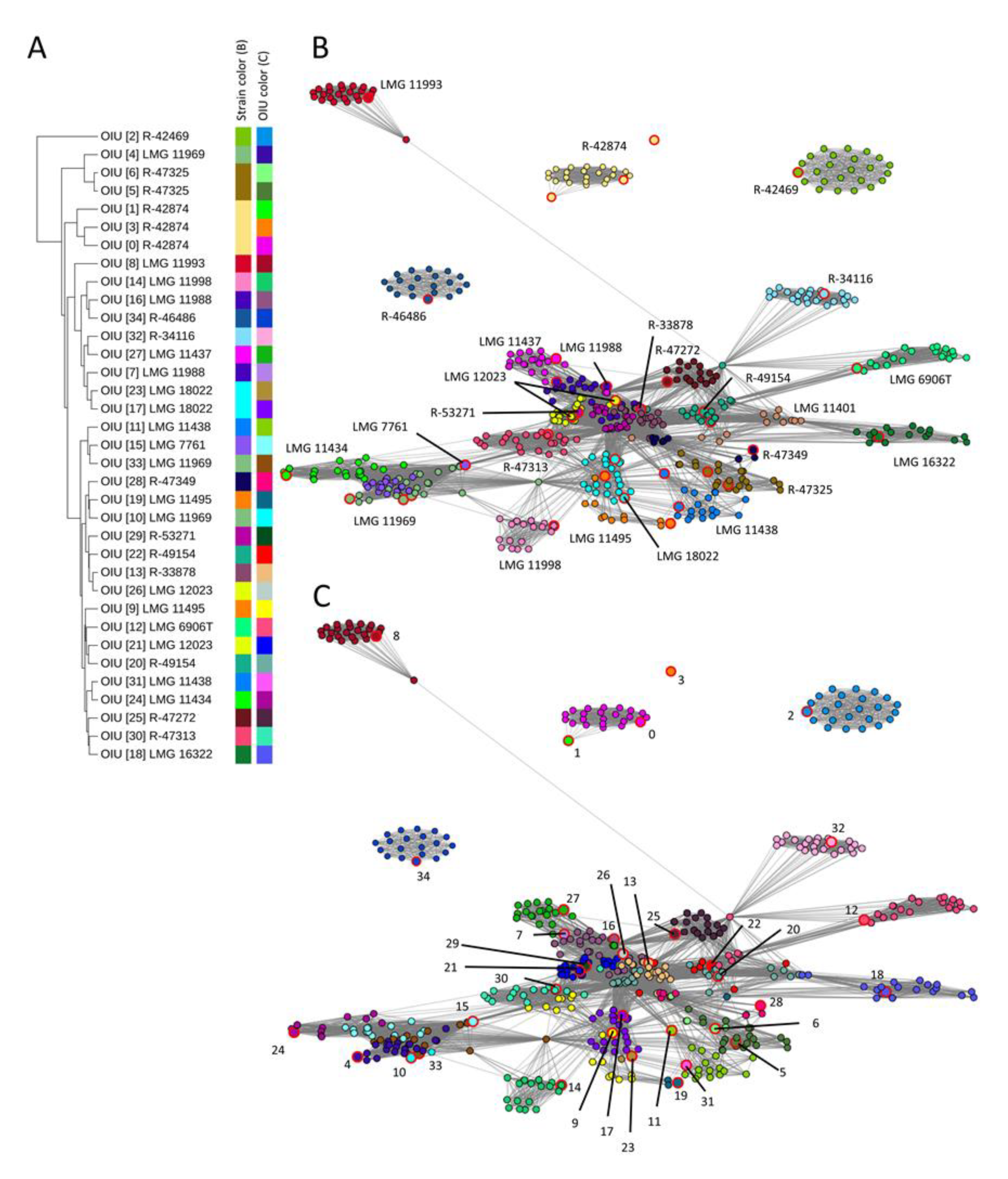
3.1. Network Analysis Clustered OIUs Representing the Same L. brevis Strains Together
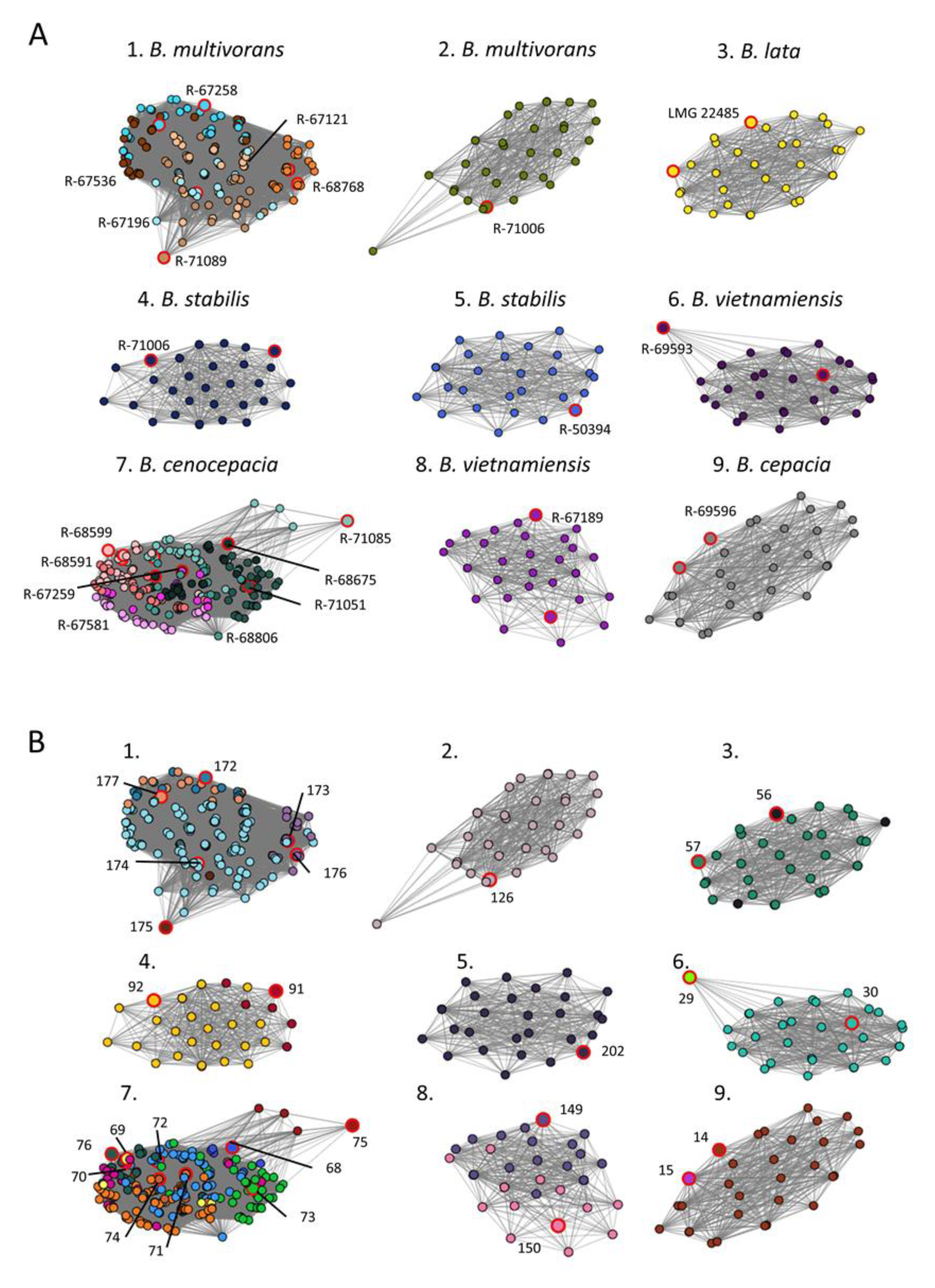
3.2. Network Analysis Differentiated OIUs Representing Closely Related Burkholderia Cepacia Complex Species
3.3. Dereplication of a Set of Soil Isolates
3.4. Genomic Analysis of Soil Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Carvalho, C.C.C.R. Whole cell biocatalysts: Essential workers from Nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- BCC Research. Microbial Products: Technologies, Applications and Global Markets; Report Code: BIO086D; BCC Research: Wellesley, MA, USA, 2018. [Google Scholar]
- Anteneh, Y.S.; Franco, C.M.M. Whole Cell Actinobacteria as Biocatalysts. Front. Microbiol. 2019, 10, 77. [Google Scholar] [CrossRef]
- Zhang, L.; Sandrin, T.R. Maximizing the Taxonomic Resolution of MALDI-TOF-MS-Based Approaches to Bacterial Characterization: From Culture Conditions Through Data Analysis. In Applications of Mass Spectrometry in Microbiology; Springer International Publishing: Cham, Switzerland, 2016; pp. 147–181. [Google Scholar]
- Giraud-Gatineau, A.; Texier, G.; Garnotel, E.; Raoult, D.; Chaudet, H. Insights Into Subspecies Discrimination Potentiality From Bacteria MALDI-TOF Mass Spectra by Using Data Mining and Diversity Studies. Front. Microbiol. 2020, 11, 1931. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef]
- Maier, T.; Klepel, S.; Renner, U.; Kostrzewa, M. Fast and reliable MALDI-TOF MS–based microorganism identification. Nat. Chem. Biol. 2006, 3. [Google Scholar] [CrossRef]
- Dubois, D.; Grare, M.; Prere, M.-F.; Segonds, C.; Marty, N.; Oswald, E. Performances of the Vitek MS Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry System for Rapid Identification of Bacteria in Routine Clinical Microbiology. J. Clin. Microbiol. 2012, 50, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- Rahi, P.; Prakash, O.; Shouche, Y.S. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass-Spectrometry (MALDI-TOF MS) Based Microbial Identifications: Challenges and Scopes for Microbial Ecologists. Front. Microbiol. 2016, 7, 1359. [Google Scholar] [CrossRef] [PubMed]
- Huschek, D.; Witzel, K. Rapid dereplication of microbial isolates using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: A mini-review. J. Adv. Res. 2019, 19, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Kashtan, N.; Roggensack, S.E.; Rodrigue, S.; Thompson, J.W.; Biller, S.J.; Coe, A.; Ding, H.; Marttinen, P.; Malmstrom, R.R.; Stocker, R.; et al. Single-Cell Genomics Reveals Hundreds of Coexisting Subpopulations in Wild Prochlorococcus. Science 2014, 344, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, A.F.; O Wertheim, J.; Barone, L.; Gentile, N.; Krizanc, D.; Cohan, F.M. Speedy speciation in a bacterial microcosm: New species can arise as frequently as adaptations within a species. ISME J. 2013, 7, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Spitaels, F.; Wieme, A.D.; Vandamme, P. MALDI-TOF MS as a Novel Tool for Dereplication and Characterization of Microbiota in Bacterial Diversity Studies. In Applications of Mass Spectrometry in Microbiology; Springer International Publishing: Cham, Switzerland, 2016; pp. 235–256. [Google Scholar]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2013, 32, 188–217. [Google Scholar] [CrossRef]
- Dumolin, C.; Aerts, M.; Verheyde, B.; Schellaert, S.; Vandamme, T.; Van Der Jeugt, F.; De Canck, E.; Cnockaert, M.; Wieme, A.D.; Cleenwerck, I.; et al. Introducing SPeDE: High-Throughput Dereplication and Accurate Determination of Microbial Diversity from Matrix-Assisted Laser Desorption–Ionization Time of Flight Mass Spectrometry Data. mSystems 2019, 4, e00437-19. [Google Scholar] [CrossRef] [PubMed]
- Christner, M.; Dressler, D.; Andrian, M.; Reule, C.; Petrini, O. Identification of Shiga-Toxigenic Escherichia coli outbreak isolates by a novel data analysis tool after matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. PLoS ONE 2017, 12, e0182962. [Google Scholar] [CrossRef]
- Ghyselinck, J.; Van Hoorde, K.; Hoste, B.; Heylen, K.; De Vos, P. Evaluation of MALDI-TOF MS as a tool for high-throughput dereplication. J. Microbiol. Methods 2011, 86, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-Cell MALDI-TOF MS Versus 16S rRNA Gene Analysis for Identification and Dereplication of Recurrent Bacterial Isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef] [PubMed]
- Oberle, M.; Wohlwend, N.; Jonas, D.; Maurer, F.P.; Jost, G.; Tschudin-Sutter, S.; Vranckx, K.; Egli, A. The Technical and Biological Reproducibility of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Based Typing: Employment of Bioinformatics in a Multicenter Study. PLoS ONE 2016, 11, e0164260. [Google Scholar] [CrossRef] [PubMed]
- Purves, K.; Macintyre, L.; Brennan, D.; Hreggviðsson, G.; Kuttner, E. Using Molecular Networking for Microbial Secondary Metabolite Bioprospecting. Metabolites 2016, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A.; et al. Molecular Networking as a Dereplication Strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Vanhellemont, M.; Verheyen, K.; Baeten, L. Relating changes in understorey diversity to environmental drivers in an ancient forest in northern Belgium. Plant Ecol. Evol. 2014, 147, 22–32. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Hagberg, A.A.; Schult, D.A.; Swart, P.J. Exploring Network Structure, Dynamics, and Function using NetworkX. In Proceedings of the 7th Python in Science Conference, Pasadena, CA, USA, 19–24 August 2008; pp. 11–15. [Google Scholar]
- Bokeh Development Team. Bokeh: Python Library for Interactive Visualization. Available online: https://bokeh.pydata.org/en/latest/ (accessed on 1 December 2020).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Arnold, R.J.; Karty, J.A.; Ellington, A.D.; Reilly, J.P. Monitoring the growth of a bacteria culture by MALDI-MS of whole cells. Anal. Chem. 1999, 71, 1990–1996. [Google Scholar] [CrossRef]
- Ruelle, V.; El Moualij, B.; Zorzi, W.; Ledent, P.; De Pauw, E. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2013–2019. [Google Scholar] [CrossRef]
- Giebel, R.; Worden, C.; Rust, S.; Kleinheinz, G.; Robbins, M.; Sandrin, T. Microbial Fingerprinting using Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry (MALDI-TOF MS). Adv. Appl. Microbiol. 2010, 71, 149–184. [Google Scholar] [PubMed]
- Wieme, A.D.; Spitaels, F.; Aerts, M.; De Bruyne, K.; Van Landschoot, A.; Vandamme, P. Effects of Growth Medium on Matrix-Assisted Laser Desorption–Ionization Time of Flight Mass Spectra: A Case Study of Acetic Acid Bacteria. Appl. Environ. Microbiol. 2014, 80, 1528–1538. [Google Scholar] [CrossRef]
- Christensen, J.J.; Dargis, R.; Hammer, M.; Justesen, U.S.; Nielsen, X.C.; Kemp, M. The Danish MALDI-TOF MS Study Group Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Analysis of Gram-Positive, Catalase-Negative Cocci Not Belonging to the Streptococcus or Enterococcus Genus and Benefits of Database Extension. J. Clin. Microbiol. 2012, 50, 1787–1791. [Google Scholar] [CrossRef] [PubMed]
- Stafsnes, M.H.; Dybwad, M.; Brunsvik, A.; Bruheim, P. Large scale MALDI-TOF MS based taxa identification to identify novel pigment producers in a marine bacterial culture collection. Antonie Leeuwenhoek 2012, 103, 603–615. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, N.; Xu, B.; Hao, H.; Kang, L.; Zheng, Y.; Jiang, Y.; Jiang, H. Identification and Cluster Analysis of Streptococcus pyogenes by MALDI-TOF Mass Spectrometry. PLoS ONE 2012, 7, e47152. [Google Scholar] [CrossRef] [PubMed]

| R-69749 | R-69927 | R-69980 | R-70006 | R-70199 | R-70211 | R-70036 | R-69658 | R-69781 | R-69955 | R-69643 | R-69776 | R-70025 | R-69608 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-69749 | 100 | 99.6 | 99.6 | 99.6 | 99.6 | 99.6 | 89.9 | 89.9 | 90.0 | 89.9 | 90.8 | 90.9 | 89.9 | 90.8 |
| R-69927 | 100 | 99.6 | 99.5 | 99.7 | 99.6 | 89.8 | 89.8 | 89.8 | 89.8 | 90.0 | 90.0 | 89.8 | 90.1 | |
| R-69980 | 100 | 99.6 | 99.6 | 99.7 | 89.9 | 90.0 | 90.0 | 90.0 | 90.1 | 90.0 | 90.0 | 90.0 | ||
| R-70006 | 100 | 99.7 | 99.7 | 89.8 | 89.9 | 89.8 | 89.9 | 90.1 | 90.0 | 89.9 | 90.1 | |||
| R-70199 | 100 | 99.7 | 89.7 | 89.9 | 89.8 | 89.8 | 90.1 | 90.0 | 89.7 | 90.0 | ||||
| R-70211 | 100 | 89.9 | 89.9 | 89.9 | 89.9 | 89.9 | 89.9 | 89.9 | 89.9 | |||||
| R-70036 | 100 | 98.0 | 98.0 | 98.0 | 95.3 | 95.3 | 95.1 | 95.3 | ||||||
| R-69658 | 100 | 100 | 100 | 95.6 | 95.6 | 95.4 | 95.5 | |||||||
| R-69781 | 100 | 100 | 95.6 | 95.6 | 95.4 | 95.5 | ||||||||
| R-69955 | 100 | 95.6 | 95.6 | 95.4 | 95.5 | |||||||||
| R-69643 | 100 | 99.0 | 98.7 | 99.0 | ||||||||||
| R-69776 | 100 | 98.8 | 99.0 | |||||||||||
| R-70025 | 100 | 98.8 | ||||||||||||
| R-69608 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumolin, C.; Peeters, C.; De Canck, E.; Boon, N.; Vandamme, P. Network Analysis Based on Unique Spectral Features Enables an Efficient Selection of Genomically Diverse Operational Isolation Units. Microorganisms 2021, 9, 416. https://doi.org/10.3390/microorganisms9020416
Dumolin C, Peeters C, De Canck E, Boon N, Vandamme P. Network Analysis Based on Unique Spectral Features Enables an Efficient Selection of Genomically Diverse Operational Isolation Units. Microorganisms. 2021; 9(2):416. https://doi.org/10.3390/microorganisms9020416
Chicago/Turabian StyleDumolin, Charles, Charlotte Peeters, Evelien De Canck, Nico Boon, and Peter Vandamme. 2021. "Network Analysis Based on Unique Spectral Features Enables an Efficient Selection of Genomically Diverse Operational Isolation Units" Microorganisms 9, no. 2: 416. https://doi.org/10.3390/microorganisms9020416
APA StyleDumolin, C., Peeters, C., De Canck, E., Boon, N., & Vandamme, P. (2021). Network Analysis Based on Unique Spectral Features Enables an Efficient Selection of Genomically Diverse Operational Isolation Units. Microorganisms, 9(2), 416. https://doi.org/10.3390/microorganisms9020416






