Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Measurement of SCFA Production by GC-MS
2.3. Tolerance to Gastric Stress
2.4. Immunomodulation Assay
2.5. In Vitro Epithelial Barrier Model
2.6. Enteroendocrine Cell Line and GLP-1 Induction
2.7. Statistical Analysis
3. Results
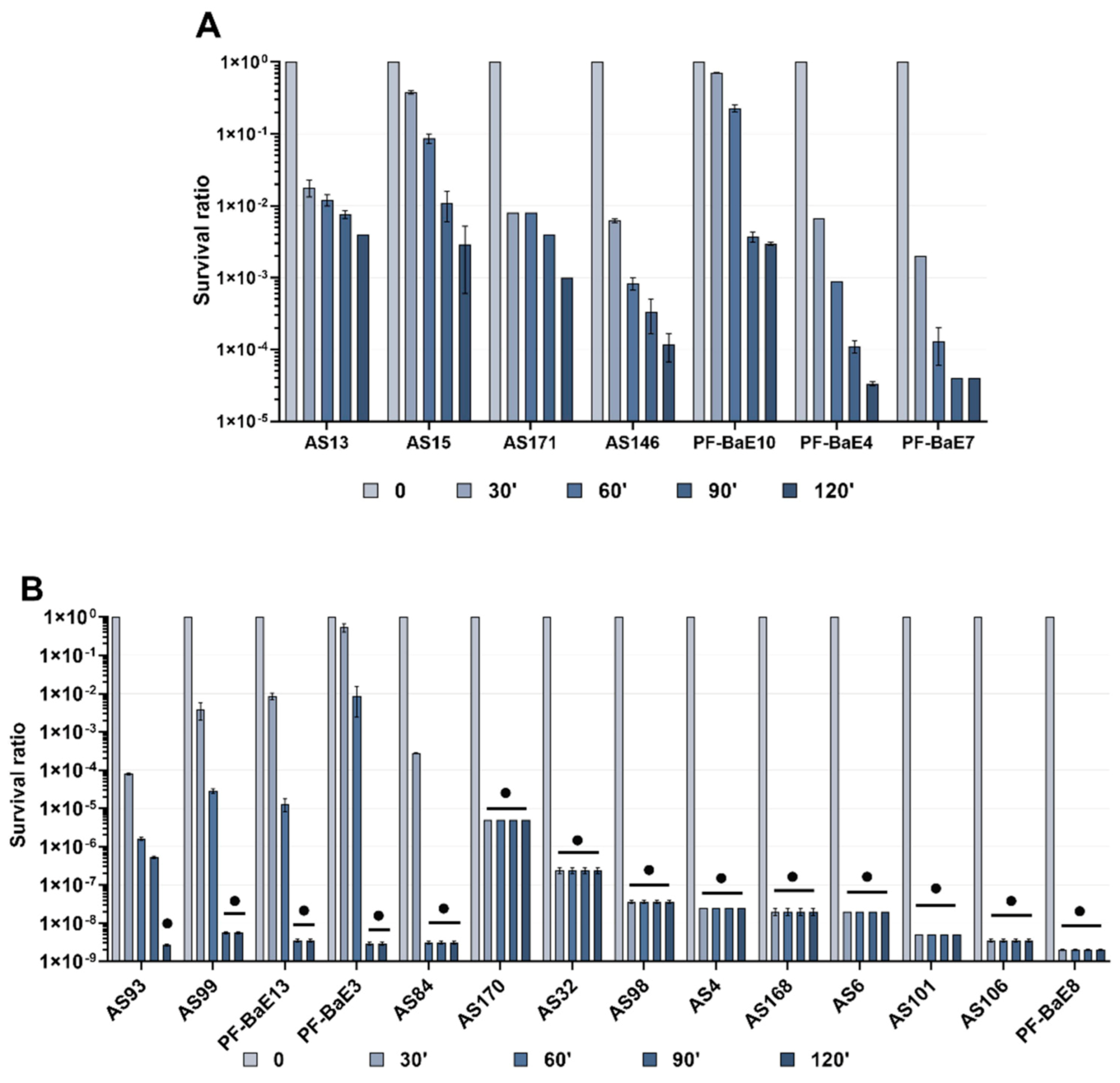
3.1. Strain-Dependent Survival to Gastric Stress
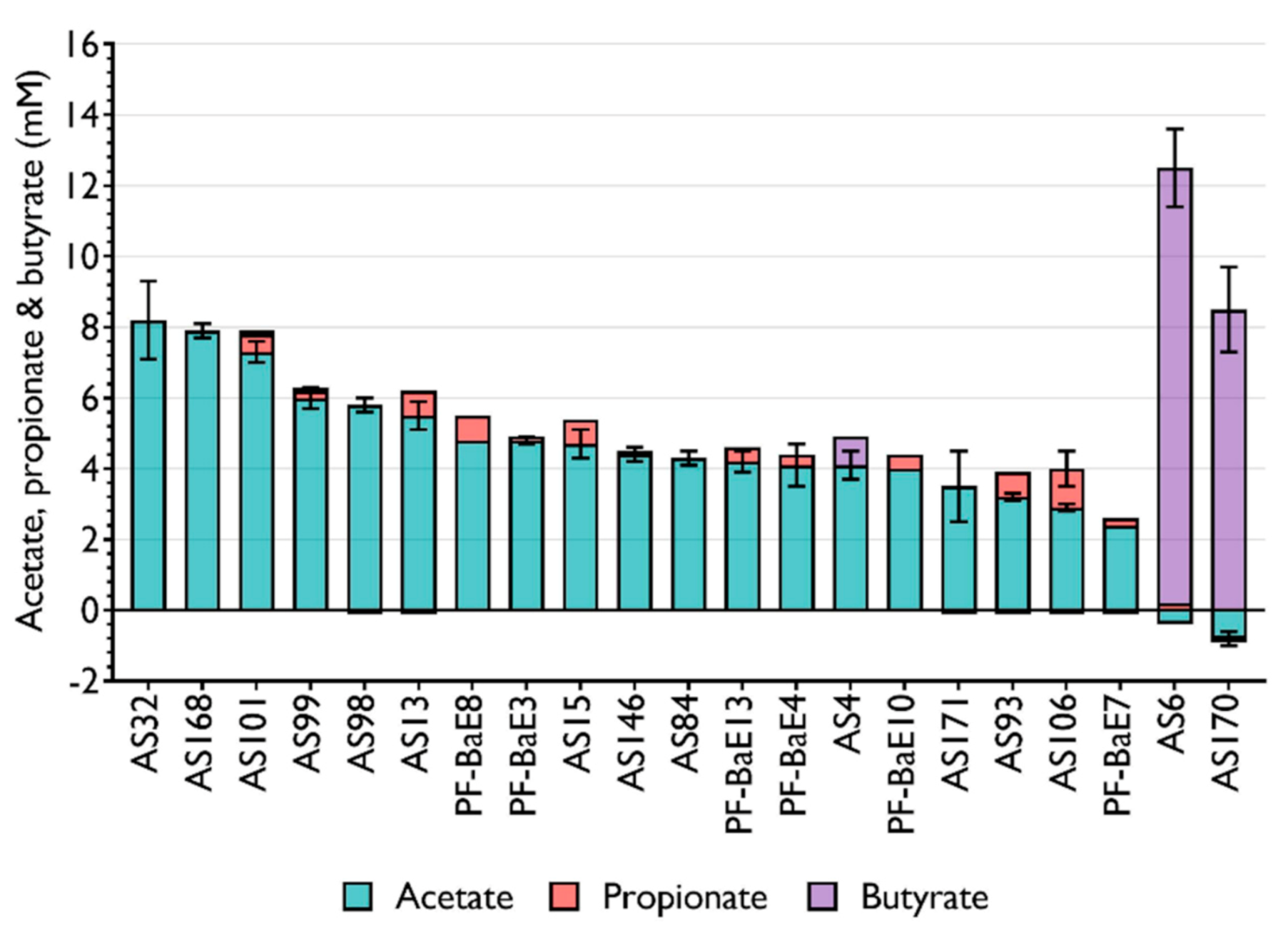
3.2. Production of SCFA by the Selected Bacterial Strains
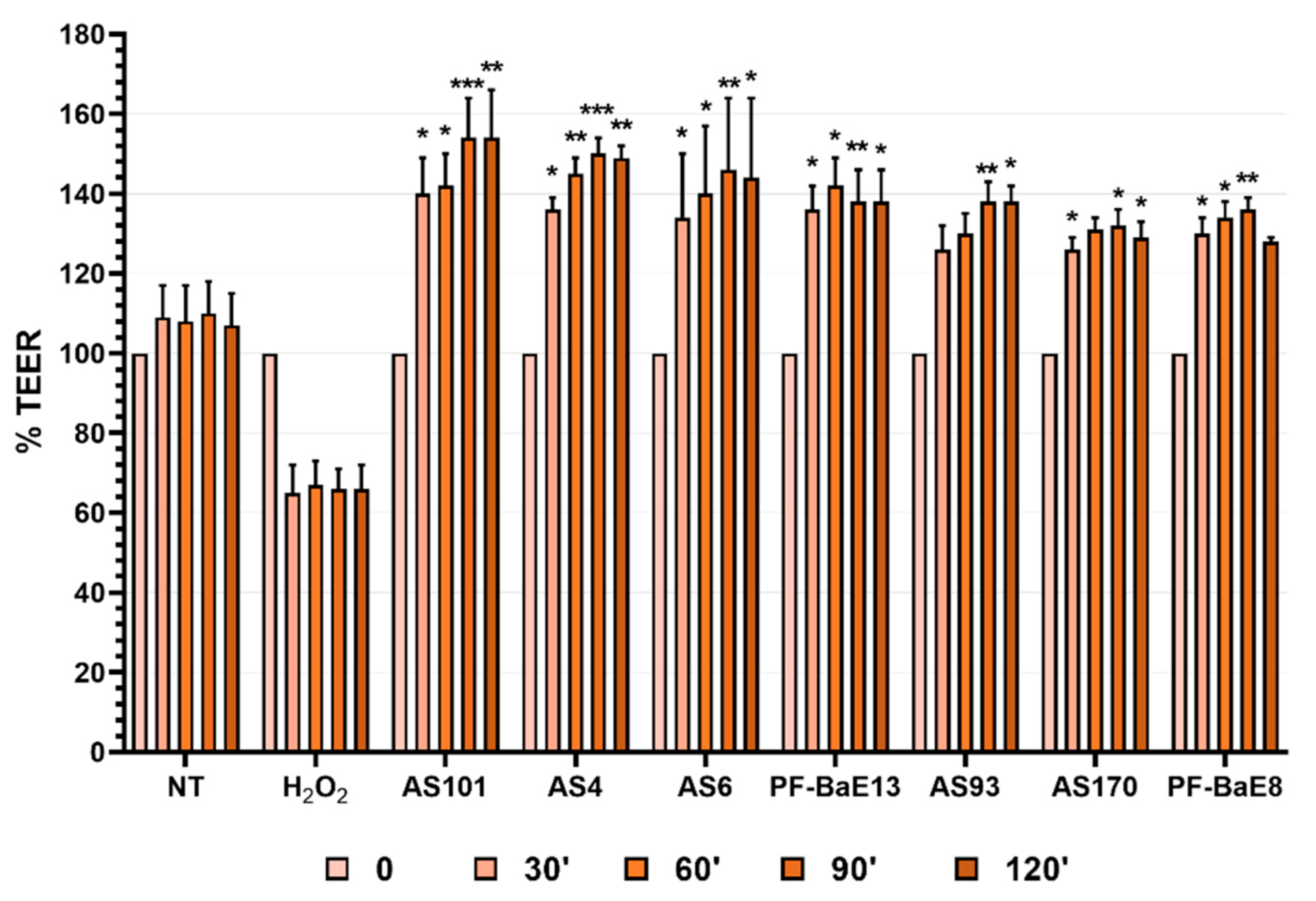
3.3. Ability of the Strains to Strengthen the Epithelial Barrier
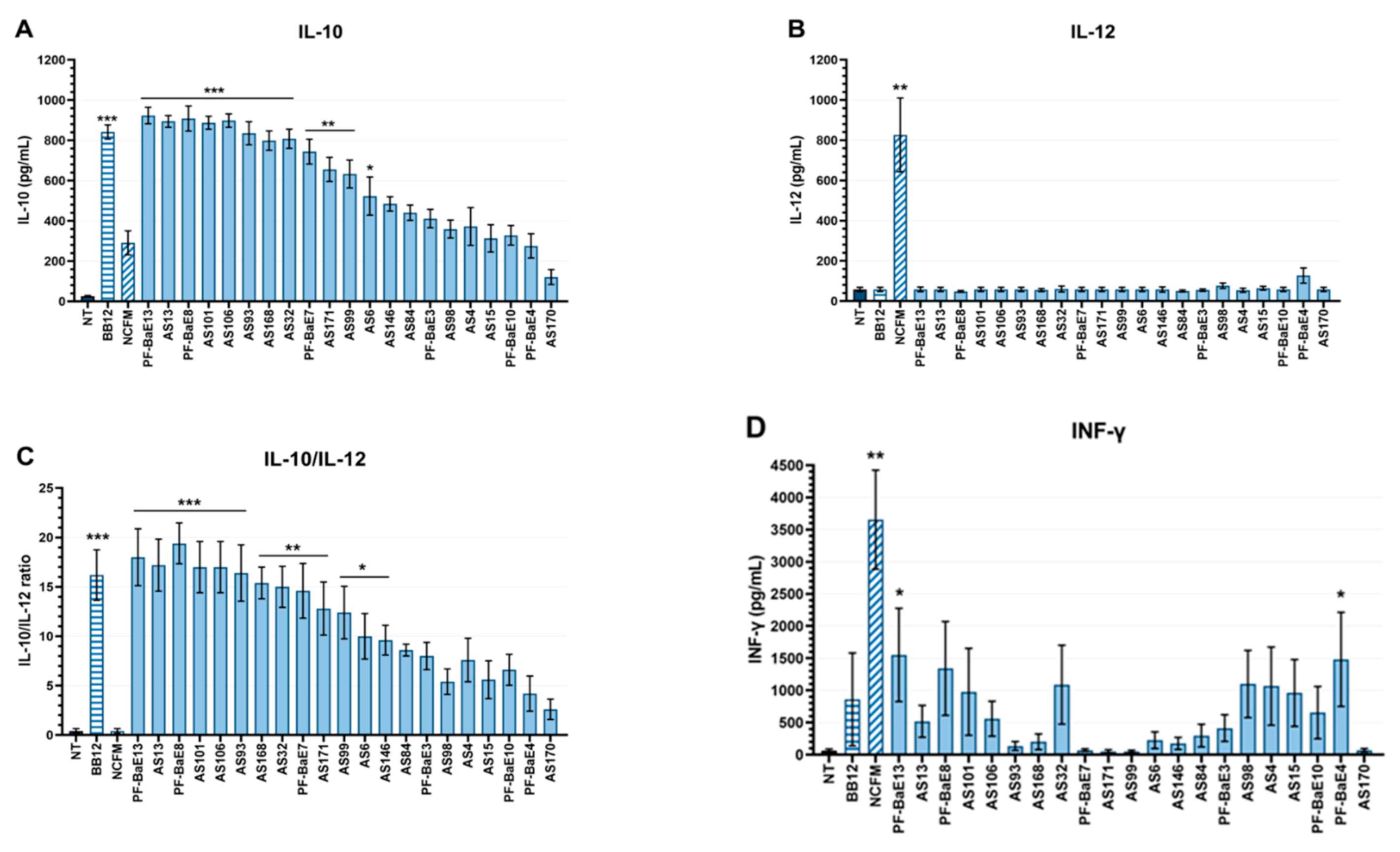
3.4. Immunomodulation by the Selected Strains
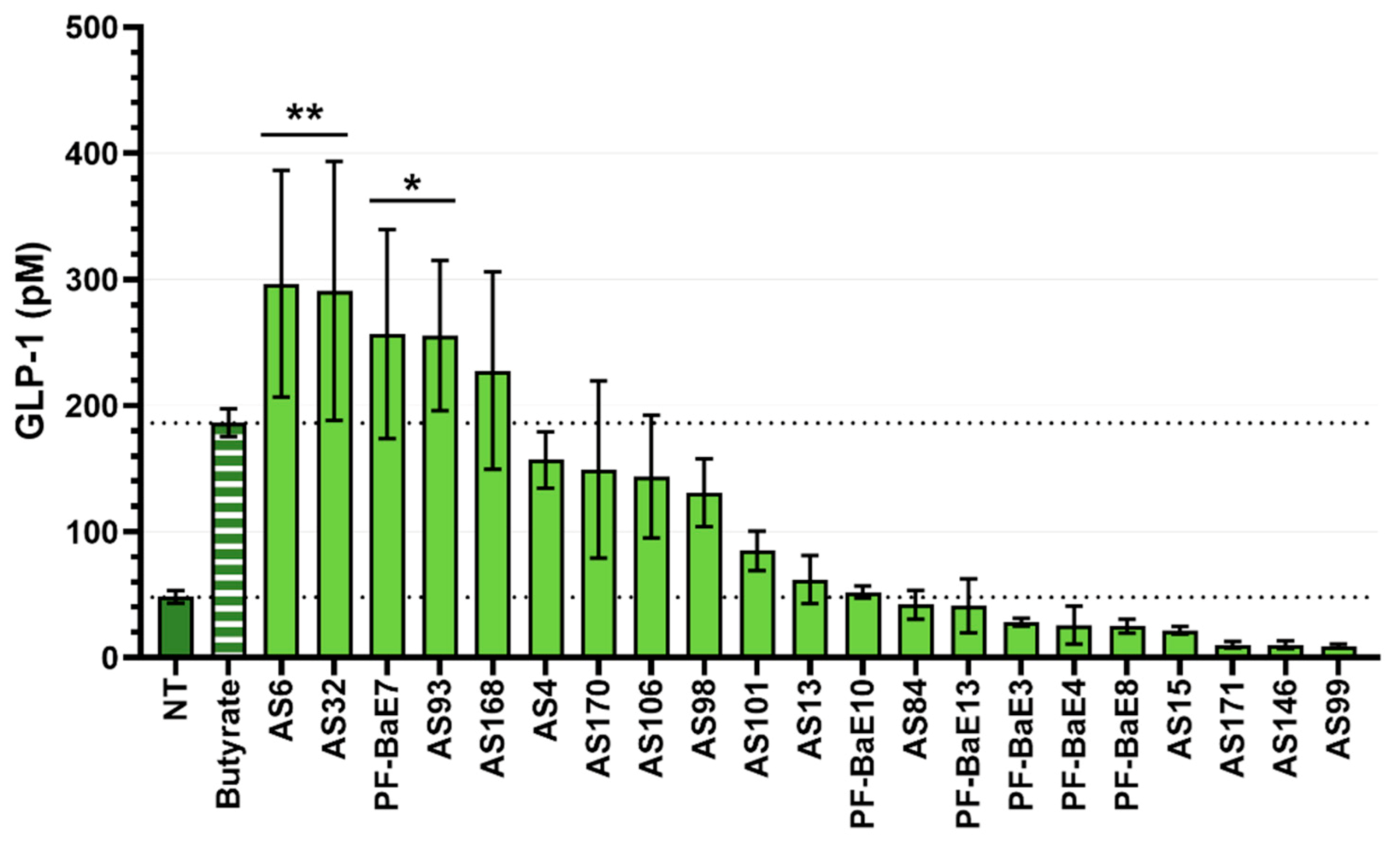
3.5. Ability of the Strains to Induce the Secretion of GLP-1
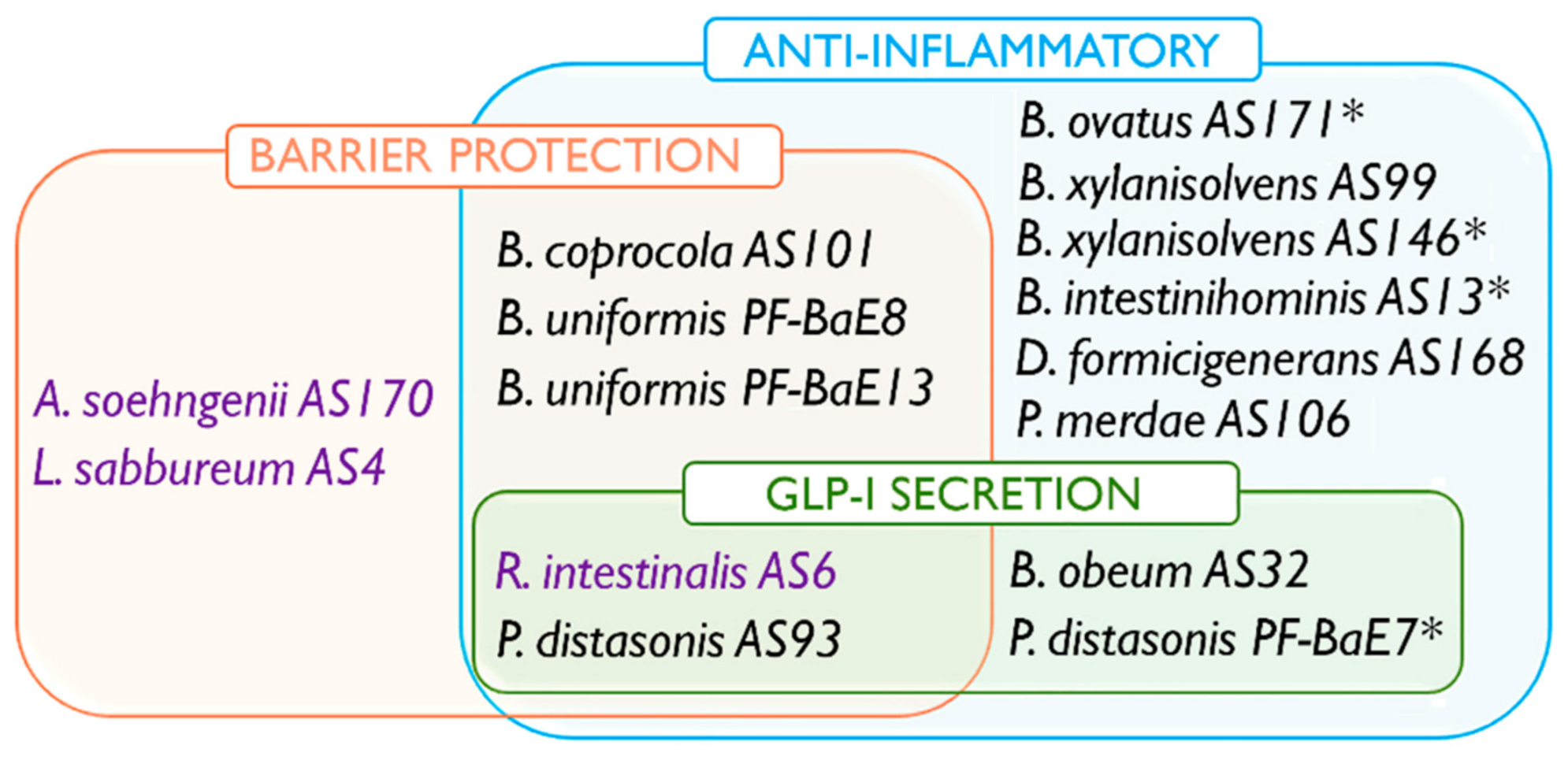
3.6. Combination of Beneficial Properties in Next Generation Probiotic Candidates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Bosch, T.C.G.; Cryan, J.F.; Gilbert, S.F.; Goodnight, C.J.; Lloyd, E.A.; et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems 2016, 1. [Google Scholar] [CrossRef]
- Simon, J.-C.; Marchesi, J.R.; Mougel, C.; Selosse, M.-A. Host-Microbiota Interactions: From Holobiont Theory to Analysis. Microbiome 2019, 7. [Google Scholar] [CrossRef]
- van de Guchte, M.; Blottière, H.M.; Doré, J. Humans as Holobionts: Implications for Prevention and Therapy. Microbiome 2018, 6, 81. [Google Scholar] [CrossRef]
- FAQ: Human Microbiome; American Academy of Microbiology FAQ Reports; American Society for Microbiology: Washington DC, USA, 2013.
- Hadrich, D. New EU Projects Delivering Human Microbiome Applications. Future Sci. OA 2020, 6, FSO474. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An Integrated Catalog of Reference Genes in the Human Gut Microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of Previously Uncultured Members of the Human Gut Microbiota by Culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The Microbiota in Adaptive Immune Homeostasis and Disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Surette, M.; Bercik, P. The Interplay between the Intestinal Microbiota and the Brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Mazmanian, S.K. Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host Microbe 2015, 17, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Takeda, K. Manipulation of Epithelial Integrity and Mucosal Immunity by Host and Microbiota-Derived Metabolites. Eur. J. Immunol. 2020, 50, 921–931. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T. Role of Intestinal Bacteria in Nutrient Metabolism. JPEN J. Parenter. Enteral Nutr. 1997, 21, 357–365. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A.; Tillisch, K.; Gupta, A. Gut/Brain Axis and the Microbiota. J. Clin. Invest. 2015, 125, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The Gut-Liver Axis and the Intersection with the Microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity Alters Gut Microbial Ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced Diversity of Faecal Microbiota in Crohn’s Disease Revealed by a Metagenomic Approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-Microbe Interactions Have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal Microbiota in Patients with Nonalcoholic Fatty Liver Disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis Gut Microbiota Associated with Inflammation and Impaired Mucosal Immune Function in Intestine of Humans with Non-Alcoholic Fatty Liver Disease. Sci. Rep. 2015, 5, 8096. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The Gut Microbiota-Brain Axis in Behaviour and Brain Disorders. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef]
- Chen, Z.; Maqbool, J.; Sajid, F.; Hussain, G.; Sun, T. Human Gut Microbiota and Its Association with Pathogenesis and Treatments of Neurodegenerative Diseases. Microb. Pathog. 2020, 150, 104675. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low Counts of Faecalibacterium Prausnitzii in Colitis Microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Shetty, S.A.; Zuffa, S.; Bui, T.P.N.; Aalvink, S.; Smidt, H.; De Vos, W.M. Reclassification of Eubacterium Hallii as Anaerobutyricum Hallii Gen. Nov., Comb. Nov., and Description of Anaerobutyricum Soehngenii Sp. Nov., a Butyrate and Propionate-Producing Bacterium from Infant Faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 3741–3746. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium Prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral Treatment with Eubacterium Hallii Improves Insulin Sensitivity in Db/Db Mice. NPJ Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Cano, P.G.; Santacruz, A.; Trejo, F.M.; Sanz, Y. Bifidobacterium CECT 7765 Improves Metabolic and Immunological Alterations Associated with Obesity in High-Fat Diet-Fed Mice. Obesity 2013, 21, 2310–2321. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Lee, Y.-S.; Kim, Y.; Lee, S.-H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.-S.; Lim, H.S.; Kim, M.-S.; et al. Gut Commensal Bacteroides Acidifaciens Prevents Obesity and Improves Insulin Sensitivity in Mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides Distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells 2020, 9, 2104. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides Distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering Probiotic Microorganisms: In Vitro, in Vivo, Genetic and Omics Approaches. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Saez-Lara, M.J.; Gomez-Llorente, C.; Plaza-Diaz, J.; Gil, A. The Role of Probiotic Lactic Acid Bacteria and Bifidobacteria in the Prevention and Treatment of Inflammatory Bowel Disease and Other Related Diseases: A Systematic Review of Randomized Human Clinical Trials. Biomed. Res. Int. 2015, 2015, 505878. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Richards, D.M.; Rahimi, E.F.; Krill, J.T.; Jelinek, K.A.; DuPont, A.W. Systematic Review of Randomized Controlled Trials of Probiotics, Prebiotics, and Synbiotics in Inflammatory Bowel Disease. Clin. Exp. Gastroenterol. 2014, 7, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Angelakis, E.; Paul, M.; Armougom, F.; Leibovici, L.; Raoult, D. Comparative Meta-Analysis of the Effect of Lactobacillus Species on Weight Gain in Humans and Animals. Microb. Pathog. 2012, 53, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Alard, J.; Peucelle, V.; Boutillier, D.; Breton, J.; Kuylle, S.; Pot, B.; Holowacz, S.; Grangette, C. New Probiotic Strains for Inflammatory Bowel Disease Management Identified by Combining in Vitro and in Vivo Approaches. Benef. Microbes 2018, 9, 317–331. [Google Scholar] [CrossRef]
- Zaylaa, M.; Al Kassaa, I.; Alard, J.; Peucelle, V.; Boutillier, D.; Desramaut, J.; Dabboussi, F.; Pot, B.; Grangette, C. Probiotics in IBD: Combining in Vitro and in Vivo Models for Selecting Strains with Both Anti-Inflammatory Potential as Well as a Capacity to Restore the Gut Epithelial Barrier. J. Funct. Foods 2018, 47, 304–315. [Google Scholar] [CrossRef]
- Bourgin, M.; Labarthe, S.; Kriaa, A.; Lhomme, M.; Gérard, P.; Lesnik, P.; Laroche, B.; Maguin, E.; Rhimi, M. Exploring the Bacterial Impact on Cholesterol Cycle: A Numerical Study. Front. Microbiol. 2020, 11, 1121. [Google Scholar] [CrossRef]
- Laurans, L.; Venteclef, N.; Haddad, Y.; Chajadine, M.; Alzaid, F.; Metghalchi, S.; Sovran, B.; Denis, R.G.P.; Dairou, J.; Cardellini, M.; et al. Genetic Deficiency of Indoleamine 2,3-Dioxygenase Promotes Gut Microbiota-Mediated Metabolic Health. Nat. Med. 2018, 24, 1113–1120. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A Standardised Static in Vitro Digestion Method Suitable for Food - an International Consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Foligne, B.; Nutten, S.; Grangette, C.; Dennin, V.; Goudercourt, D.; Poiret, S.; Dewulf, J.; Brassart, D.; Mercenier, A.; Pot, B. Correlation between in Vitro and in Vivo Immunomodulatory Properties of Lactic Acid Bacteria. World J. Gastroenterol. 2007, 13, 236–243. [Google Scholar] [CrossRef]
- Burns, P.; Alard, J.; Hrdỳ, J.; Boutillier, D.; Páez, R.; Reinheimer, J.; Pot, B.; Vinderola, G.; Grangette, C. Spray-Drying Process Preserves the Protective Capacity of a Breast Milk-Derived Bifidobacterium Lactis Strain on Acute and Chronic Colitis in Mice. Sci. Rep. 2017, 7, 43211. [Google Scholar] [CrossRef]
- Chantret, I.; Rodolosse, A.; Barbat, A.; Dussaulx, E.; Brot-Laroche, E.; Zweibaum, A.; Rousset, M. Differential Expression of Sucrase-Isomaltase in Clones Isolated from Early and Late Passages of the Cell Line Caco-2: Evidence for Glucose-Dependent Negative Regulation. J. Cell Sci. 1994, 107 Pt 1, 213–225. [Google Scholar]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Rindi, G.; Grant, S.G.; Yiangou, Y.; Ghatei, M.A.; Bloom, S.R.; Bautch, V.L.; Solcia, E.; Polak, J.M. Development of Neuroendocrine Tumors in the Gastrointestinal Tract of Transgenic Mice. Heterogeneity of Hormone Expression. Am. J. Pathol. 1990, 136, 1349–1363. [Google Scholar] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Schmidt, C. The Startup Bugs. Nat. Biotechnol. 2013, 31, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Rijkers, G.T.; de Vos, W.M.; Brummer, R.-J.; Morelli, L.; Corthier, G.; Marteau, P. Health Benefits and Health Claims of Probiotics: Bridging Science and Marketing. Br. J. Nutr. 2011, 106, 1291–1296. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; O’Toole, P.W. When Regulation Challenges Innovation: The Case of the Genus Lactobacillus. Trends Food Sci. Technol. 2017, 66, 187–194. [Google Scholar] [CrossRef]
- Saldanha, L.G. US Food and Drug Administration Regulations Governing Label Claims for Food Products, Including Probiotics. Clin. Infect. Dis. 2008, 46 (Suppl. 2), S119–S121, discussion S144–S151. [Google Scholar] [CrossRef]
- Douillard, F.P.; de Vos, W.M. Biotechnology of Health-Promoting Bacteria. Biotechnol. Adv. 2019, 37, 107369. [Google Scholar] [CrossRef]
- Almeida, D.; Machado, D.; Andrade, J.C.; Mendo, S.; Gomes, A.M.; Freitas, A.C. Evolving Trends in Next-Generation Probiotics: A 5W1H Perspective. Crit. Rev. Food Sci. Nutr. 2020, 60, 1783–1796. [Google Scholar] [CrossRef]
- Drucker, D.J. Mechanisms of Action and Therapeutic Application of Glucagon-like Peptide-1. Cell Metab. 2018, 27, 740–756. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia Spp.: A Marker of Health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant Gut Microbiota Markers for the Classification of Obesity-Related Metabolic Abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Van Hul, M. Gut Microbiota and Obesity: Causally Linked? Expert Rev. Gastroenterol. Hepatol. 2020, 14, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Indiani, C.M.D.S.P.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood Obesity and Firmicutes/Bacteroidetes Ratio in the Gut Microbiota: A Systematic Review. Child. Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef]
- Tan, H.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Novel Strains of Bacteroides Fragilis and Bacteroides Ovatus Alleviate the LPS-Induced Inflammation in Mice. Appl. Microbiol. Biotechnol. 2019, 103, 2353–2365. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Bäckhed, F. Signals from the Gut Microbiota to Distant Organs in Physiology and Disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A Purified Membrane Protein from Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Kverka, M.; Zakostelska, Z.; Klimesova, K.; Sokol, D.; Hudcovic, T.; Hrncir, T.; Rossmann, P.; Mrazek, J.; Kopecny, J.; Verdu, E.F.; et al. Oral Administration of Parabacteroides Distasonis Antigens Attenuates Experimental Murine Colitis through Modulation of Immunity and Microbiota Composition. Clin. Exp. Immunol. 2011, 163, 250–259. [Google Scholar] [CrossRef] [PubMed]
| Strain | Species | Growth Medium | Origin |
|---|---|---|---|
| AS4 | Lachnoanaerobaculum saburreum | BHI-YS | Healthy adult feces |
| AS6 | Roseburia intestinalis | BHI-YS | Healthy adult feces |
| AS13 | Barnesiella intestinihominis | BHI-YS | Healthy adult feces |
| AS15 | Bacteroides vulgatus | BHI-YS | Newborn fecal samples |
| AS32 | Blautia obeum | BHI-YS | Healthy adult feces |
| AS84 | Bacteroides thetaiotaomicron | BHI-YS | Healthy adult feces |
| AS93 | Parabacteroides distasonis | BHI-YS | Healthy adult feces |
| AS98 | Bacteroides massiliensis | BHI-YS | Healthy adult feces |
| AS99 | Bacteroides xylanisolvens | BHI-YS | Healthy adult feces |
| AS101 | Bacteroides coprocola | BHI-YS | Healthy adult feces |
| AS106 | Parabacteroides merdae | BHI-YS | Healthy adult feces |
| AS146 | Bacteroides xylanisolvens | BHI-YS | Healthy adult feces |
| AS168 | Dorea formicigenerans | BHI-YS | Healthy adult feces |
| AS170 | Anaerobutyricum soehngenii DSMZ17630 | M104S | Healthy infant feces |
| AS171 | Bacteroides ovatus | BHI-YS | Healthy adult feces |
| PF-BaE3 | Bacteroides caccae | BHI-YS | Newborn fecal samples |
| PF-BaE4 | Bacteroides fragilis | BHI-YS | Newborn fecal samples |
| PF-BaE7 | Parabacteroides distasonis | BHI-YS | Newborn fecal samples |
| PF-BaE8 | Bacteroides uniformis | BHI-YS | Newborn fecal samples |
| PF-BaE10 | Bacteroides vulgatus | BHI-YS | Newborn fecal samples |
| PF-BaE13 | Bacteroides uniformis | BHI-YS | Newborn fecal samples |
| NCFM | Lactobacillus acidophilus | MRS | DuPont™ Danisco-Madison, USA |
| BB12 | Bifidobacterium animalis subsp. lactis | MRS Cystein | Dietary origin (Chr Hansen, Hoesholm, Denmark) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Peucelle, V.; Desramaut, J.; Boudebbouze, S.; Croyal, M.; Waligora-Dupriet, A.-J.; Rhimi, M.; Grangette, C.; et al. Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria. Microorganisms 2021, 9, 565. https://doi.org/10.3390/microorganisms9030565
Cuffaro B, Assohoun ALW, Boutillier D, Peucelle V, Desramaut J, Boudebbouze S, Croyal M, Waligora-Dupriet A-J, Rhimi M, Grangette C, et al. Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria. Microorganisms. 2021; 9(3):565. https://doi.org/10.3390/microorganisms9030565
Chicago/Turabian StyleCuffaro, Bernardo, Aka L. W. Assohoun, Denise Boutillier, Véronique Peucelle, Jérémy Desramaut, Samira Boudebbouze, Mikael Croyal, Anne-Judith Waligora-Dupriet, Moez Rhimi, Corinne Grangette, and et al. 2021. "Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria" Microorganisms 9, no. 3: 565. https://doi.org/10.3390/microorganisms9030565
APA StyleCuffaro, B., Assohoun, A. L. W., Boutillier, D., Peucelle, V., Desramaut, J., Boudebbouze, S., Croyal, M., Waligora-Dupriet, A.-J., Rhimi, M., Grangette, C., & Maguin, E. (2021). Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria. Microorganisms, 9(3), 565. https://doi.org/10.3390/microorganisms9030565







