Gut Microbiome of a Multiethnic Community Possessed No Predominant Microbiota
Abstract
1. Introduction
2. Materials and Methods
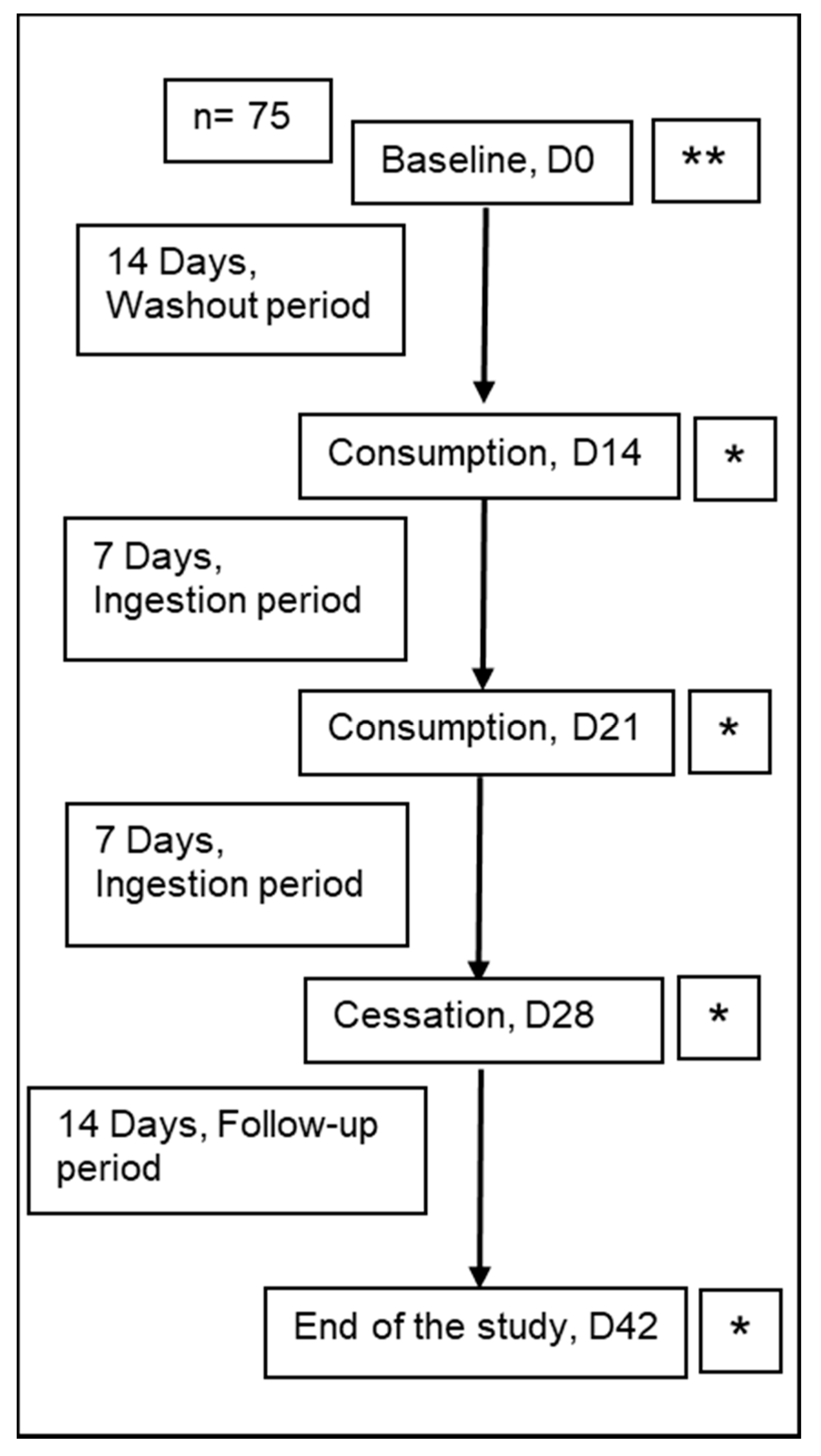
2.1. Study Design
2.2. Food Frequency Questionnaire
2.3. Faecal Sample Collection and DNA Extraction
2.4. 16s rRNA DNA Sequencing
2.5. Bioinformatics Analysis and Clustering
2.6. Faecal Water Cytokines Analysis
2.7. Statistical Analysis
3. Results
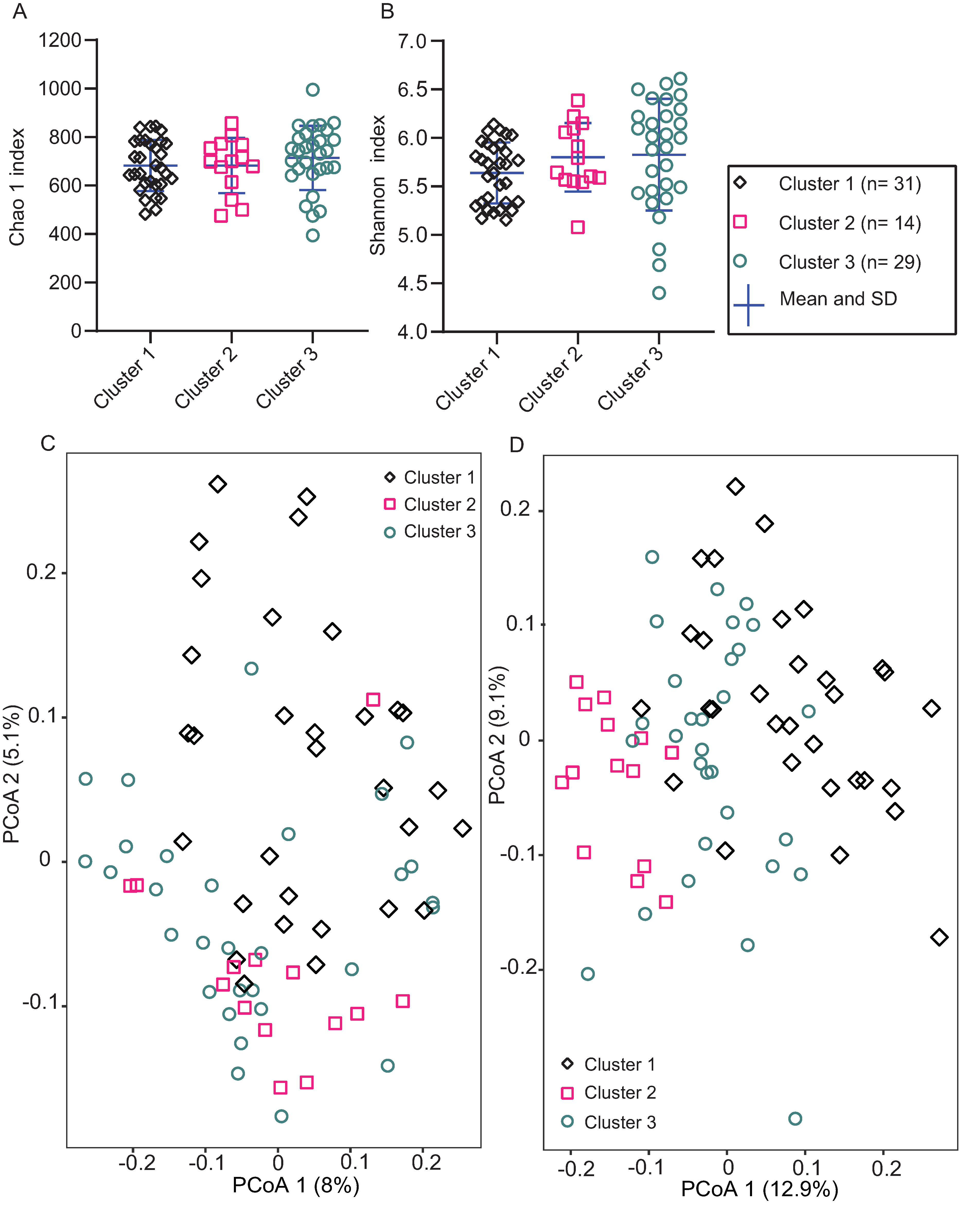
3.1. Clustering of Basal GI Microbiome
3.2. Dietary Composition and Habit
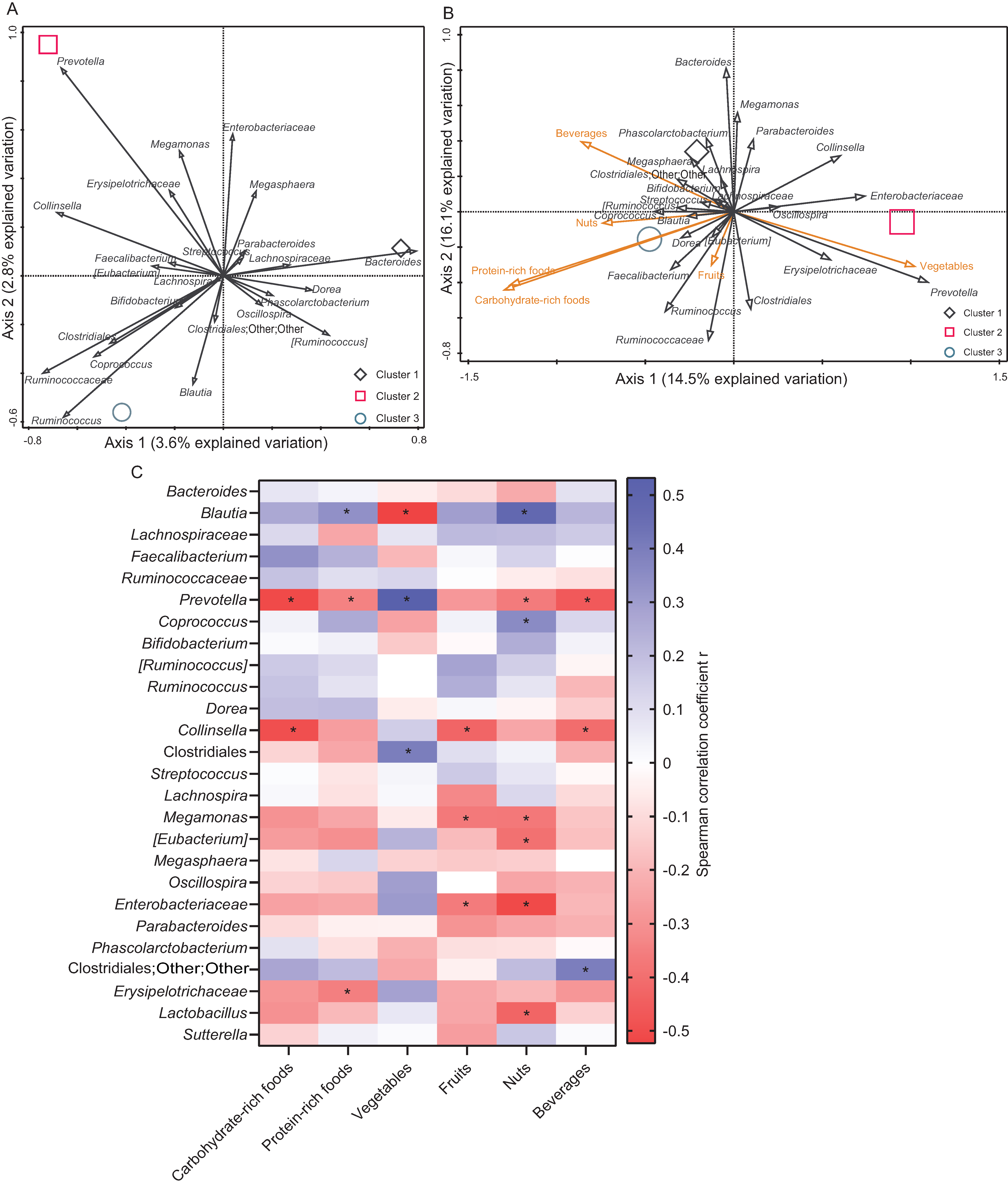
3.3. Correlation between Diet and Microbiota
3.4. Effects of Ethnicities, Genders and Types of Bristol Stool Scale in Association with GI Microbiome
3.5. Biodiversity of GI Microbiome
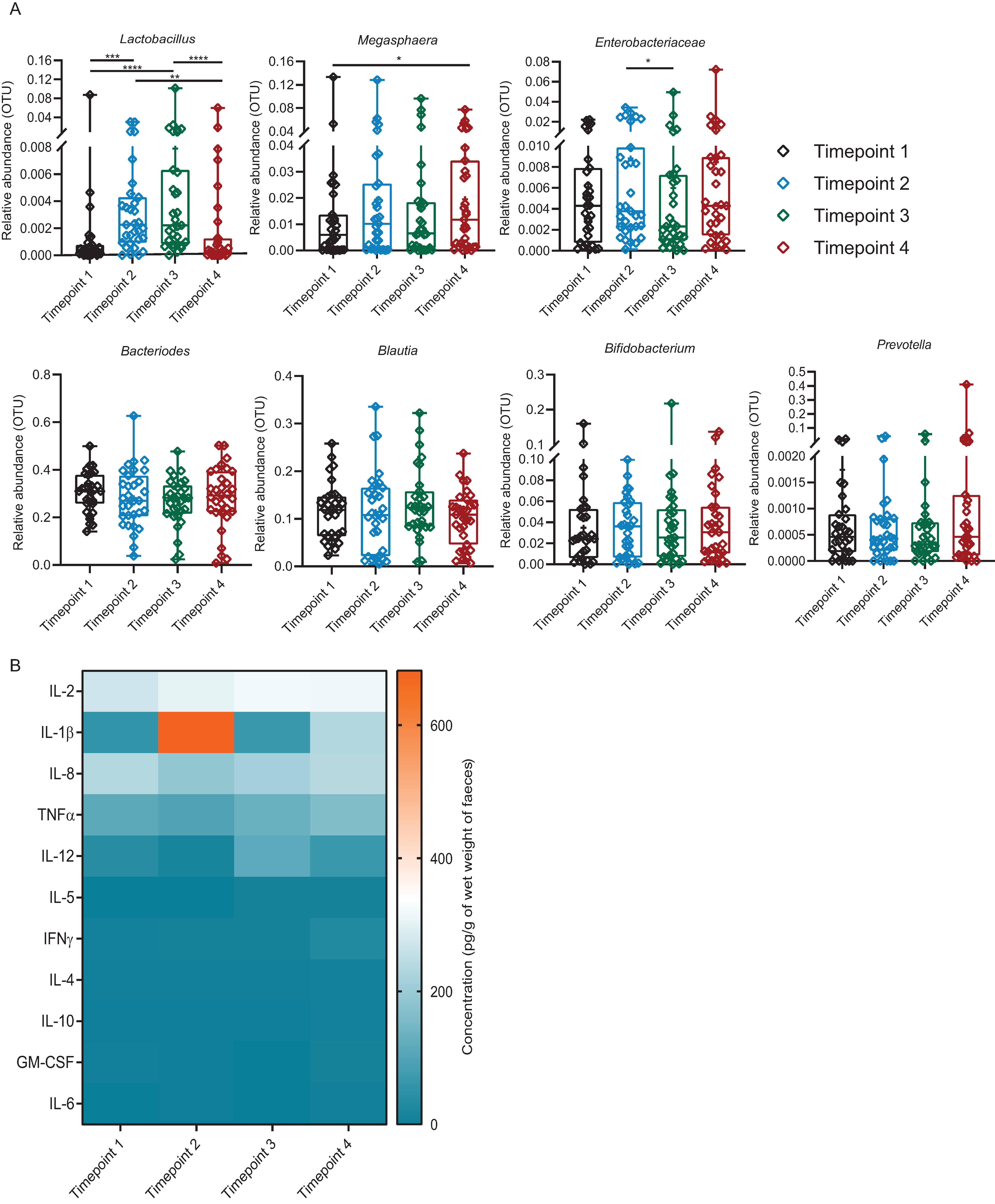
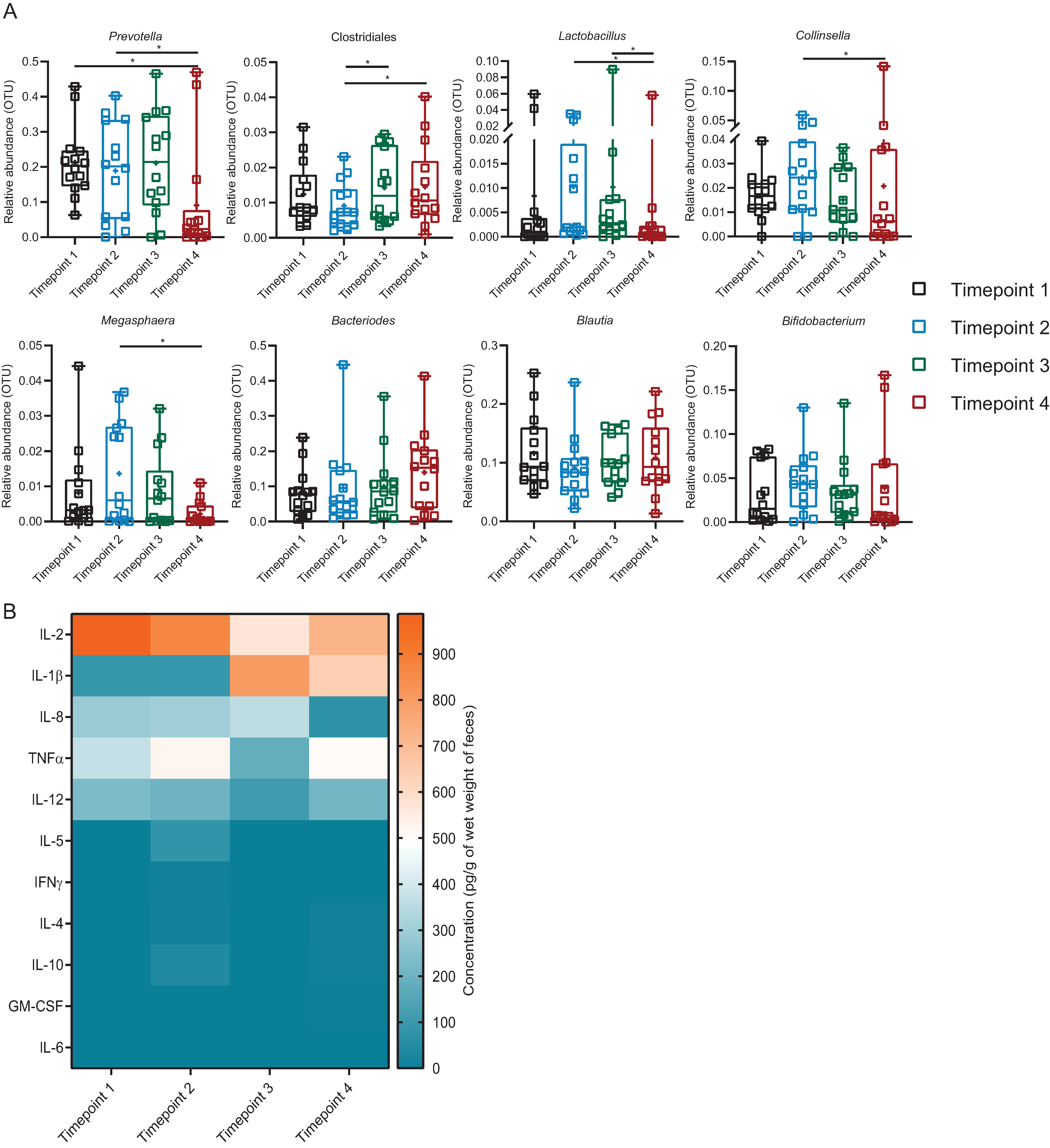
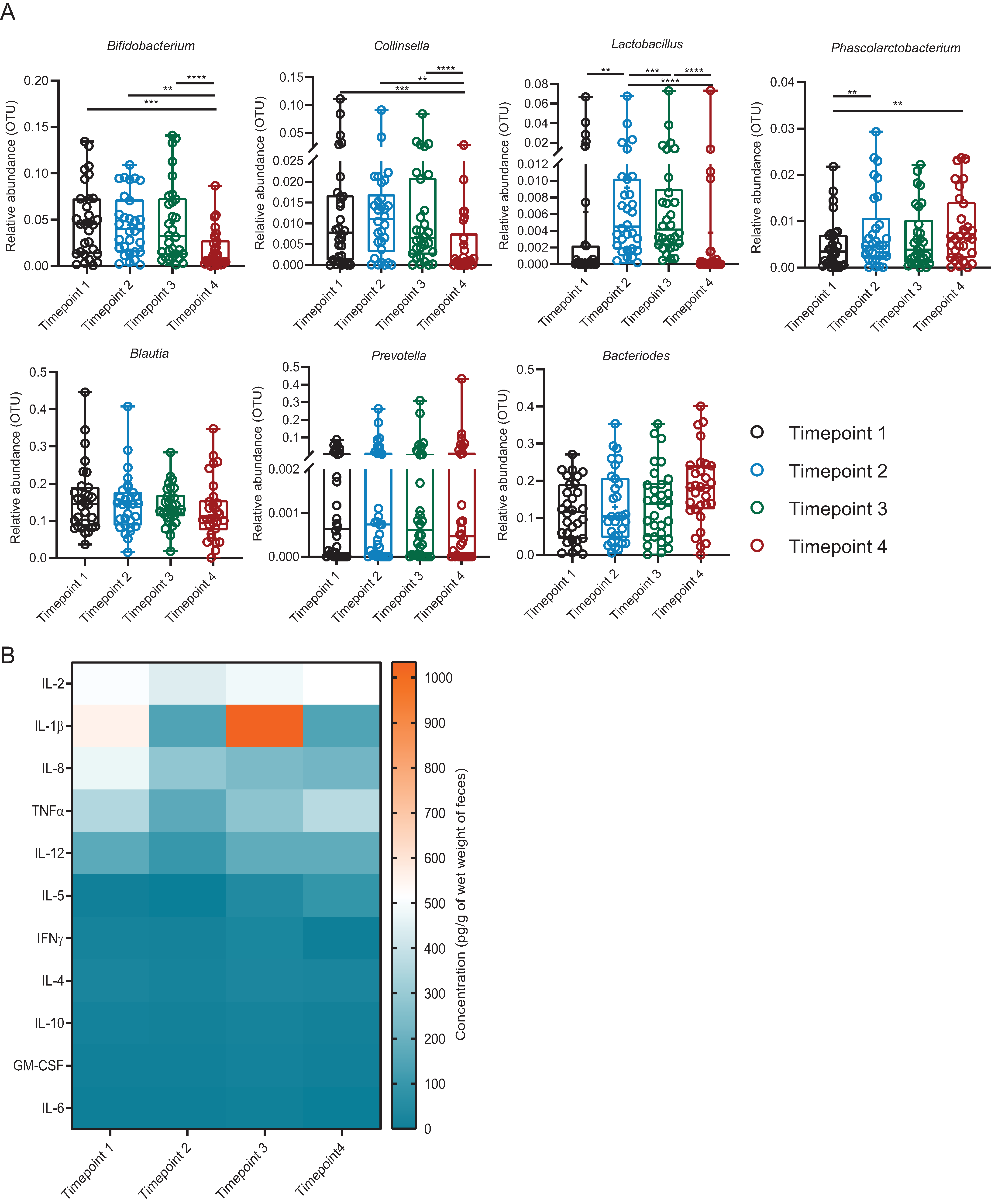
3.6. Introduction of Exogenous Lactobacillus: Cluster 1
3.7. Introduction of Exogenous Lactobacillus: Cluster 2
3.8. Introduction of Exogenous Lactobacillus: Cluster 3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GRAS | Generally regarded as safe |
| IL | Interleukin |
| TNFα | Tumour Necrosis Factor alpha |
| GM-CSF | Granulocyte-macrophage Colony-Stimulating Factor |
| IFNɣ | Interferon gamma |
| BMI | Body Mass Index |
| OTU | Operational Taxonomic Unit |
| NAFLD | Non-Alcoholic Fatty Liver Disease |
| HALE | Healthy Life Expectancy |
| CFU | Colony Forming Unit |
| API | Analytical Profile Index |
| FFQ | Food Frequency Questionnaire |
| PBS | Phosphate-Buffered Saline |
| TE | Tris Ethylenediaminetetraacetic acid |
| HT1 | Hybridisation Buffer |
| NaOH | Sodium hydroxide |
| PCR | Polymerase Chain Reaction |
| QIIME | Quantitative Insights Into Microbial Ecology |
| JSD | Jensen-Shannon distance |
| PAM | Partitioning Around medoids |
| CH | Calinski-Harabasz |
| Si | Silhouette coefficient |
| db-RDA | distance based Redundancy Analysis |
| PCoA | principal coordinate analysis |
| PMSF | Phenyl-methyl-sulfonyl fluoride |
| BSA | Bovine Serum Albumin |
| LLOQ | lower limit of quantification |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| ANOVA | Analysis of Variance |
References
- Mulligan, C.M.; Friedman, J.E. Maternal modifiers of the infant gut microbiota: Metabolic consequences. J. Endocrinol. 2017, 235, R1–R12. [Google Scholar] [CrossRef]
- Requena, T.; Martinez-Cuesta, M.C.; Peláez, C. Diet and microbiota linked in health and disease. Food Funct. 2018, 9, 688–704. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, V.K.; Arrieta, M.-C. Host–microbiome intestinal interactions during early life: Considerations for atopy and asthma development. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 138–148. [Google Scholar] [CrossRef]
- Ruohtula, T.; De Goffau, M.C.; Nieminen, J.K.; Honkanen, J.; Siljander, H.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Ilonen, J.; Niemelä, O.; et al. Maturation of Gut Microbiota and Circulating Regulatory T Cells and Development of IgE Sensitization in Early Life. Front. Immunol. 2019, 10, 2494. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nat. Cell Biol. 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016, 24, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Soty, M.; Gautier-Stein, A.; Rajas, F.; Mithieux, G. Gut-Brain Glucose Signaling in Energy Homeostasis. Cell Metab. 2017, 25, 1231–1242. [Google Scholar] [CrossRef]
- Khine, W.W.T.; Rahayu, E.S.; See, T.Y.; Kuah, S.; Salminen, S.; Nakayama, J.; Lee, Y.-K. Indonesian children fecal microbiome from birth until weaning was different from microbiomes of their mothers. Gut Microbes 2020, 12, 1–19. [Google Scholar] [CrossRef]
- Khine, W.W.T.; Zhang, Y.; GoIe, G.J.Y.; Wong, M.S.; Liong, M.; Lee, Y.Y.; Cao, H.; Lee, Y.-K. Gut microbiome of pre-adolescent children of two ethnicities residing in three distant cities. Sci. Rep. 2019, 9, 7831. [Google Scholar] [CrossRef]
- Nakayama, J.; Watanabe, K.; Jiang, J.; Matsuda, K.; Chao, S.-H.; Haryono, P.; La-Ongkham, O.; Sarwoko, M.-A.; Sujaya, I.N.; Zhao, L.; et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015, 5, 8397. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.-J.; Zhang, W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; Van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; De Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.P. Pairwiseadonis: Pairwise Multilevel Comparison Using ADONIS. R Package Version 0.3. 2019. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 20 December 2020).
- Lewis, S.J.; Heaton, K.W. Stool Form Scale as a Useful Guide to Intestinal Transit Time. Scand. J. Gastroenterol. 1997, 32, 920–924. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Lee, Y.-S.; Kim, Y.; Lee, S.-H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.-S.; Lim, H.S.; Kim, M.-S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Zhao, F.; Liu, W.; Lv, R.; Khine, W.W.T.; Han, J.; Sun, Z.; Lee, Y.-K.; Zhang, H. Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes 2020, 12, 1–20. [Google Scholar] [CrossRef]
- Khine, W.; Ang, X.; Chan, Y.; Lee, W.; Quek, S.; Tan, S.; Teo, H.; Teo, J.; Lau, Q.; Lee, Y.-K. Recovery of Lactobacillus casei strain Shirota (LcS) from faeces of healthy Singapore adults after intake of fermented milk. Benef. Microbes 2019, 10, 721–728. [Google Scholar] [CrossRef]
- Atassi, F.; Brassart, D.; Grob, P.; Graf, F.; Servin, A.L. Lactobacillusstrains isolated from the vaginal microbiota of healthy women inhibitPrevotella biviaandGardnerella vaginalisin coculture and cell culture. FEMS Immunol. Med. Microbiol. 2006, 48, 424–432. [Google Scholar] [CrossRef]
- Vuotto, C.; Barbanti, F.; Mastrantonio, P.; Donelli, G. Lactobacillus brevisCD2 inhibitsPrevotella melaninogenicabiofilm. Oral Dis. 2014, 20, 668–674. [Google Scholar] [CrossRef]
- Naidoo, N.; Van Dam, R.M.; Ng, S.; Tan, C.S.; Chen, S.; Lim, J.Y.; Chan, M.F.; Chew, L.; Rebello, S.A. Determinants of eating at local and western fast-food venues in an urban Asian population: A mixed methods approach. Int. J. Behav. Nutr. Phys. Act. 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Lehtimäki, J.; Ruokolainen, L. The ecology of human microbiota: Dynamics and diversity in health and disease. Ann. N. Y. Acad. Sci. 2017, 1399, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Rigatto, K.; Gazzana, M.B.; Knorst, M.M.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. Altered Gut Microbiome Profile in Patients With Pulmonary Arterial Hypertension. Hypertension 2020, 75, 1063–1071. [Google Scholar] [CrossRef]
- Yoshida, N.; Yamashita, T.; Hirata, K.-I. Gut Microbiome and Cardiovascular Diseases. Diseases 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Wang, C.; Wang, L.; He, T.; Hu, H.; Song, J.; Cui, C.; Qiao, J.; Qing, L.; et al. Enterotype Bacteroides Is Associated with a High Risk in Patients with Diabetes: A Pilot Study. J. Diabetes Res. 2020, 2020, 1–11. [Google Scholar] [CrossRef]
- Alhinai, E.A.; Walton, G.E.; Commane, D.M. The Role of the Gut Microbiota in Colorectal Cancer Causation. Int. J. Mol. Sci. 2019, 20, 5295. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, J.; Qian, Y.; Sun, T.; Chen, H.; Chen, Z.; Sun, D.; Zhong, M.; Chen, H.; Hong, J.; et al. Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, C.G.; Jo, M.; Park, C.O.; Gwon, S.-Y.; Hwang, S.; Yi, H.C.; Lee, S.-Y.; Eom, Y.-B.; Karim, B.; et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int. J. Med Sci. 2020, 17, 145–152. [Google Scholar] [CrossRef]
- Gil-Cruz, C.; Perez-Shibayama, C.; De Martin, A.; Ronchi, F.; Van Der Borght, K.; Niederer, R.; Onder, L.; Lütge, M.; Novkovic, M.; Nindl, V.; et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 2019, 366, 881–886. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Q.; Lin, P.; Xu, R.; He, D.; Ji, W.; Bian, Y.; Shen, Y.; Li, Q.; Liu, C.; et al. Characteristics of Gut Microbiota in Patients With Rheumatoid Arthritis in Shanghai, China. Front. Cell. Infect. Microbiol. 2019, 9, 369. [Google Scholar] [CrossRef]
- Fernández-Tomé, S.; Marin, A.C.; Moreno, L.O.; Baldan-Martin, M.; Mora-Gutiérrez, I.; Lanas-Gimeno, A.; Moreno-Monteagudo, J.A.; Santander, C.; Sánchez, B.; Chaparro, M.; et al. Immunomodulatory Effect of Gut Microbiota-Derived Bioactive Peptides on Human Immune System from Healthy Controls and Patients with Inflammatory Bowel Disease. Nutrients 2019, 11, 2605. [Google Scholar] [CrossRef]
- Lin, C.-H.; Chen, C.-C.; Chiang, H.-L.; Liou, J.-M.; Chang, C.-M.; Lu, T.-P.; Chuang, E.Y.; Tai, Y.-C.; Cheng, C.; Lin, H.-Y.; et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 2019, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Viitasalo, L.; Kurppa, K.; Ashorn, M.; Saavalainen, P.; Huhtala, H.; Ashorn, S.; Mäki, M.; Ilus, T.; Kaukinen, K.; Iltanen, S. Microbial Biomarkers in Patients with Nonresponsive Celiac Disease. Dig. Dis. Sci. 2018, 63, 3434–3441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lukiw, W.J. Microbiome-Mediated Upregulation of MicroRNA-146a in Sporadic Alzheimer’s Disease. Front. Neurol. 2018, 9, 145. [Google Scholar] [CrossRef]
- Larsen, J.M. The immune response toPrevotellabacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Chan, M.H.; Chen, Z.; Lam, E.W.H.; Wong, P.Y.; Ngai, C.M.; Chan, P.K.S.; Hui, M. The human oral cavity microbiota composition during acute tonsillitis: A cross-sectional survey. BMC Oral Health 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Dong, T.S.; Katzka, W.; Lagishetty, V.; Luu, K.; Hauer, M.; Pisegna, J.; Jacobs, J.P. A Microbial Signature Identifies Advanced Fibrosis in Patients with Chronic Liver Disease Mainly Due to NAFLD. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Fu, B.C.; Hullar, M.A.J.; Randolph, T.W.; Franke, A.A.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Shepherd, J.A.; Madeleine, M.M.; Le Marchand, L.; et al. Associations of plasma trimethylamine N-oxide, choline, carnitine, and betaine with inflammatory and cardiometabolic risk biomarkers and the fecal microbiome in the Multiethnic Cohort Adiposity Phenotype Study. Am. J. Clin. Nutr. 2020, 111, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.P.; Cruz, Á.A.; Xavier, M.T.; Stöcker, A.; Carvalho-Filho, P.; Miranda, P.M.; Meyer, R.J.; Soledade, K.R.; Gomes-Filho, I.S.; Trindade, S.C. Prevotella intermedia and periodontitis are associated with severe asthma. J. Periodontol. 2020, 91, 46–54. [Google Scholar] [CrossRef]
- Human Development Report 2019, Yearly Human Development Reports UNDP. Available online: http://hdr.undp.org/en/content/2019-human-development-index-ranking (accessed on 1 July 2020).
- Healthy Life Expectancy (HALE) Data by WHO Region. Available online: https://apps.who.int/gho/data/view.main.HALEXREGv?lang=en (accessed on 1 July 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khine, W.W.T.; Teo, A.H.T.; Loong, L.W.W.; Tan, J.J.H.; Ang, C.G.H.; Ng, W.; Lee, C.N.; Zhu, C.; Lau, Q.C.; Lee, Y.-K. Gut Microbiome of a Multiethnic Community Possessed No Predominant Microbiota. Microorganisms 2021, 9, 702. https://doi.org/10.3390/microorganisms9040702
Khine WWT, Teo AHT, Loong LWW, Tan JJH, Ang CGH, Ng W, Lee CN, Zhu C, Lau QC, Lee Y-K. Gut Microbiome of a Multiethnic Community Possessed No Predominant Microbiota. Microorganisms. 2021; 9(4):702. https://doi.org/10.3390/microorganisms9040702
Chicago/Turabian StyleKhine, Wei Wei Thwe, Anna Hui Ting Teo, Lucas Wee Wei Loong, Jarett Jun Hao Tan, Clarabelle Geok Hui Ang, Winnie Ng, Chuen Neng Lee, Congju Zhu, Quek Choon Lau, and Yuan-Kun Lee. 2021. "Gut Microbiome of a Multiethnic Community Possessed No Predominant Microbiota" Microorganisms 9, no. 4: 702. https://doi.org/10.3390/microorganisms9040702
APA StyleKhine, W. W. T., Teo, A. H. T., Loong, L. W. W., Tan, J. J. H., Ang, C. G. H., Ng, W., Lee, C. N., Zhu, C., Lau, Q. C., & Lee, Y.-K. (2021). Gut Microbiome of a Multiethnic Community Possessed No Predominant Microbiota. Microorganisms, 9(4), 702. https://doi.org/10.3390/microorganisms9040702






