The Transcriptional Regulatory Network of Corynebacterium pseudotuberculosis
Abstract
1. Introduction
2. Gene Co-Expression Networks and Transcriptional Regulatory Networks
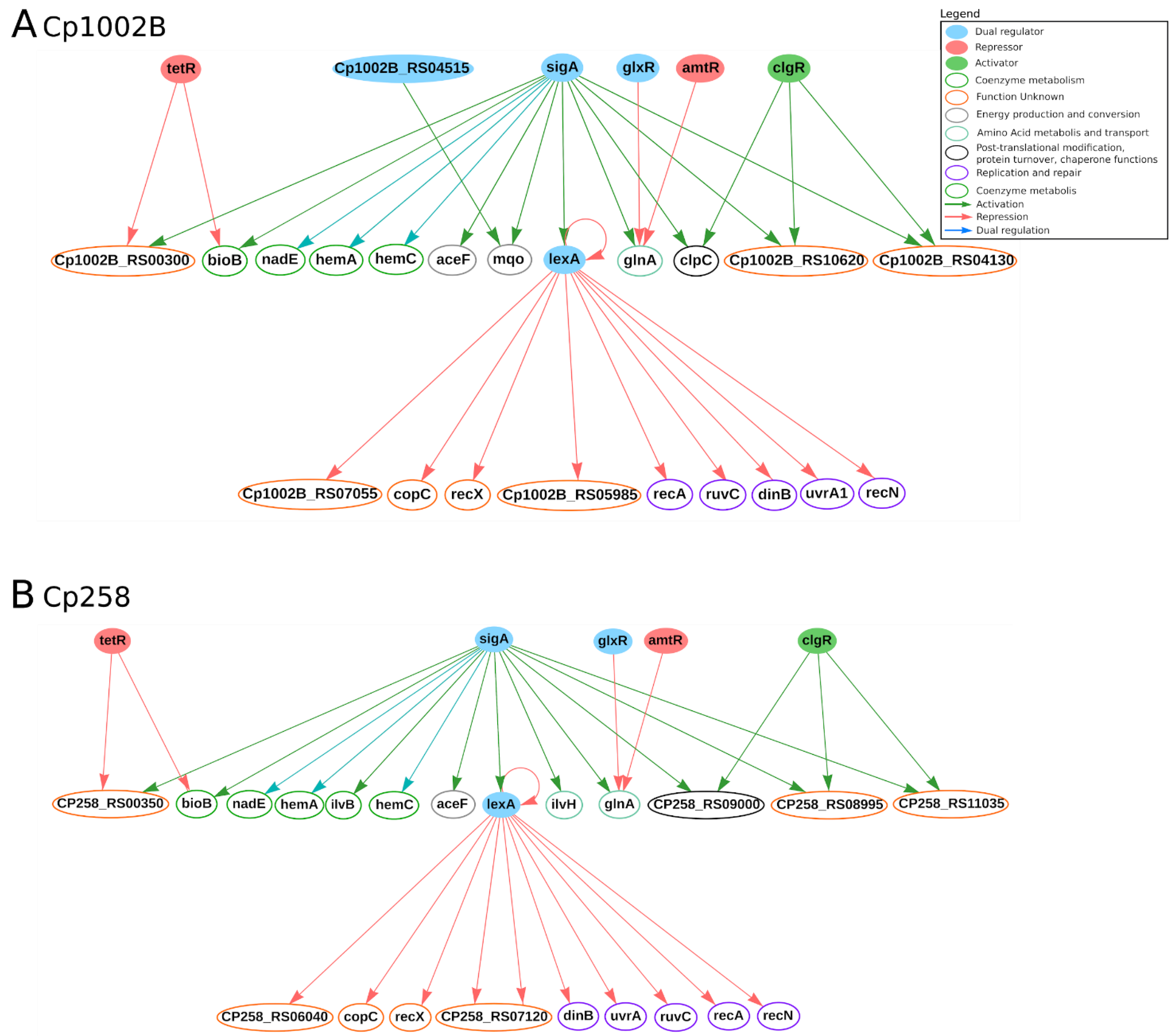
3. Regulators of Gene Expression
3.1. Two-Component Systems
3.2. Transcription Factors
3.3. Metalloregulation: Iron Uptake
3.4. Response to Osmotic, Thermal and Acid Stress
3.5. Sigma Factors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- González Plaza, J.J. Small RNAs as fundamental players in the transference of information during bacterial infectious diseases. Front. Mol. Biosci. 2020, 7, 101. [Google Scholar] [CrossRef]
- Bervoets, I.; Charlier, D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: Opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol. Rev. 2019, 43, 304–339. [Google Scholar] [CrossRef]
- Baumbach, J.; Brinkrolf, K.; Czaja, L.F.; Rahmann, S.; Tauch, A. CoryneRegNet: An ontology-based data warehouse of corynebacterial transcription factors and regulatory networks. BMC Genomics 2006, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M.; Lang, B.; Aravind, L. Methods to reconstruct and compare transcriptional regulatory networks. Methods Mol. Biol. 2009, 541, 163–180. [Google Scholar]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein—Nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Garner, M.M.; Revzin, A. The use of gel electrophoresis to detect and study nucleic acid—Protein interactions. Trends Biochem. Sci. 1986, 11, 395–396. [Google Scholar] [CrossRef]
- Horak, C.E.; Snyder, M. ChIP-chip: A genomic approach for identifying transcription factor binding sites. Guide Yeast Genet. Mol. Cell Biol. Part B 2002, 350, 469–483. [Google Scholar]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M.; Stefano, G.B. Comparing bioinformatic gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Bähler, J. RNA-seq: From technology to biology. Cell. Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Pan, W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics 2002, 18, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, A.; Simon, R.M. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J. Natl. Cancer Inst. 2007, 99, 147–157. [Google Scholar] [CrossRef]
- Furey, T.S. ChIP-seq and beyond: New and improved methodologies to detect and characterize protein—DNA interactions. Nat. Rev. Genetics 2012, 13, 840–852. [Google Scholar] [CrossRef]
- Park, P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genetics 2009, 10, 669–680. [Google Scholar] [CrossRef]
- Buck, M.J.; Lieb, J.D. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 2004, 83, 349–360. [Google Scholar] [CrossRef]
- Santos-Zavaleta, A.; Salgado, H.; Gama-Castro, S.; Sánchez-Pérez, M.; Gómez-Romero, L.; Ledezma-Tejeida, D.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Muñiz-Rascado, L.J.; Peña-Loredo, P.; et al. RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019, 47, D212–D220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Stülke, J. SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 2018, 46, D743–D748. [Google Scholar] [CrossRef]
- Ibarra-Arellano, M.A.; Campos-González, A.I.; Treviño-Quintanilla, L.G.; Tauch, A.; Freyre-González, J.A. Abasy Atlas: A comprehensive inventory of systems, global network properties and systems-level elements across bacteria. Database 2016, 2016. [Google Scholar] [CrossRef]
- Parise, M.T.D.; Parise, D.; Kato, R.B.; Pauling, J.K.; Tauch, A.; de Azevedo, V.A.C.; Baumbach, J. CoryneRegNet 7, the reference database and analysis platform for corynebacterial gene regulatory networks. Sci. Data 2020, 7, 142. [Google Scholar] [CrossRef]
- Kreikemeyer, B.; McIver, K.S.; Podbielski, A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen—Host interactions. Trends Microbiol. 2003, 11, 224–232. [Google Scholar] [CrossRef]
- Pauling, J.; Röttger, R.; Neuner, A.; Salgado, H.; Collado-Vides, J.; Kalaghatgi, P.; Azevedo, V.; Tauch, A.; Pühler, A.; Baumbach, J. On the trail of EHEC/EAEC—Unraveling the gene regulatory networks of human pathogenic Escherichia coli bacteria. Integr. Biol. 2012, 4, 728–733. [Google Scholar] [CrossRef]
- Galagan, J.E.; Minch, K.; Peterson, M.; Lyubetskaya, A.; Azizi, E.; Sweet, L.; Gomes, A.; Rustad, T.; Dolganov, G.; Glotova, I.; et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 2013, 499, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.R.G.; Li, Y.; Wang, L.; Sintsova, A.; van Bakel, H.; Tian, S.; Navarre, W.W.; Xia, B.; Liu, J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 5154–5159. [Google Scholar] [CrossRef]
- Gonzalo-Asensio, J.; Mostowy, S.; Harders-Westerveen, J.; Huygen, K.; Hernández-Pando, R.; Thole, J.; Behr, M.; Gicquel, B.; Martín, C. PhoP: A missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE 2008, 3, e3496. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Ayala, D.A.; Tilleman, L.; van Nieuwerburgh, F.; Deforce, D.; Palomino, J.C.; Vandamme, P.; Gonzalez-Y-Merchand, J.A.; Martin, A. The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 2017, 7, 17665. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.-G.; Kim, K.K.; Lim, Y.-J.; Choi, J.-A.; Cho, S.-N.; Park, C.; Song, C.-H. Characterisation of genes differentially expressed in macrophages by virulent and attenuated Mycobacterium tuberculosis through RNA-Seq analysis. Sci. Rep. 2019, 9, 4027. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 2931. [Google Scholar] [CrossRef] [PubMed]
- Danielli, A.; Amore, G.; Scarlato, V. Built shallow to maintain homeostasis and persistent infection: Insight into the transcriptional regulatory network of the gastric human pathogen Helicobacter pylori. PLoS Pathog. 2010, 6, e1000938. [Google Scholar] [CrossRef] [PubMed]
- Dorella, F.A.; Pacheco, L.G.C.; Oliveira, S.C.; Miyoshi, A.; Azevedo, V. Corynebacterium pseudotuberculosis: Microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet. Res. 2006, 37, 201–218. [Google Scholar] [CrossRef]
- Yeruham, I.; Friedman, S.; Perl, S.; Elad, D.; Berkovich, Y.; Kalgard, Y. A herd level analysis of a Corynebacterium pseudotuberculosis outbreak in a dairy cattle herd. Vet. Dermatol. 2004, 15, 315–320. [Google Scholar] [CrossRef]
- Silva, A.; Schneider, M.P.C.; Cerdeira, L.; Barbosa, M.S.; Ramos, R.T.J.; Carneiro, A.R.; Santos, R.; Lima, M.; D’Afonseca, V.; Almeida, S.S.; et al. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with Bovine Mastitis. J. Bacteriol. 2011, 193, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Spier, S.J.; Azevedo, V. Corynebacterium pseudotuberculosis infection in horses: Increasing frequency and spread to new regions of North America. Equine Vet. Educ. 2017, 29, 436–439. [Google Scholar] [CrossRef]
- Selim, S.A. Oedematous skin disease of buffalo in Egypt. J. Vet. Med. B Infect. Dis. Vet. Public Health 2001, 48, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Windsor, P.A.; Bush, R.D. Caseous lymphadenitis: Present and near forgotten from persistent vaccination? Small Rumin. Res. 2016, 142, 6–10. [Google Scholar] [CrossRef]
- Brum, A.A.; de Rezende, A.F.S.; Brilhante, F.S.; Collares, T.; Begnine, K.; Seixas, F.K.; Collares, T.V.; Dellagostin, O.A.; Azevedo, V.; Santos, A.; et al. Recombinant esterase from Corynebacterium pseudotuberculosis in DNA and subunit recombinant vaccines partially protects mice against challenge. J. Med. Microbiol. 2017, 66, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Droppa-Almeida, D.; Franceschi, E.; Padilha, F.F. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinform. Biol. Insights 2018, 12, 117793221875533. [Google Scholar] [CrossRef]
- Pinto, A.C.; de Sá, P.H.C.G.; Ramos, R.T.J.; Barbosa, S.; Barbosa, H.P.M.; Ribeiro, A.C.; Silva, W.M.; Rocha, F.S.; Santana, M.P.; de Paula Castro, T.L.; et al. Differential transcriptional profile of Corynebacterium pseudotuberculosis in response to abiotic stresses. BMC Genomics 2014, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Gomide, A.C.P.; de Sá, P.G.; Cavalcante, A.L.Q.; de Jesus Sousa, T.; Gomes, L.G.R.; Ramos, R.T.J.; Azevedo, V.; Silva, A.; Folador, A.R.C. Heat shock stress: Profile of differential expression in Corynebacterium pseudotuberculosis biovar Equi. Gene 2018, 645, 124–130. [Google Scholar] [CrossRef]
- Gomide, A.C.P.; Ibraim, I.C.; Alves, J.T.C.; de Sá, P.G.; de Oliveira Silva, Y.R.; Santana, M.P.; Silva, W.M.; Folador, E.L.; Mariano, D.C.B.; de Paula Castro, T.L.; et al. Transcriptome analysis of Corynebacterium pseudotuberculosis biovar Equi in two conditions of the environmental stress. Gene 2018, 677, 349–360. [Google Scholar] [CrossRef]
- Ibraim, I.C.; Parise, M.T.D.; Parise, D.; Sfeir, M.Z.T.; de Paula Castro, T.L.; Wattam, A.R.; Ghosh, P.; Barh, D.; Souza, E.M.; Góes-Neto, A.; et al. Transcriptome profile of Corynebacterium pseudotuberculosis in response to iron limitation. BMC Genomics 2019, 20, 663. [Google Scholar] [CrossRef]
- McKean, S.C.; Davies, J.K.; Moore, R.J. Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis, is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology 2007, 153, 2203–2211. [Google Scholar] [CrossRef] [PubMed]
- Billington, S.J.; Esmay, P.A.; Glenn Songer, J.; Helen Jost, B. Identification and role in virulence of putative iron acquisition genes from Corynebacterium pseudotuberculosis. FEMS Microbiol. Lett. 2002, 208, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; da Costa, M.P.; Almeida, S.; Hassan, S.S.; Jamal, S.B.; Oliveira, A.; Folador, E.L.; Rocha, F.; de Abreu, V.A.C.; Dorella, F.; et al. C. pseudotuberculosis Phop confers virulence and may be targeted by natural compounds. Integr. Biol. 2014, 6, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Turkarslan, S.; Peterson, E.J.R.; Rustad, T.R.; Minch, K.J.; Reiss, D.J.; Morrison, R.; Ma, S.; Price, N.D.; Sherman, D.R.; Baliga, N.S. A comprehensive map of genome-wide gene regulation in Mycobacterium tuberculosis. Sci. Data 2015, 2, 150010. [Google Scholar] [CrossRef]
- Luo, F.; Yang, Y.; Zhong, J.; Gao, H.; Khan, L.; Thompson, D.K.; Zhou, J. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinform. 2007, 8, 299. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Long, X.; You, W.; Zhong, Y.; Wang, M.; Tao, H.; Lin, S.; He, H. Construction and Analysis of gene co-expression networks in Escherichia coli. Cells 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Brinkrolf, K.; Brune, I.; Tauch, A. The transcriptional regulatory network of the amino acid producer Corynebacterium glutamicum. J. Biotechnol. 2007, 129, 191–211. [Google Scholar] [CrossRef]
- Kohl, T.A.; Tauch, A. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: Detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J. Biotechnol. 2009, 143, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Schneper, L.; Kumari, H.; Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.R.; Smoot, L.M.; Migliaccio, C.A.L.; Virtaneva, K.; Sturdevant, D.E.; Porcella, S.F.; Federle, M.J.; Adams, G.J.; Scott, J.R.; Musser, J.M. Virulence control in group A Streptococcus by a two-component gene regulatory system: Global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 2002, 99, 13855–13860. [Google Scholar] [CrossRef]
- Franco, E.F.; Rana, P.; Queiroz Cavalcante, A.L.; da Silva, A.L.; Cybelle Pinto Gomide, A.; Carneiro Folador, A.R.; Azevedo, V.; Ghosh, P.; Ramos, R.T.J. Co-expression networks for causal gene identification based on RNA-Seq data of Corynebacterium pseudotuberculosis. Genes 2020, 11, 794. [Google Scholar] [CrossRef] [PubMed]
- Nalluri, J.J.; Barh, D.; Azevedo, V.; Ghosh, P. miRsig: A consensus-based network inference methodology to identify pan-cancer miRNA-miRNA interaction signatures. Sci. Rep. 2017, 7, 39684. [Google Scholar] [CrossRef] [PubMed]
- Nalluri, J.J.; Rana, P.; Barh, D.; Azevedo, V.; Dinh, T.N.; Vladimirov, V.; Ghosh, P. Determining causal miRNAs and their signaling cascade in diseases using an influence diffusion model. Sci. Rep. 2017, 7, 8133. [Google Scholar] [CrossRef]
- Chen, W.-H.; Lu, G.; Chen, X.; Zhao, X.-M.; Bork, P. OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017, 45, D940–D944. [Google Scholar] [CrossRef]
- Padilla, L.; Morbach, S.; Krämer, R.; Agosin, E. Impact of heterologous expression of Escherichia coli UDP-glucose pyrophosphorylase on trehalose and glycogen synthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2004, 70, 3845–3854. [Google Scholar] [CrossRef]
- Anishetty, S.; Pulimi, M.; Pennathur, G. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput. Biol. Chem. 2005, 29, 368–378. [Google Scholar] [CrossRef]
- Sharp, P.M.; Mitchell, K.J. Corynebacterium glutamicum arginyl-tRNA synthetase. Mol. Microbiol. 1993, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- de Bruin, O.M.; Duplantis, B.N.; Ludu, J.S.; Hare, R.F.; Nix, E.B.; Schmerk, C.L.; Robb, C.S.; Boraston, A.B.; Hueffer, K.; Nano, F.E. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 2011, 157, 3483–3491. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Lv, X.; Liu, Y.; Li, J.; Lu, W.; Du, G.; Liu, L. Metabolic engineering of Corynebacterium glutamicum S9114 based on whole-genome sequencing for efficient n-acetylglucosamine synthesis. Synth. Syst. Biotechnol. 2019, 4, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Boyle, D.S.; Donachie, W.D. mraY is an essential gene for cell growth in Escherichia coli. J. Bacteriol. 1998, 180, 6429–6432. [Google Scholar] [CrossRef]
- Oguiza, J.A.; Malumbres, M.; Eriani, G.; Pisabarro, A.; Mateos, L.M.; Martin, F.; Martín, J.F. A gene encoding arginyl-tRNA synthetase is located in the upstream region of the lysA gene in Brevibacterium lactofermentum: Regulation of argS-lysA cluster expression by arginine. J. Bacteriol. 1993, 175, 7356–7362. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cracan, V.; Padovani, D.; Banerjee, R. IcmF is a fusion between the radical B12 enzyme isobutyryl-CoA mutase and its G-protein chaperone. J. Biol. Chem. 2010, 285, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Mizoguchi, H.; Shiraishi, N.; Obayashi, M.; Nakagawa, S.; Imai, J.-I.; Watanabe, S.; Ota, T.; Ikeda, M. Transcriptome analysis of acetate metabolism in Corynebacterium glutamicum using a newly developed metabolic array. Biosci. Biotechnol. Biochem. 2002, 66, 1337–1344. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, M.; Zhang, H.; Lei, J.; Jin, R.; Xu, S.; Bao, J.; Zhang, L.; Wang, H. Purification and characterization of Mycobacterium tuberculosis indole-3-glycerol phosphate synthase. Biochemistry 2006, 71, S38–S43. [Google Scholar] [CrossRef] [PubMed]
- Peters-Wendisch, P.; Stolz, M.; Etterich, H.; Kennerknecht, N.; Sahm, H.; Eggeling, L. Metabolic engineering of Corynebacterium glutamicum for l-serine production. Appl. Environ. Microbiol. 2005, 71, 7139–7144. [Google Scholar] [CrossRef]
- Lim, C.J.; Hwang, W.; Park, E.H.; Fuchs, J.A. Cyclic AMP-dependent expression of the Escherichia coli serC-aroA operon. Biochim. Biophys. Acta 1994, 1218, 250–253. [Google Scholar] [CrossRef]
- Ikeda, M.; Wachi, M.; Jung, H.K.; Ishino, F.; Matsuhashi, M. The Escherichia coli mraY gene encoding UDP-n-acetylmuramoyl-pentapeptide: Undecaprenyl-phosphate phospho-n-acetylmuramoyl-pentapeptide transferase. J. Bacteriol. 1991, 173, 1021–1026. [Google Scholar] [CrossRef]
- Burkovski, A. Cell envelope of corynebacteria: Structure and influence on pathogenicity. ISRN Microbiol. 2013, 2013, 935736. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.; Price, N.D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 17845–17850. [Google Scholar] [CrossRef]
- Moraes, G.L.; Gomes, G.C.; de Monteiro Sousa, P.R.; Alves, C.N.; Govender, T.; Kruger, H.G.; Maguire, G.E.M.; Lamichhane, G.; Lameira, J. Structural and functional features of enzymes of Mycobacterium tuberculosis peptidoglycan biosynthesis as targets for drug development. Tuberculosis 2015, 95, 95–111. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [PubMed]
- Pao, G.M.; Saier, M.H., Jr. Response regulators of bacterial signal transduction systems: Selective domain shuffling during evolution. J. Mol. Evol. 1995, 40, 136–154. [Google Scholar] [CrossRef]
- Blanco, A.G.; Sola, M.; Gomis-Rüth, F.X.; Coll, M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002, 10, 701–713. [Google Scholar] [CrossRef]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- López-Goñi, I.; Guzmán-Verri, C.; Manterola, L.; Sola-Landa, A.; Moriyón, I.; Moreno, E. Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet. Microbiol. 2002, 90, 329–339. [Google Scholar] [CrossRef]
- Matsushita, M.; Janda, K.D. Histidine kinases as targets for new antimicrobial agents. Bioorg. Med. Chem. 2002, 10, 855–867. [Google Scholar] [CrossRef]
- Cardona, P.J.; Asensio, J.G.; Arbués, A.; Otal, I.; Lafoz, C.; Gil, O.; Caceres, N.; Ausina, V.; Gicquel, B.; Martin, C. Extended safety studies of the attenuated live tuberculosis vaccine SO2 based on phoP mutant. Vaccine 2009, 27, 2499–2505. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Ann. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef]
- Sorger-Herrmann, U.; Taniguchi, H.; Wendisch, V.F. Regulation of the pstSCAB operon in Corynebacterium glutamicum by the regulator of acetate metabolism RamB. BMC Microbiol. 2015, 15, 113. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, M.A.; Cianciotto, N.P. A Legionella pneumophila Peptidyl-Prolyl cis-trans Isomerase Present in Culture Supernatants is necessary for optimal growth at low temperatures. Appl. Environ. Microbiol. 2008, 74, 1634–1638. [Google Scholar] [CrossRef]
- Göthel, S.F.; Scholz, C.; Schmid, F.X.; Marahiel, M.A. Cyclophilin and Trigger factor from Bacillus subtilis catalyze in Vitro protein folding and are necessary for viability under starvation conditions†. Biochemistry 1998, 37, 13392–13399. [Google Scholar] [CrossRef] [PubMed]
- Unal, C.M.; Steinert, M. Microbial Peptidyl-Prolyl cis/trans Isomerases (PPIases): Virulence factors and potential alternative drug targets. Microbiol. Mol. Biol. Rev. 2014, 78, 544–571. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Choi, E.; Cho, Y.-J.; Nam, D.; Lee, J.; Lee, E.-J. The Salmonella virulence protein MgtC promotes phosphate uptake inside macrophages. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Lucchini, S.; Thompson, A.; Rhen, M.; Hinton, J.C.D. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 2003, 47, 103–118. [Google Scholar] [CrossRef]
- Heithoff, D.M.; Shimp, W.R.; House, J.K.; Xie, Y.; Weimer, B.C.; Sinsheimer, R.L.; Mahan, M.J. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS Pathog. 2012, 8, e1002647. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Bibb, L.A.; Kunkle, C.A.; Schmitt, M.P. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 2007, 75, 2421–2431. [Google Scholar] [CrossRef] [PubMed]
- Frunzke, J.; Gätgens, C.; Brocker, M.; Bott, M. Control of heme homeostasis in Corynebacterium glutamicum by the two-component system HrrSA. J. Bacteriol. 2011, 193, 1212–1221. [Google Scholar] [CrossRef]
- Keppel, M.; Hünnefeld, M.; Filipchyk, A.; Viets, U.; Davoudi, C.-F.; Krüger, A.; Mack, C.; Pfeifer, E.; Polen, T.; Baumgart, M.; et al. HrrSA orchestrates a systemic response to heme and determines prioritization of terminal cytochrome oxidase expression. Nucleic Acids Res. 2020, 48, 6547–6562. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J.W. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004, 2, 57–65. [Google Scholar] [CrossRef]
- Browning, D.F.; Butala, M.; Busby, S.J.W. Bacterial transcription factors: Regulation by pick “N” Mix. J Mol. Biol. 2019, 431, 4067–4077. [Google Scholar] [CrossRef]
- Merchant, S.S.; Helmann, J.D. Elemental economy: Microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 2012, 60, 91–210. [Google Scholar]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Fu, M.; Su, H.; Su, Z.; Yin, Z.; Jin, J.; Wang, L.; Zhang, Q.; Xu, X. Transcriptome analysis of Corynebacterium pseudotuberculosis-infected spleen of dairy goats. Microb. Pathog. 2020, 147, 104370. [Google Scholar] [CrossRef]
- Kunkle, C.A.; Schmitt, M.P. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 2005, 187, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Trost, E.; Ott, L.; Schneider, J.; Schröder, J.; Jaenicke, S.; Goesmann, A.; Husemann, P.; Stoye, J.; Dorella, F.A.; Rocha, F.S.; et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics 2010, 11, 728. [Google Scholar] [CrossRef]
- Dorella, F.A.; Estevam, E.M.; Pacheco, L.G.C.; Guimarães, C.T.; Lana, U.G.P.; Gomes, E.A.; Barsante, M.M.; Oliveira, S.C.; Meyer, R.; Miyoshi, A.; et al. In vivo insertional mutagenesis in Corynebacterium pseudotuberculosis: An efficient means to identify DNA sequences encoding exported proteins. Appl. Environ. Microbiol. 2006, 72, 7368–7372. [Google Scholar] [CrossRef]
- Ribeiro, D.; de Rocha, F.S.; Leite, K.M.C.; de Soares, S.C.; Silva, A.; Portela, R.W.D.; Meyer, R.; Miyoshi, A.; Oliveira, S.C.; Azevedo, V.; et al. An iron-acquisition-deficient mutant of Corynebacterium pseudotuberculosis efficiently protects mice against challenge. Vet. Res. 2014, 45, 28. [Google Scholar] [CrossRef][Green Version]
- Henderson, A.G.; Ehre, C.; Button, B.; Abdullah, L.H.; Cai, L.-H.; Leigh, M.W.; DeMaria, G.C.; Matsui, H.; Donaldson, S.H.; Davis, C.W.; et al. Cystic fibrosis airway secretions exhibit mucin hyperconcentration and increased osmotic pressure. J. Clin. Invest. 2014, 124, 3047–3060. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.M. Bacterial responses to osmotic challenges. J. Gen. Physiol. 2015, 145, 381–388. [Google Scholar] [CrossRef]
- Janakiraman, A.; Lesser, C.F. How to manage stress: Lessons from an intracellular pathogen. Virulence 2017, 8, 359–361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vázquez-Torres, A. Bacterial stress responses during host infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 2010, 6, 364. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.R.; Mooij, M.J.; Reen, F.J.; Lesouhaitier, O.; O’Gara, F. A new regulator of pathogenicity (bvlR) is required for full virulence and tight microcolony formation in Pseudomonas aeruginosa. Microbiology 2014, 160, 1488–1500. [Google Scholar] [CrossRef]
- Sasindran, S.J.; Saikolappan, S.; Dhandayuthapani, S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2007, 2, 619–630. [Google Scholar] [CrossRef]
- Choi, S.H.; Baumler, D.J.; Kaspar, C.W. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157: H7. Appl. Environ. Microbiol. 2000, 66, 3911–3916. [Google Scholar] [CrossRef]
- Calhoun, L.N.; Kwon, Y.M. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: A review. J. Appl. Microbiol. 2011, 110, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Das Gupta, T.; Bandyopadhyay, B.; Das Gupta, S.K. Modulation of DNA-binding activity of Mycobacterium tuberculosis HspR by chaperones. Microbiology 2008, 154, 484–490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Ann. Rev. Genetics 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Bandyopadhyay, B.; Das Gupta, T.; Roy, D.; Das Gupta, S.K. DnaK dependence of the mycobacterial stress-responsive regulator HspR is mediated through its hydrophobic C-terminal tail. J. Bacteriol. 2012, 194, 4688–4697. [Google Scholar] [CrossRef][Green Version]
- Ishikawa, M.; Okamoto-Kainuma, A.; Matsui, K.; Takigishi, A.; Kaga, T.; Koizumi, Y. Cloning and characterization of clpB in Acetobacter pasteurianus NBRC 3283. J. Biosci. Bioeng. 2010, 110, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Chastanet, A.; Derre, I.; Nair, S.; Msadek, T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004, 186, 1165–1174. [Google Scholar] [CrossRef]
- Engels, S.; Ludwig, C.; Schweitzer, J.-E.; Mack, C.; Bott, M.; Schaffer, S. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 2005, 57, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Doig, P.; Boriack-Sjodin, P.A.; Dumas, J.; Hu, J.; Itoh, K.; Johnson, K.; Kazmirski, S.; Kinoshita, T.; Kuroda, S.; Sato, T.-O.; et al. Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping. Bioorg. Med. Chem. 2014, 22, 6256–6269. [Google Scholar] [CrossRef]
- Zhang, W.; Jones, V.C.; Scherman, M.S.; Mahapatra, S.; Crick, D.; Bhamidi, S.; Xin, Y.; McNeil, M.R.; Ma, Y. Expression, essentiality, and a microtiter plate assay for mycobacterial GlmU, the bifunctional glucosamine-1-phosphate acetyltransferase and n-acetylglucosamine-1-phosphate uridyltransferase. Int. J. Biochemistry Cell Biol. 2008, 40, 2560–2571. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, H.J.; Kikuchi, Y.; Kitagawa, W.; Nikoh, N.; Fukatsu, T.; Lee, B.L. Bacterial cell wall synthesis gene uppP is required for Burkholderia colonization of the Stinkbug Gut. Appl. Environ. Microbiol. 2013, 79, 4879–4886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, Y.; Peters, J.M.; Gross, C.A.; Garner, E.C.; Helmann, J.D. Depletion of undecaprenyl pyrophosphate phosphatases disrupts cell envelope biogenesis in Bacillus subtilis. J. Bacteriol. 2016, 198, 2925–2935. [Google Scholar] [CrossRef] [PubMed]
- Braibant, M.; Guilloteau, L.; Zygmunt, M.S. Functional characterization of Brucella melitensis NorMI, an efflux pump belonging to the multidrug and toxic compound extrusion family. Antimicrob. Agents Chemother. 2002, 46, 3050–3053. [Google Scholar] [CrossRef][Green Version]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782. [Google Scholar] [CrossRef] [PubMed]
- Spirig, T.; Weiner, E.M.; Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011, 82, 1044–1059. [Google Scholar] [CrossRef]
- Paterson, G.K.; Mitchell, T.J. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 2006, 8, 145–153. [Google Scholar] [CrossRef]
- Paget, M.S. Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 2015, 5, 1245–1265. [Google Scholar] [CrossRef]
- Davis, M.C.; Kesthely, C.A.; Franklin, E.A.; MacLellan, S.R. The essential activities of the bacterial sigma factor. Can. J. Microbiol. 2017, 63, 89–99. [Google Scholar] [CrossRef]
- Adcock, I.M.; Caramori, G. Transcription factors. Asthma COPD 2009, 373–380. [Google Scholar]
- Feklístov, A.; Sharon, B.D.; Darst, S.A.; Gross, C.A. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014, 68, 357–376. [Google Scholar] [CrossRef]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005, 69, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.T.; Mitra, A. Regulation of Escherichia coli pathogenesis by alternative sigma factor N. EcoSal Plus 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Pátek, M.; Nešvera, J. Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 2011, 154, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Busche, T.; Patschkowski, T.; Niehaus, K.; Pátek, M.; Kalinowski, J.; Wendisch, V.F. Physiological roles of sigma factor SigD in Corynebacterium glutamicum. BMC Microbiol. 2017, 17, 158. [Google Scholar] [CrossRef]
- Ruiz, J.C.; D’Afonseca, V.; Silva, A.; Ali, A.; Pinto, A.C.; Santos, A.R.; Rocha, A.A.M.C.; Lopes, D.O.; Dorella, F.A.; Pacheco, L.G.C.; et al. Evidence for reductive genome evolution and lateral acquisition of virulence functions in two Corynebacterium pseudotuberculosis strains. PLoS ONE 2011, 6, e18551. [Google Scholar] [CrossRef]
- Helmann, J.D.; Chamberlin, M.J. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988, 57, 839–872. [Google Scholar] [CrossRef]
- Wu, S.; Howard, S.T.; Lakey, D.L.; Kipnis, A.; Samten, B.; Safi, H.; Gruppo, V.; Wizel, B.; Shams, H.; Basaraba, R.J.; et al. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol. Microbiol. 2004, 51, 1551–1562. [Google Scholar] [CrossRef]
- Rodriguez Ayala, F.; Bartolini, M.; Grau, R. The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: An old friend with new functions. Front. Microbiol. 2020, 11, 228. [Google Scholar] [CrossRef]
- Pacheco, L.; Castro, T.; Carvalho, R.; Moraes, P.; Dorella, F.; Carvalho, N.; Slade, S.; Scrivens, J.; Feelisch, M.; Meyer, R.; et al. A role for Sigma Factor σE in Corynebacterium pseudotuberculosis resistance to nitric oxide/peroxide stress. Front. Microbiol. 2012, 3, 126. [Google Scholar] [CrossRef][Green Version]
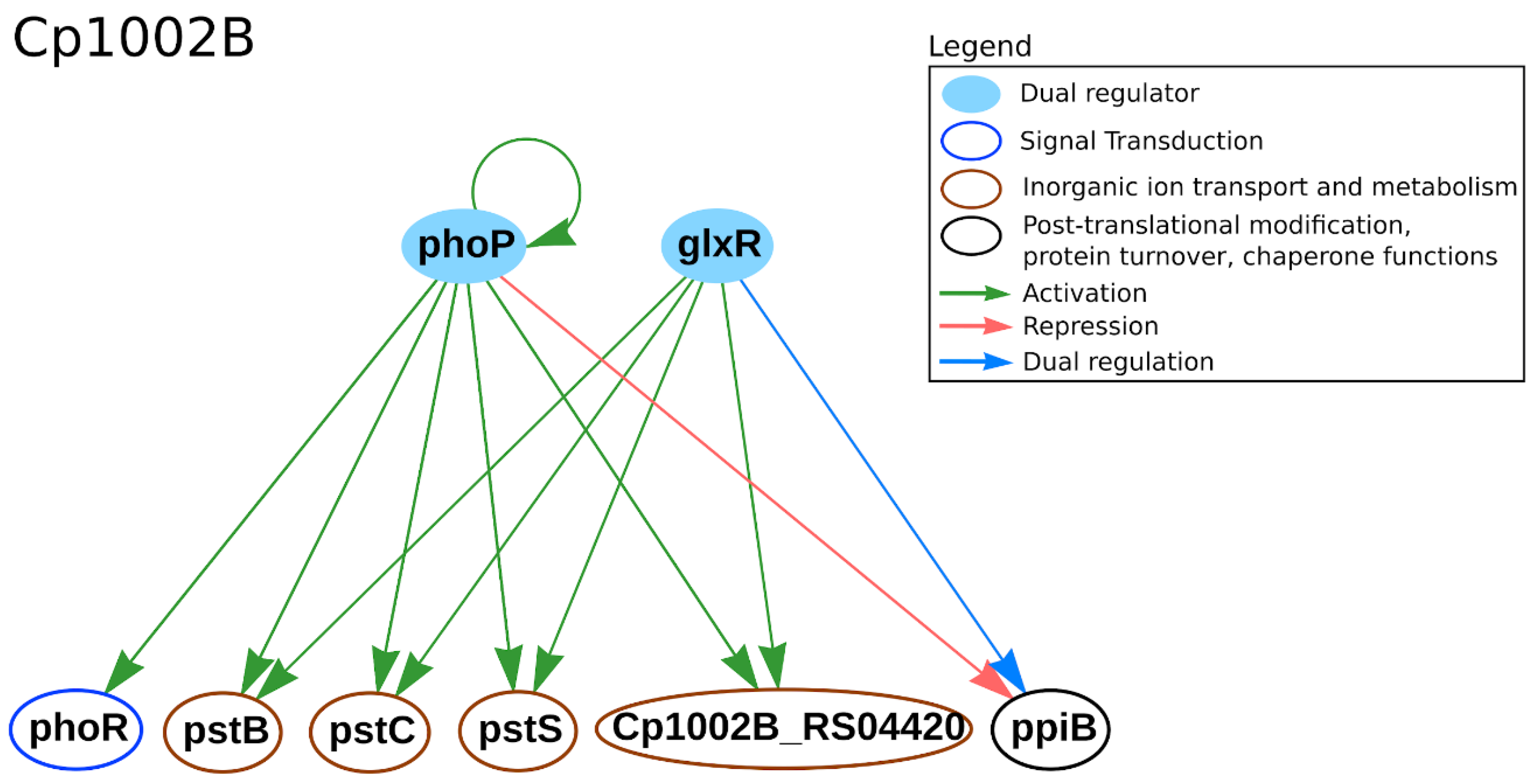
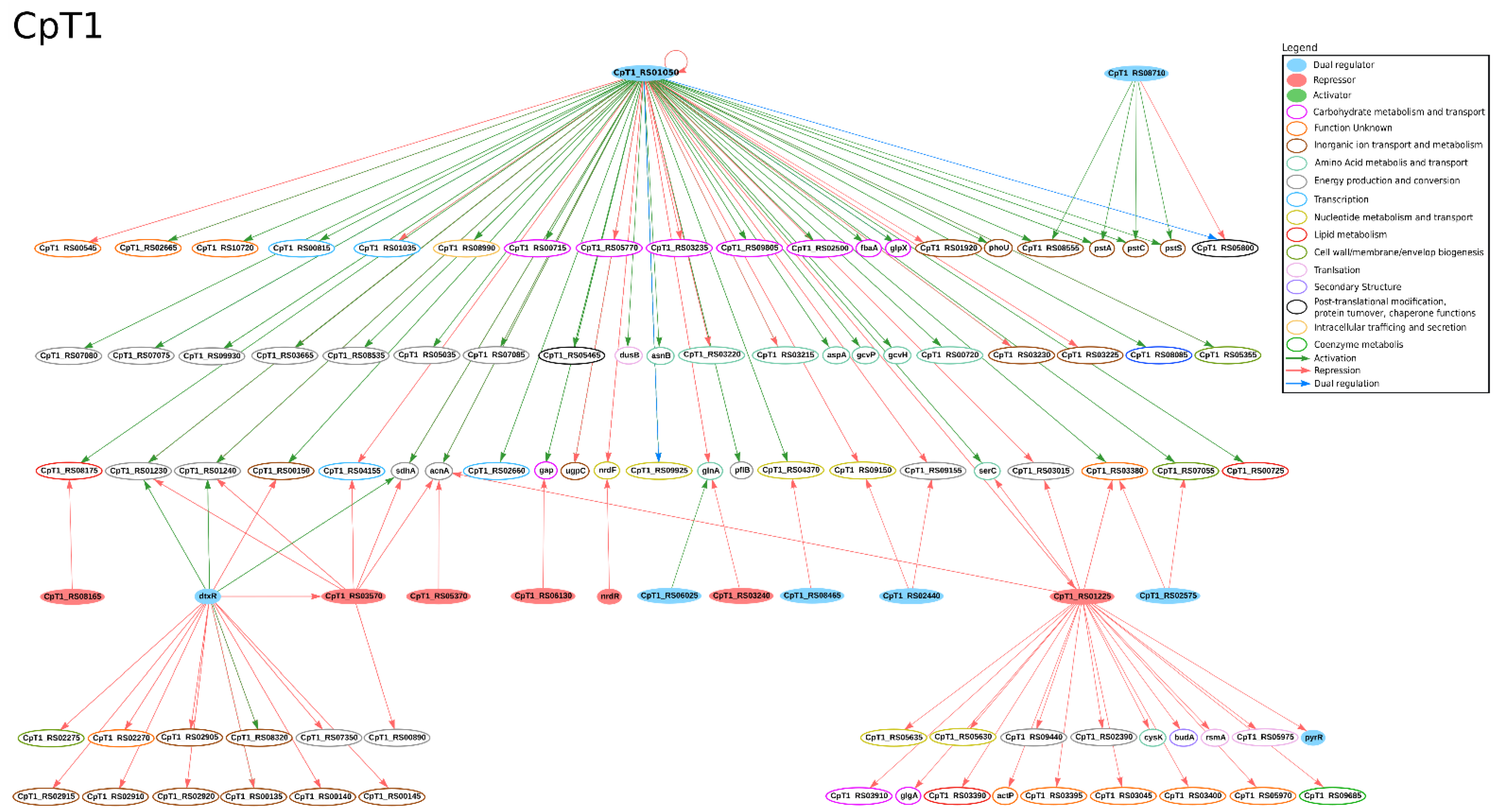
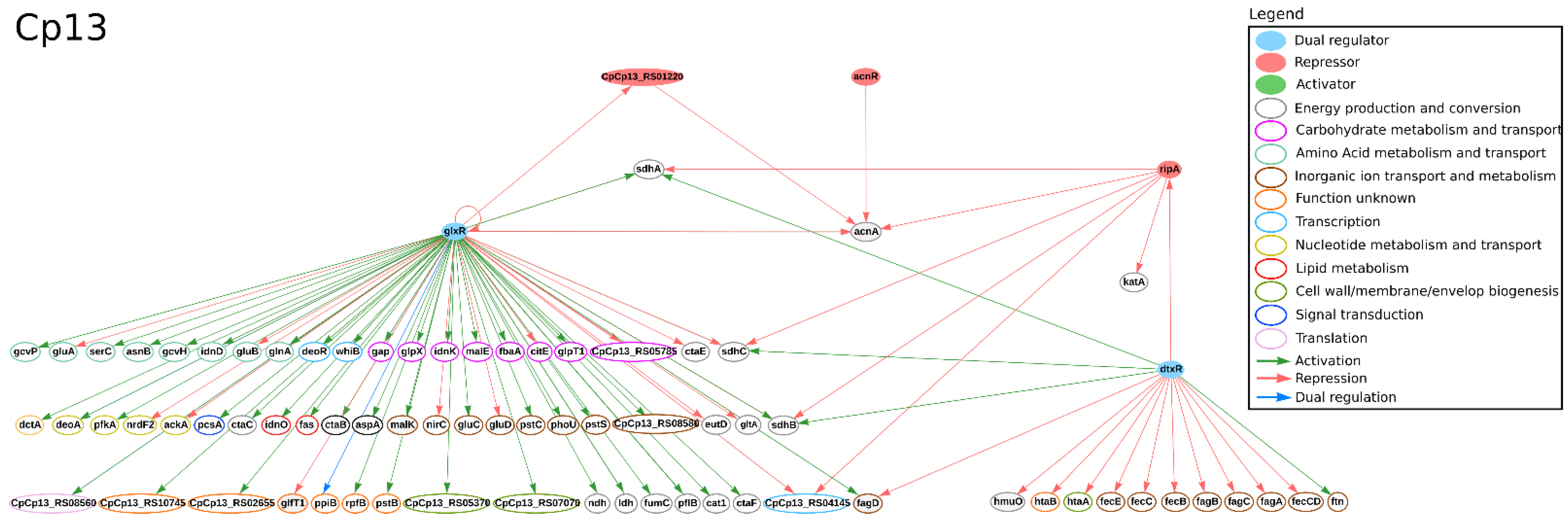
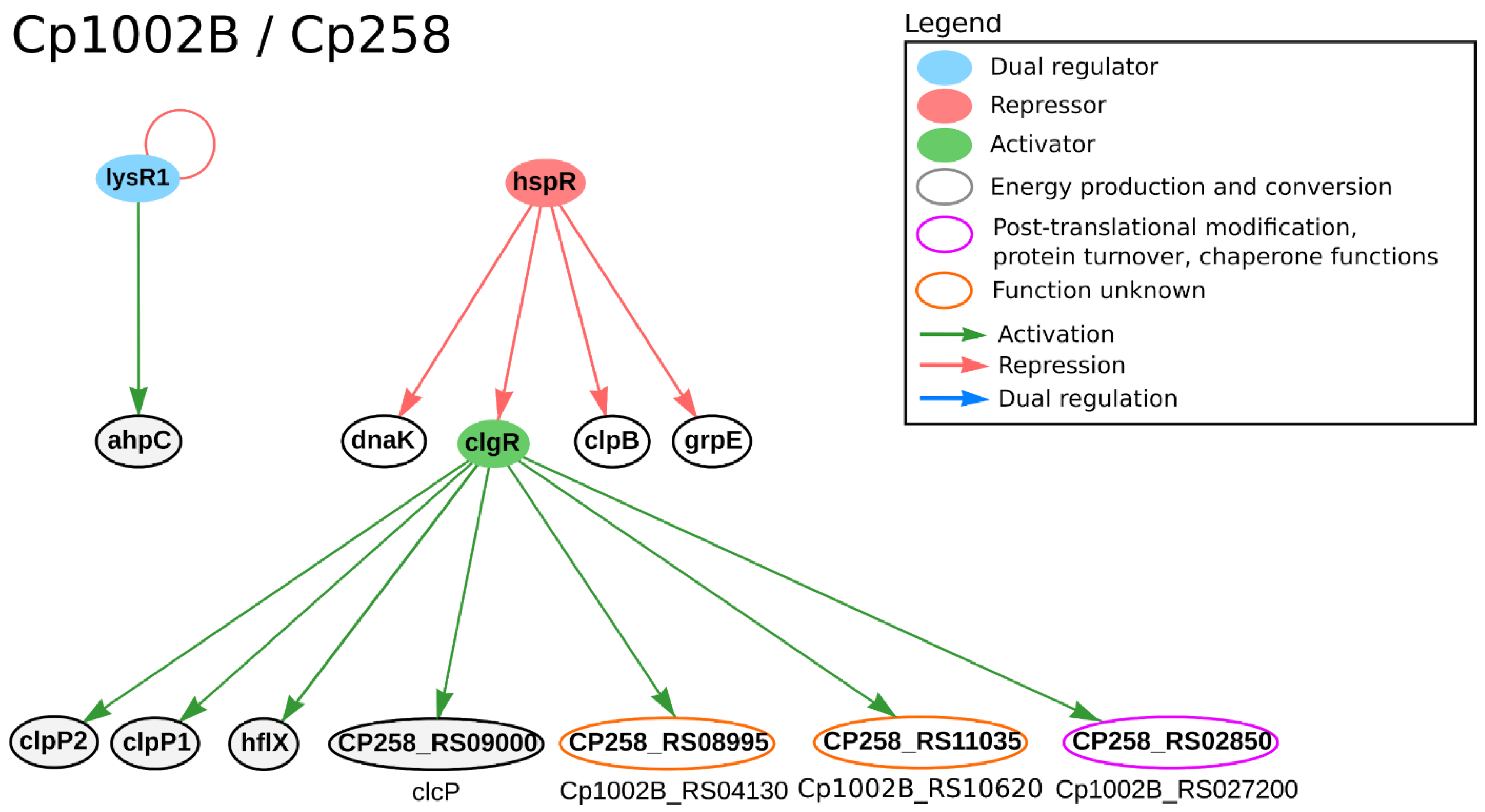
| Strain | Technology | GCN from All Genes | GCN from DEGs | Reference | ||
|---|---|---|---|---|---|---|
| Genes | Interactions | Genes | Interactions | |||
| Cp13 | Ion Proton | 2113 | 86,367 | 63 | 46 | [43] |
| T1 | Ion Proton | 2093 | 107,202 | 93 | 98 | [43] |
| 1002 | SOLiD | 2091 | 6682 | 168 | 155 | [40] |
| 258 | SOLiD | 2064 | 9376 | 139 | 165 | [41,42] |
| Sigma Factor | Product | Osmotic Stress | Thermic Stress | Acid Stress | |||
|---|---|---|---|---|---|---|---|
| Fold-Change | DEG | Fold-Change | DEG | Fold-Change | DEG | ||
| sigA | RNA polymerase sigma factor SigA (essential housekeeping sigma factor) | 2.1889 | Yes | 1.4903 | No | 0.9232 | No |
| sigB | RNA polymerase sigma factor SigB (non-essential SigA-like) | 0.6348 | No | 0.9044 | No | 2.9154 | Yes |
| sigC | RNA polymerase sigma factor SigC (ECF family) | 0.4675 | No | 0.8031 | No | 1.7238 | No |
| sigD | RNA polymerase sigma factor SigD (ECF family) | 1.5437 | No | 1.2891 | No | 0.8654 | No |
| sigE | RNA polymerase sigma factor SigE (ECF family) | 0.5483 | No | 0.9356 | No | 2.5244 | Yes |
| sigH | RNA polymerase sigma factor SigH (ECF family) | 1.8401 | No | 1.7864 | No | 3.5832 | Yes |
| sigK | RNA polymerase sigma factor SigK (ECF family) | 1.5887 | No | 1.7415 | No | 1.6199 | No |
| sigM | RNA polymerase sigma factor SigM (ECF family) | 4.7414 | Yes | 3.5593 | Yes | 4.4934 | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parise, D.; Teixeira Dornelles Parise, M.; Pinto Gomide, A.C.; Figueira Aburjaile, F.; Bentes Kato, R.; Salgado-Albarrán, M.; Tauch, A.; Ariston de Carvalho Azevedo, V.; Baumbach, J. The Transcriptional Regulatory Network of Corynebacterium pseudotuberculosis. Microorganisms 2021, 9, 415. https://doi.org/10.3390/microorganisms9020415
Parise D, Teixeira Dornelles Parise M, Pinto Gomide AC, Figueira Aburjaile F, Bentes Kato R, Salgado-Albarrán M, Tauch A, Ariston de Carvalho Azevedo V, Baumbach J. The Transcriptional Regulatory Network of Corynebacterium pseudotuberculosis. Microorganisms. 2021; 9(2):415. https://doi.org/10.3390/microorganisms9020415
Chicago/Turabian StyleParise, Doglas, Mariana Teixeira Dornelles Parise, Anne Cybelle Pinto Gomide, Flávia Figueira Aburjaile, Rodrigo Bentes Kato, Marisol Salgado-Albarrán, Andreas Tauch, Vasco Ariston de Carvalho Azevedo, and Jan Baumbach. 2021. "The Transcriptional Regulatory Network of Corynebacterium pseudotuberculosis" Microorganisms 9, no. 2: 415. https://doi.org/10.3390/microorganisms9020415
APA StyleParise, D., Teixeira Dornelles Parise, M., Pinto Gomide, A. C., Figueira Aburjaile, F., Bentes Kato, R., Salgado-Albarrán, M., Tauch, A., Ariston de Carvalho Azevedo, V., & Baumbach, J. (2021). The Transcriptional Regulatory Network of Corynebacterium pseudotuberculosis. Microorganisms, 9(2), 415. https://doi.org/10.3390/microorganisms9020415










