Abstract
Mutual relationships with symbionts play a crucial role in the evolution and ecology of plant-feeding hemipteran insects. However, there was no specific dominant bacterium observed in soft scales (Coccidae) in the previous studies, it is still unclear whether soft scales have specific primary symbionts. In this study, a nuclear ribosomal internal transcribed spacer (ITS)gene fragment was used to analyze the diversity of fungal communities in 28 Coccidae species based on next-generation sequencing (NGS). Furthermore, samples from different developmental stages of Ceroplastes japonicus were sequenced to illustrate the dynamics of fungal community. Our results showed that Coccidae-associated Ophiocordyceps fungi (COF) were prevalent in all 28 tested species with high relative abundance. Meanwhile, the first and second instars of C. japonicus, two important stages for growth and development, had high relative abundance of COF, while the relative abundances in other stages were low, ranging from 0.68% to 2.07%. The result of fluorescent in situ hybridization showed that the COF were widely present in hemolymph and vertically transmitted from mother to offspring. Our study confirms that the COF have intimate associations with the growth and development of soft scales, and provides new evidence to support that COF are primary fungal symbionts for Coccidae.
1. Introduction
Due to the lack of various essential amino acids in the plant sap, hemipteran insects usually harbor some obligate bacterial symbionts, which provide essential nutrition to complete their diet [1,2]. For examples, Buchnera in aphids, Sulcia in cicadas, Tremblaya in mealybugs, and Uzinura in armored scale insects have been reported in many studies [3,4,5,6,7,8]. Normally, these obligate symbionts own extreme genome reduction including several essential amino acid pathways, and provide amino acids and vitamins for insect hosts [6,9]. However, except for the mealybugs and armored scale insects, it has been very ambiguous whether soft scales harbor certain obligate symbionts, which play important roles in growth and development of hosts.
Soft scale insects (Hemiptera: Coccoidea: Coccidae), an economically important plant-feeding group [10], is the third largest family of scale insects, with 1282 species described across 176 genera in the world [11]. Among them, Ceroplastes rubens, Parasaissetia nigra, Saissetia coffeae, Saissetia oleae, and Coccus hesperidum are well-known polyphagous pests infested on crops and ornamental plants around the world [12]. The newly hatched nymph of soft scales hunts for the appropriate feeding location and becomes immobile after that. Meanwhile, the nymph begins to secrete wax and honeydew, which provides a medium for the growth of some fungi. The small body size and high complexity of morphological identification of soft scales have hindered the progress on the studies of diversity of microbial symbionts for a long time. Flavobacteria (Bacteroidetes) associated with scale insects (superfamily Coccoidea), since 150–250 million years ago, has been considered a potential obligate symbiont in the family Coccidae [9,13]. However, by high-throughput sequencing of bacterial 16S gene, Gomez-Polo et al. (2017) [14] showed that only negligible copy numbers of Flavobacteria were found, and there was no specific dominant bacterium in all seven Coccidae species they sampled. Instead, Gomez-Polo et al. (2017) [14] reported that six healthy soft scale species were dominated by Ophiocordyceps-allied fungus (OAF) (Hypocreales, Ophiocordycipitaceae), and vertical transmission of OAF from mother to offspring was observed in two soft scale species using Fluorescence in situ hybridization (FISH), which indicated the possibility of Ophiocordyceps being obligate fungal symbionts for soft scales.
Ophiocordycipitaceae was proposed as a new fungal family by Sung et al. (2007) [15]. After that, the species number in this family increased to 358, according to the record of Catalogue of Life [16]. Many fungi of Ophiocordycipitaceae have clavate asci with thickening apices and fusiform ascospores [17]. The dominant hosts of the Ophiocordycipitaceae are arthropods, as well as some nematodes and other fungi [18]. Most Ophiocordycipitaceae species are considered as arthropod or nematode pathogens [18], such as the medicinal fungus Ophiocordyceps sinensis [19], the zombie-ant fungus Ophiocordyceps unilateralis [20,21], and endoparasitic nematophagous fungus Hirsutella rhossiliensis [22]. For scale insects, some Ophiocordyceps species have also been identified and observed in the fat body and hemolymph of Kermes quercus (Coccoidea, Kermesidae) [23] and Kerria lacca (Coccoidea, Tachardiidae) [24], though their function for hosts is not clear. These indicate a potential lifestyle switch from insect pathogens to mutualists might occur for the Ophiocordyceps species in soft scales.
Some rare possible mutualistic (or at least nonpathogenic) relationships between Ophiocordyceps species and their hosts have been reported in some arthropods, especially in Hemiptera [4]. These studies have investigated an evolutionary process from a prokaryotic symbiosis to a eukaryotic one in certain hemipteran groups. In two species of eared leafhoppers, an endoparasitic Ophiocordyceps fungus was observed to replaces obligate symbiont Sulcia and Nasuia-allied co-symbiont in an ancestral lineage [25]. The same situation also occurs in Japanese cicada. Matsuura et al. (2018) [4] reported that cicada-parasitizing Ophiocordyceps fungi replaced obligate symbiont Hodgkinia in 15 species, although the obligate symbiont Sulcia still remained in all 24 species. In addition, two types of symbionts (bacterium Sulcia and Ophiocordyceps symbionts) are simultaneously present in four Deltocephalinae leafhoppers, and form “dual bacterial-fungal symbiosis” [26]. These eukaryotic symbionts might have similar nutritional roles with the obligate bacterial symbionts, which support the growth of hosts feeding solely on nutritionally deficient plant sap. Considering that most Ophiocordyceps species are pathogens, this group provides a model system for understanding evolutionary shift from pathogens to possible mutualists associated with their insect hosts.
In the past, based on some short bacterial gene fragments, researchers attempted to reveal symbiont communities in insect species [27,28] as well as determine the evolution relationship between obligate symbionts and their hosts [29,30]. With the development of next generation sequencing (NGS), this technique has been widely used in characterization of symbiont communities in soil, water, and insects [31,32,33]. Based on the results of six tested species that Gomez-Polo et al. (2017) [14] reported, Ophiocordyceps fungi might be the primary symbionts in soft scales. However, more tested species were needed to confirm that Coccidae-associated Ophiocordyceps fungi (COF) are primary symbionts in Coccidae. Three conditions should be satisfied to determine that a symbiont is a primary symbiont for insect hosts: the first one is that the symbiont is of strict vertical transmission from mother to offspring; the second one is the presence of a symbiont in the majority of species in a single phylogenetic clade; the last one is that the symbiont plays a functional role for insect hosts. To confirm that the COF were primary symbionts in Coccidae, we attempted to demonstrate (1) that the COF should be observed in eggs using the FISH and NGS technique, which indicated vertical transmission; (2) that the COF should be prevalent and dominant in the fungal communities of 28 tested Coccidae species; (3) that the COF should be present in different developmental stages, and had development-related abundance dynamics reflecting functional roles.
2. Materials and Methods
2.1. Insect Samples
To determine whether the Ophiocordyceps were widespread in soft scales, we collected 61 samples from 28 common soft scale species in this study. The samples were field-caught on 28 host plant species from 15 localities in southern China. The information on host plant, locality, and collection date were recorded for each sample (Supplementary Materials Table S1). To show changes of the microbiome profiles associated with Ophiocordyceps at different host developmental stages, Ceroplastes japonicus was reared on gardenias at 25 °C under long daylight condition (16L:8D) from June to September, 2018, in the laboratory. In total, 25 samples were collected, including the egg, newly hatched nymph (newN), first instar nymph (star1), second instar female nymph (star2F), second instar male nymph (star2M), third instar female nymph (star3F), prepupa, pupa, and male adult (adultM) (Figure 1, Table S1). For male individuals, the stages of prepupa and pupa occurred after the second instar, then became male adult. For female individuals, however, they became female adults after the third instar. The stage of female adult was not included due to insufficient individual numbers. All samples were preserved in 95% ethanol at −80 °C for molecular analysis. All species were identified based on morphological characters [34,35] and molecular identification (Table S1). In the field, soft scales are occasionally parasitized by parasitoid wasps or fungi. Hence, to select healthy individuals for experiments, we observed and filtrated healthy individuals as samples under a stereoscopic microscope (Nikon SMZ18) after removing wax with a dissecting needle (Figure 2).
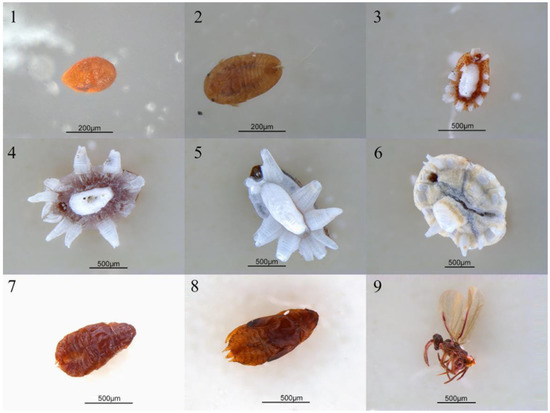
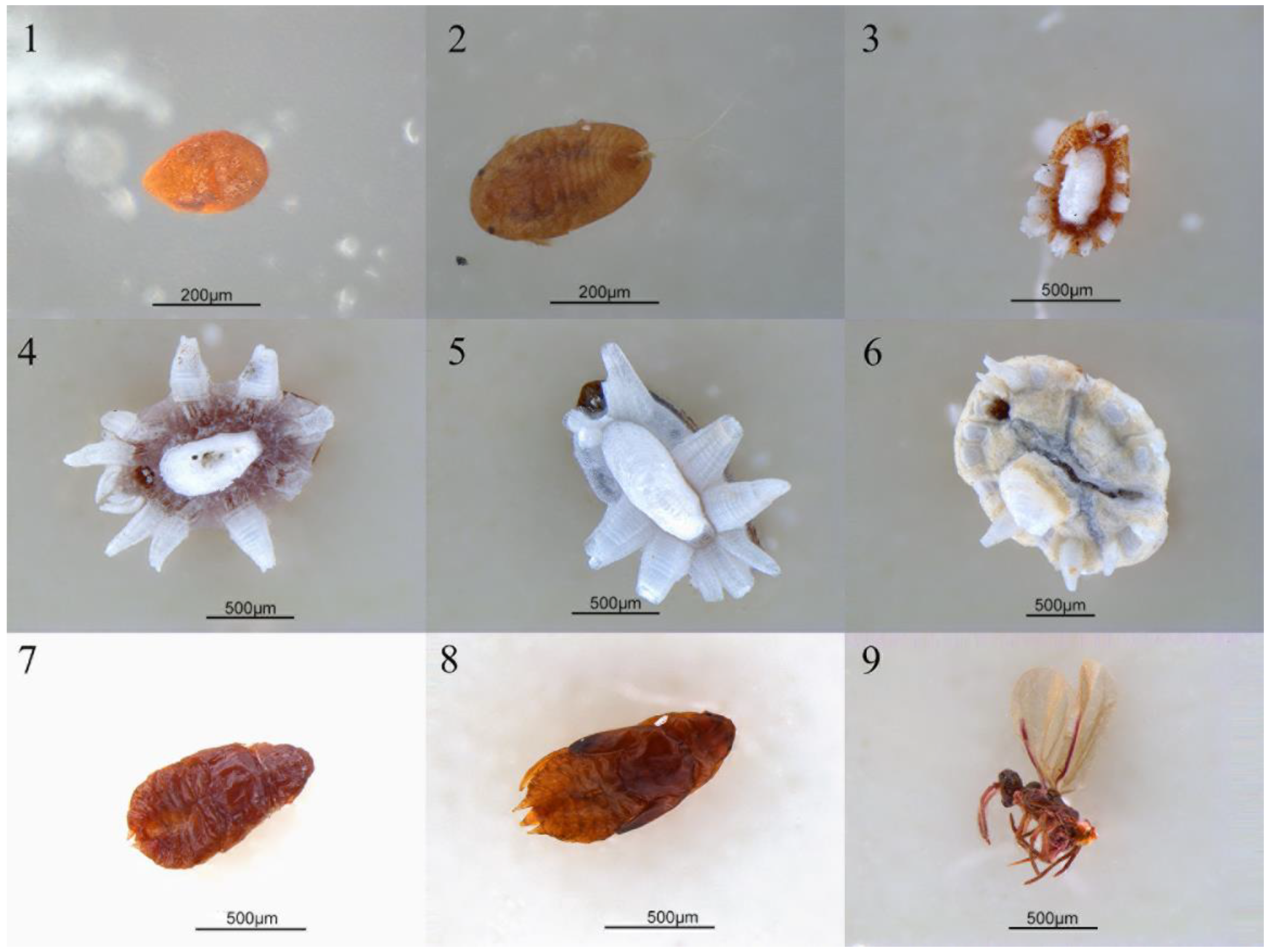
Figure 1.
The photos of Ceroplastes japonicus at nine developmental stages. 1. Egg; 2. new nymph; 3. first instar nymph; 4. second instar female nymph; 5. second instar male nymph; 6. third instar female nymph; 7. prepupa; 8. pupa; 9. male adult.

Figure 2.
The microphotographs of 28 Coccidae species without wax in this study. 1. Ceroplastes floridensis, 2. Ceroplastes rubens, 3. Ceroplastes cirripediformis, 4. Ceroplastes ceriferus, 5. Ceroplastes pseudoceriferus, 6. Ceroplastes japonicus, 7. Ceroplastes rusci, 8. Ceroplastes kunmingensis, 9. Ceroplastes sp., 10. Dicyphococcus ficicola, 11. Dicyphococcus sp., 12. Ericerus pela, 13. Rhodococcus sariuoni, 14. Megapulvinaria maxima, 15. Saissetia coffeae, 16. Protopulvinaria pyriformis, 17. Saissetia oleae, 18. Saissetia miranda, 19. Saissetia sp., 20. Pulvinaria aurantii, 21. Parasaissetia sp., 22. Eucalymnatus tessellatus, 23. Takahashia japonica, 24. Coccus praetermissus, 25. Pulvinaria psidii, 26. Coccus viridis, 27. Coccus hesperidum, 28. Coccus celatus.
2.2. DNA Extraction, PCR, Cloning, and Sequencing
All individuals without wax were surface-sterilized to avoid contamination from external microbes with 95% alcohol for 1 min, 3% sodium hypochlorite solution for 3 min, and washed three times with distilled water. The DNeasy Blood and Tissue Kit (Qiagen, Dalian, China) was used to extract genomic DNA from whole insect, following the manufacture’s instruction. For most samples, DNA extraction was performed in a pool of 1–5 individuals. For some samples with very tiny body sizes, 10 or more individuals were used. The COI and internal transcribed spacer (ITS) genes of each sample were amplified using specific primers C1-1554F and C1-2342R [36], Hyp2084F and Hyp3489R, CFSZ-CS and LSU-CS-1R [4]; the PCR primers and annealing temperatures are shown in Table S2. PCR products were visualized on agarose gels, and the most intense products were sequenced bidirectionally. The samples with overlapping peaks in sequencing chromatograms were cloned and then sequenced.
2.3. Next Generation Sequencing
The symbiotic fungal communities of all samples were assessed by next generation sequencing of the ITS region of small subunit ribosomal RNA gene amplicons, using the Illumina HiSeq platform performed by Biomarker (Beijing, China) Technologies Corporation. The forward primer ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and reverse primer ITS2R (5’-GCTGCGTTCTTCATC GATGC-3’) [37] were used to generate amplicon libraries (paired-end, 2 × 250 bp). In order to get high quality data (clean reads), Trimoraic v. 0.33 [38] software was used to filter the raw tags. Forward and reverse sequences were merged into single sequences using FLASH v. 1.2.11 [39] (overlap ≥10 bp and error ration ≤0.2) for each sample. The chimera was identified and removed by UCHIME v. 4.2 [40]. After dereplication and sorting of all sequences, singletons were discarded and the remaining sequences were clustered into representative operational taxonomic units (OTUs) at 97% similarity cutoff using USEARCH (v. 10.0) [41], and 0.05% of all reads was used as a threshold to filter OTUs. The RDP Classifier v. 2.2 [42] was used to annotate OTU species classification based on the taxonomic databases Unite (Release 7.2) [43].
2.4. Fungal Diversity in Different Samples
The Chao1 richness estimator, Shannon–Wiener diversity index and Simpson diversity index were calculated in mothur v. 1.30 [44]. The Chao1 richness estimator mainly calculates the minimum number of OTUs in a sample, higher Chao1 value indicates a higher community richness. The Shannon–Wiener diversity index is the average degree of uncertainty of predicting the species of a random sample in a community. The lower the diversity a community has, the lower the Shannon value. The Simpson index is the probability that individuals from two consecutive random samples are the same species. Higher Shannon and lower Simpson values indicate a higher diversity in a community. Finally, principal coordinates analysis (PCoA) was used to visualize difference of the fungal communities at different host developmental stages.
2.5. Identification of Unclassified Otus and Phylogenetic Analyses
Some unclassified OTUs were dominant with >30% relative abundance at family level in some samples. We thought these dominant unclassified OTUs might belong to Ophiocordycipitaceae, and their unclassification might be due to having had significant sequence diversity compared to the reference sequences in the taxonomic reference databases. We obtained about 800 bp long ITS sequences representing these OTUs using the Ophiocordycipitaceae primers ITS1F and ITS4R. These ITS sequences were then used to reconstruct phylogenetic trees with known Ophiocordycipitaceae species. Forty-five ITS sequences of the Ophiocordycipitaceae were downloaded from GenBank. The phylogenetic analyses were performed to determine the phylogenetic positions of these dominant unclassified OTUs. Phylogenetic trees were reconstructed using Bayesian inference (BI) and maximum likelihood (ML) methods. The program MAFFT v. 7.313 [45] was used to carry out the multiple alignment, and ModelFinder [46] was used to infer the best-fitting evolutionary models via Bayesian information criterion (BIC). The BI analysis was inferred using MrBayes v. 3.2.6 [47] with the GTR + I + G + F model (1,000,000 generations). Meanwhile, ML analysis was performed using IQ-TREE [48,49], with the TPM3u + I + G4 + F model for 5000 ultrafastbootstraps [50]. Nectria haematococca and N. ipomoeae were selected as outgroups.
2.6. Fluorescent in Suit Hybridization
To identity the distribution of Ophiocordyceps within a soft scale body, fluorescent in suit hybridization (FISH) was performed on C. rubens and C. japonicus with specific probe Hyp760 (CY5-5′-CCTGCCTGGAGCACTCT-3′) [4]. Eggs and nymphs were used in this experiment. The samples were placed in 1.5 mL centrifuge tubes. At first, surface disinfection and cleaning were performed with 95% alcohol, then all samples were fixed in Carnoy’s solution (60% ethanol, 30% chloroform, 10% glacial) overnight at room temperature. After removing the Carnoy’s solution, they were rinsed with 100% alcohol, for three times, to ensure no reagent remains. Then samples were immersed in alcoholic 6% H2O2 solution to quench autofluorescence at room temperature for almost one week; during this time the H2O2 solution was changed once every two days. When samples were colorless, the solution was discard and samples were rinsed with 100% alcohol for three times. After rinsing the centrifuge tube with 1 mL 37 °C preheated hybridization buffer (20 mM Tris-HCl (pH 8.0), 0.9 M NaCl, 0.01% SDS, 30% formamide), the hybridization solution with probes (final concentration 10 pm/mL) was added under dark conditions, staining for 12–14 h at 37 °C. After the hybridization, the remaining probes were washed off with PBS TX containing 0.3% Triton X-100. At last, slides were made and observed under a Leica TCS SP8X DLS confocal microscope. Insects with no-probe staining were used as negatives controls.
3. Results
3.1. Symbiotic Fungal Composition in Scale Insects
In total, high-throughput sequencing yielded 3,890,910 paired reads from 61 samples of 28 soft scale species. After merging and quality filtering, 3,608,175 clear tags were selected for further analyses. At least 21,111 clean tags were obtained for each sample, with an average of 59,150 clean tags per sample. The final tags were classified into 475 OTUs representing 6 phyla, 24 classes, 50 orders, and 102 families, with an average of 39 OTUs per sample.
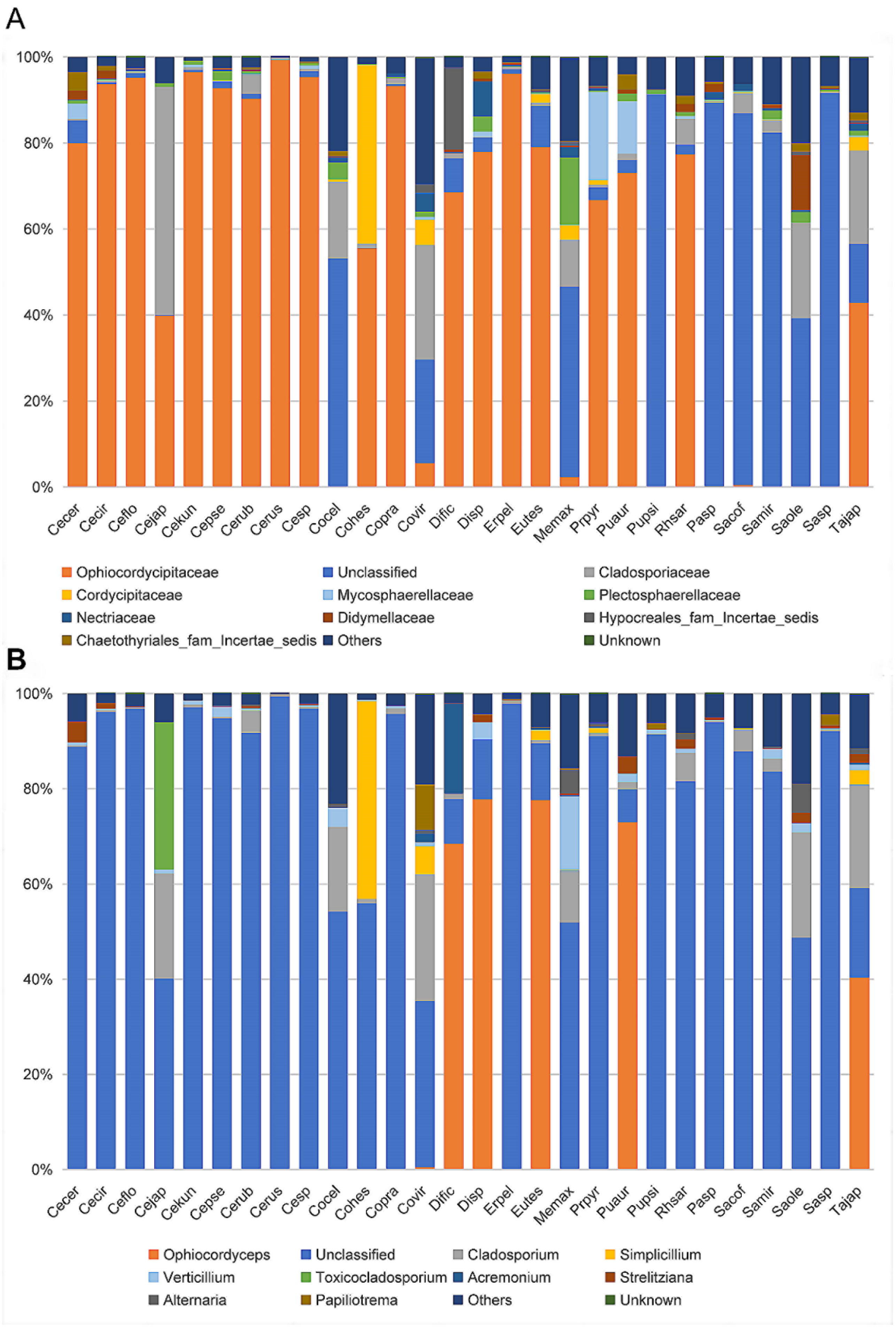
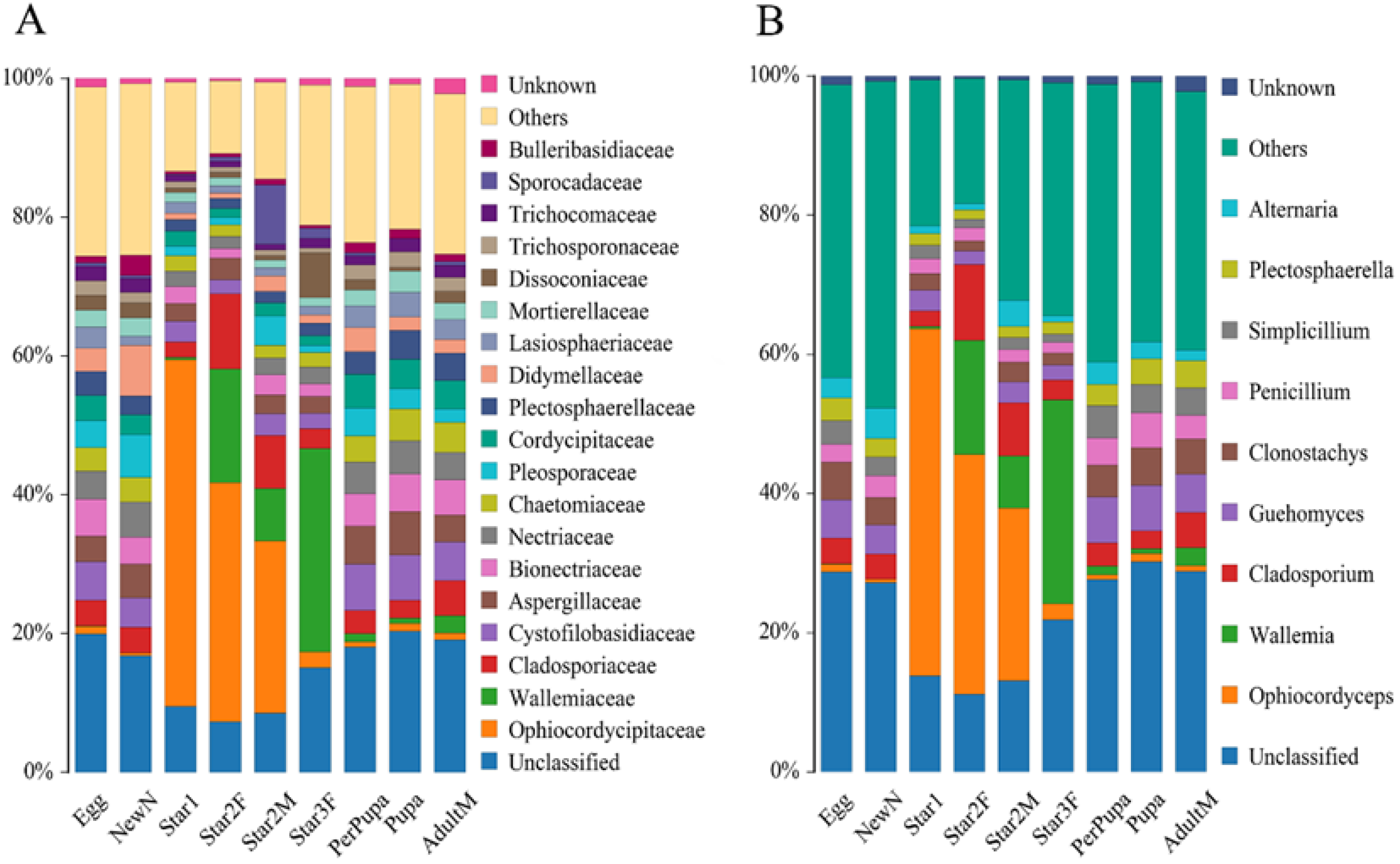
At the family level, the cluster analysis was performed on the representative OTU. Ophiocordycipitaceae species were detected in all 28 species, and the average relative abundance was 54.36% (standard deviation (SD): 39.54%) (Table S3). The relative abundances of Ophiocordycipitaceae species showed variation among 28 different host species, ranging from <0.1% in Pulvinaria psidii, Saissetia miranda, and Coccus celatus to >95% in Ceroplastes sp., C. floridensis, C. rubens, C. kunmingensis, and Ericerus pela (Table S3; Figure 3A). For eight species (Megapulvinaria maxima, Co. celatus, Parasaissetia sp., Saissetia sp., S. oleae S. coffeae, S. miranda, and Pulvinaria psidii), some unclassified OTUs were dominated with an average of 72.20% (SD: 21.10%) relative abundance, ranging from 39.16% in S. oleae to 92.51% in Saissetia sp. These unclassified OTUs were confirmed as Ophiocordyceps species in subsequent analysis. For C. japonicus and Co. viridis species, Cladosporiaceae was dominant, accounting for 53.00% and 26.65%, respectively. The relative abundance of Ophiocordycipitaceae decreased to 39.89% and 24.04% for C. japonicus and Co. viridis, respectively. For the remaining 18 species, Ophiocordycipitaceae dominated the fungal communities with averaged relative abundance of 81.87% (±15.42%). At genus level, a large number of OTUs could not be classified, the average relative abundance of unclassified sequences in all sample was 72.80% (±33.47%) (Table S3; Figure 3B). Ophiocordyceps was dominate in Dicyphococcus sp. (77.79%), Pulvinaria aurantii (73.03%), Eucalymnatus tessellatus (76.92%), Takahashia japonica (41.16%), and Dicyphococcus ficicola (69.71%). Some OTUs classified as Ophiocordycipitaceae species could not be identified at the genus level, indicating they had noteworthy sequence diversity compared to other Ophiocordycipitaceae species.
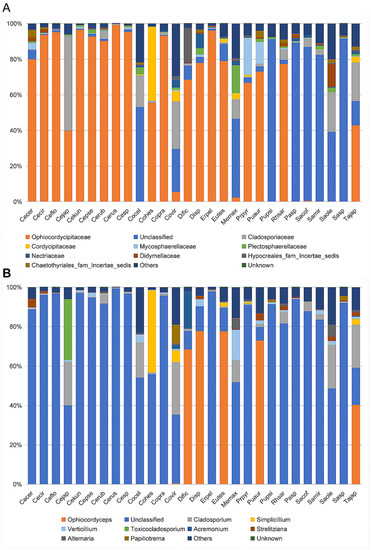
Figure 3.
Relative abundance of fungi in 28 Coccidae species at family (A) and genus (B) levels as assessed by high-throughput amplicon sequencing of the internal transcribed spacer (ITS) gene. Ceroplastes ceriferus (Cecer, 1), Ceroplastes cirripediformis (Cecir, 3), Ceroplastes, floridensis (Ceflo, 4), Ceroplastes japonicus (Cejap, 1), Ceroplastes kunmingensis (Cekun, 1), Ceroplastes pseudoceriferus (Cepse, 3), Ceroplastes rubens (Cerub, 4), Ceroplastes rusci (Cerus, 1), Ceroplastes sp. (Cesp, 3), Coccus celatus (Cocel, 1), Coccus hesperidum (Cohes, 1), Coccus praetermissus (Copra, 1), Coccus viridis (Covir, 3), Dicyphococcus ficicola (Dific, 3), Dicyphococcus sp. (Disp, 1), Ericerus pela (Erpel, 3), Eucalymnatus tessellatus (Eutes, 3), Megapulvinaria maxima (Memax, 1), protopulvinaria pyriformis (Prpyr, 3), Pulvinaria aurantii (Puaur, 1), Pulvinaria psidii (Pupsi, 1), Rhodococcus sariuoni (Rhsar, 1), Parasaissetia sp. (Pasp, 4), Saissetia coffeae (Sacof, 2), Saissetia miranda (Samir, 2), Saissetia oleae (Saole, 1), Saissetia sp. (Sasp, 5), Takahashia japonica (Tajap, 3). The number in the bracket represents the sample number of each species.
To evaluate the alpha diversity of microbial composition among 28 different species, we calculated the Chaos richness index, the Simpson, and Shannon diversity estimator (Table S3). We found that the fungal community of Megapulvinaria maxima had the highest community richness and diversity (Chaos, 78; Shannon, 2.77; Simpson = 0.12), and Ceroplastes rusci had the lowest community richness and diversity (Chaos, 18; Shannon, 0.05; Simpson, 0.99).
3.2. Identification of Dominant Unclassified OTUs
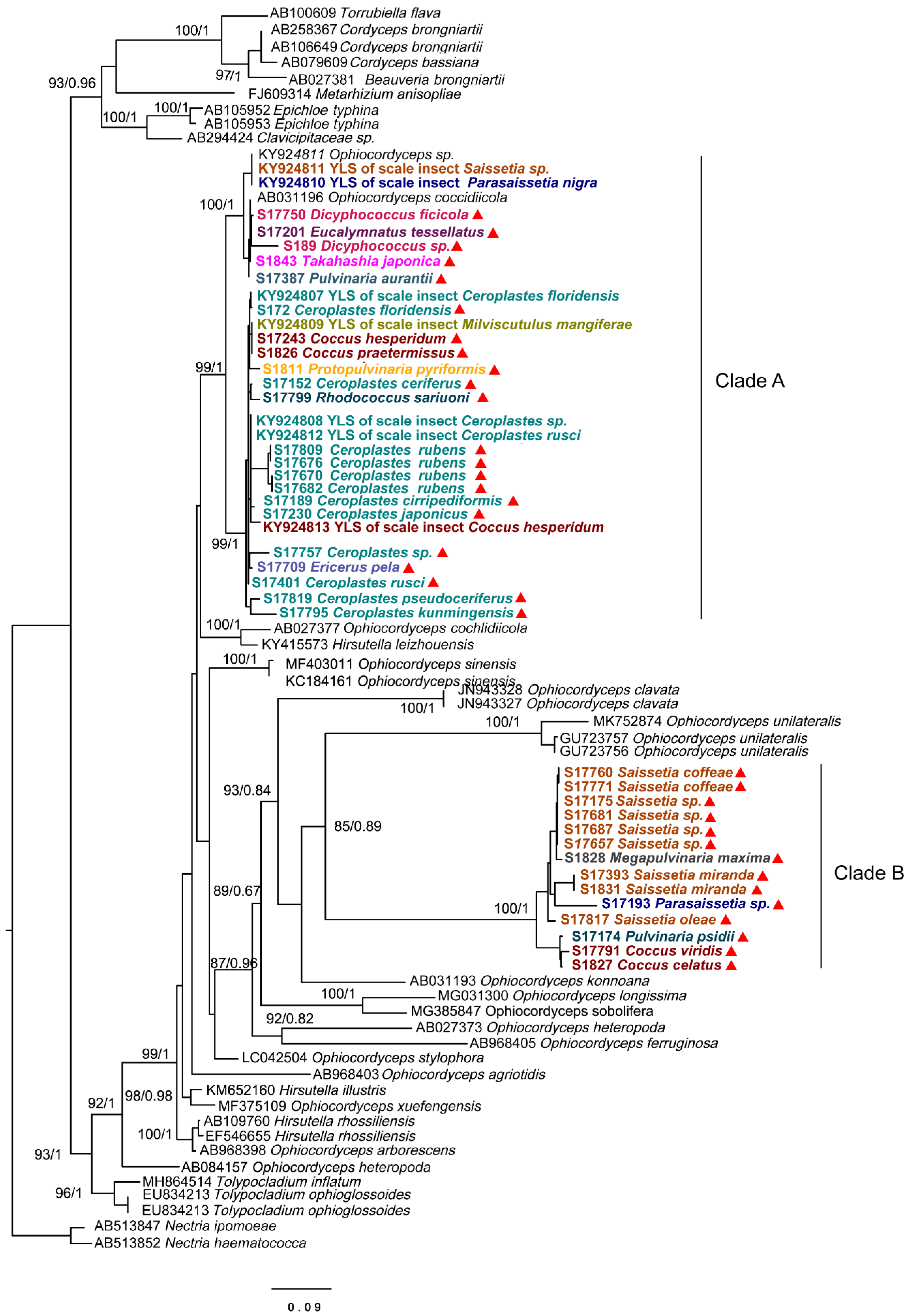
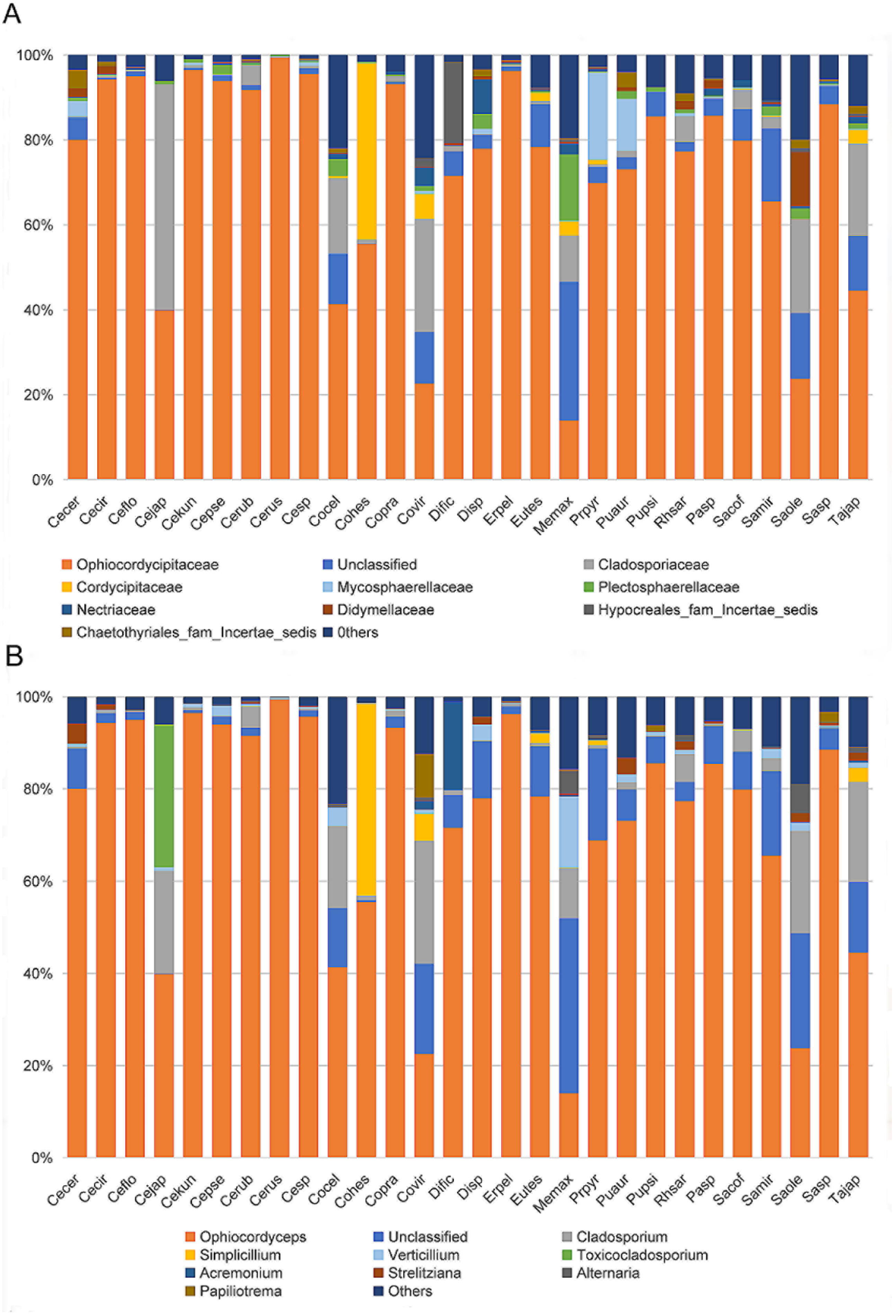
To identity the dominant unclassified OTUs, we amplified and obtained an (about) 800 bp fragment of the ITS gene from each sample using the Ophiocordycipitaceae primers ITS1F and ITS4R. By aligning these ITS sequences with the dominant OTUs sequences, we finally confirmed these ITS sequences represented those dominant unclassified OTUs. Phylogenetic analysis using ML and BI generated almost congruent tree with relatively high posterior probabilities and bootstrap values (Figure 4). The dominant unclassified OTU sequences were nested inside the Ophiocordyceps in the phylogenetic tree, which confirmed the dominant unclassified OTUs belonged to the Ophiocordyceps species. The Ophiocordyceps sequences from 28 Coccidae species formed two major clades (A and B) in the phylogenetic tree. The clade A included those Ophiocordyceps from Ceroplastes, Dicyphococcus, Eucalymnatus, Takahashia, Milviscutulus, Rhodococcus, and Ericerus species. Notably, all Ophiocordyceps sequences from seven Ceroplastes species were clustered together in the clade A. The Clade A also included a pathogenic fungus of scale insects, Ophiocordyceps coccidiicola (AB031196). Correspondingly, all Ophiocordyceps sequences from Saissetia and Parasaissetia species of our samples were included in the clade B. The Ophiocordyceps unilateralis was closed to the clade B, which was a pathogenic fungus of ants. The Ophiocordyceps sequences of the genus Coccus and Pulvinaria were scattered in the two clades. Additionally, some Ophiocordyceps species from Saissetia and Parasaissetia sampled from Ficus carica in Israel and Cyprus in Gomez-Polo et al. (2017) [14] were included in the clade A. After identifying the dominant unclassified OTUs, we replotted the relative abundance of fungi in 28 Coccidae species (Figure 5A,B). The mean relative abundance of Ophiocordycipitaceae species were 72.55% (SD: 25%), the value was the same at the genus level.
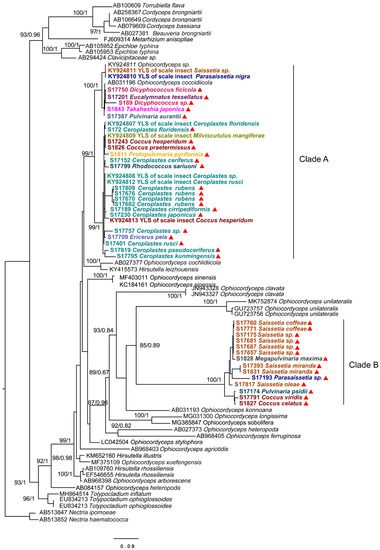
Figure 4.
Phylogenetic analysis of the Ophiocordyceps fungi based on an (about) 800 bp fragment of partial ITS gene. Bayesian inference (BI) and maximum likelihood (ML) trees had similar topology. The bootstrap support values and posterior probabilities are indicated for each node. Support values below 70 are not shown. The samples in this study are labeled with red triangles. Different colors represent species from different genus.
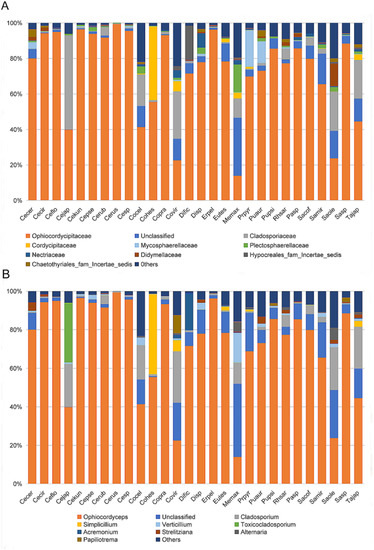
Figure 5.
Relative abundance of fungi in 28 Coccidae species at family (A) and genus (B) levels after some unclassified operational taxonomic units (OTUs) were confirmed as Ophiocordyceps species. Species Abbreviations are the same as those in Figure 3.
3.3. Abundance Dynamics of Ophiocordyceps across Different Developmental Stages of Ceroplastes japonicus
A total of 2,000,620 pairs of reads from the 25 samples were obtained. After merging and quality filtering, 1,784,292 clean tags were generated. Each sample produced at least 68,867 clean tags. High-quality sequences were clustered into 2350 OTUs, corresponded to 4 kingdoms, 16 phyla, 48 classes, 107 orders and 253 family. The average relative abundance of the total unclassified OTUs was 14.9%, the dominant unclassified OTU was from the class Agaricomycetes of phylum Basidiomycota, with mean 5.2% relative abundance. The Ophiocordyceps species was directly identified based on the taxonomic reference databases. At the family level, Ophiocordycipitaceae was observed in each sample with varied relative abundance in different developmental stages (Figure 6A, Table 1). The first instar nymph contained the highest relative abundance of Ophiocordycipitaceae, accounting for 49.91%. For the second and third instars, the relative abundance of Ophiocordycipitaceae decreased gradually with the nymph development. The relative abundance decreased to 2.05% in the third instar. The low relative abundances of Ophiocordycipitaceae were observed in stages of prepupa, pupa, and adult male. Interestingly, the second instar female nymphs had much higher relative abundance of Ophiocordycipitaceae (34.49%) than the second instar male nymphs (24.94%). At the genus level, the results were similar to those at family level (Figure 6B). Ophiocordyceps predominated in the first and second instar nymphs compared to the relative low relative abundances for other instar nymphs (Table 1). The changes in relative abundances did not mean change in absolute numbers of the Ophiocordyceps, and were a good way to show symbiotic fungal composition changes. Ophiocordyceps was found in the egg, indicating that vertical transmission occurred from the female parent to offspring.
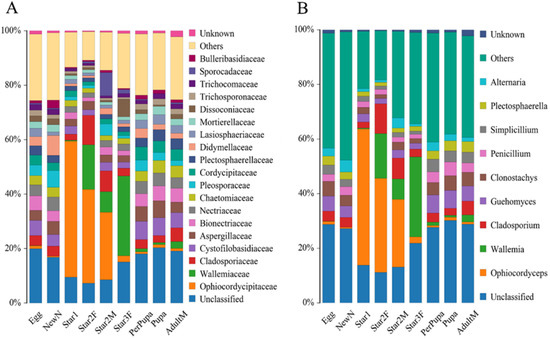
Figure 6.
Relative abundance of fungi at family level (A) and genus level (B) in Ceroplastes japonicus at nine developmental stages, including the egg, newly hatched nymph (newN), first instar nymph (star1), second instar female nymph (star2F), second instar male nymph (star2M), third instar female nymph (star3F), prepupa, pupa, and male adult (adultM).

Table 1.
The fungal alpha diversity and relative abundance of Ophiocordycipitaceae in Ceroplastes japonicus at different developmental stages (mean ± SD).
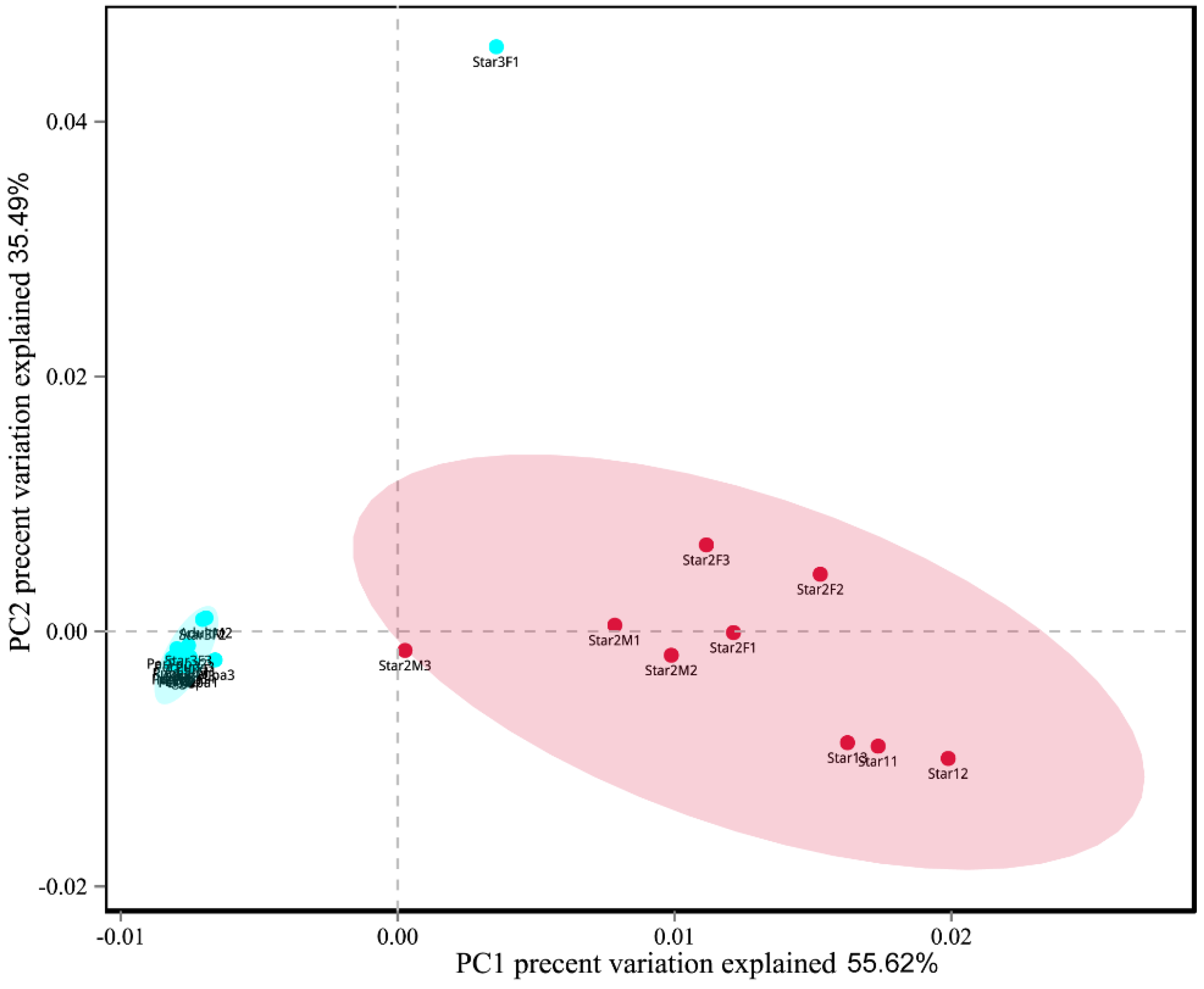
The Chaos richness indexes, the Simpson, and Shannon diversity estimators are shown in Table 1. Egg, new nymph, prepupa, pupa, and adult stage had the higher values of Shannon index and lower values of Simpson index than other stages. This result suggested higher community diversity in these developmental stages, and also indicated that Ophiocordyceps was dominant in the other life stages (instars 1–2). The Chao index of star3F was highest, and lowest in the pupa. It indicated that the greatest fungal community richness was in the third instar female nymph compared to lowest community richness in the pupa. The dissimilarity of the communities from different samples was visualized using principal component analysis (PCA) (Figure 7). We used the first two PCA dimensions as they explained 91% of the variation. The composition of fungal communities of the adult, pupa, prepupa, egg, and new nymph were similar and clustered together, whilst the fungal communities of the first and second instar nymph were in the other cluster.
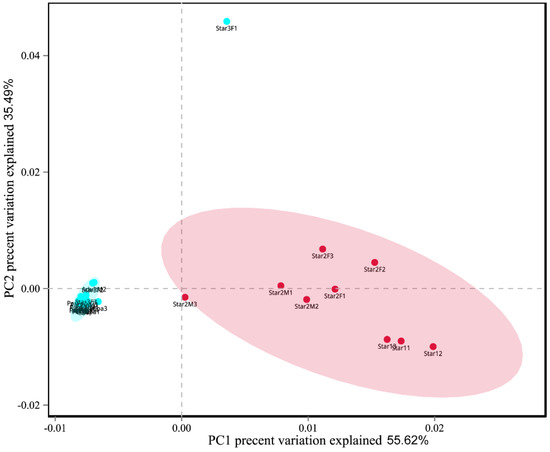
Figure 7.
Principal component analysis scatter diagram of microbial community structure of Ceroplastes japonicus among nine development stages. Each point represents a sample, different colors indicated different clusters. Abbreviations are the same as those in Figure 6.
3.4. Fluorescence in Suit Hybridization
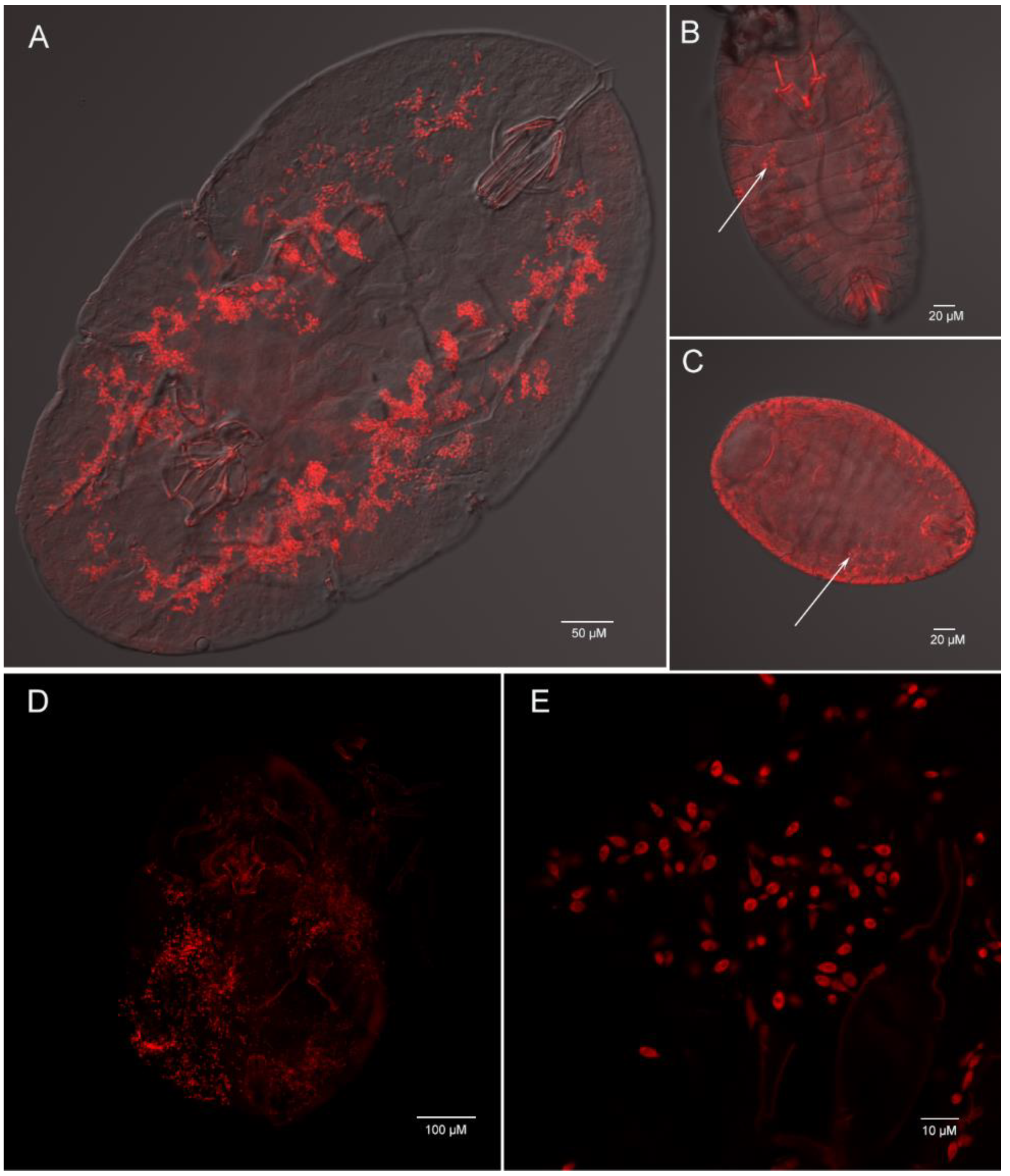
Using whole insect fluorescence in suit hybridization (FISH), we determined that Ophiocordyceps are both presented in C. rubens and C. japonicus with the lemon-shape cells (about 6–8 µm long and 1–3 µm wide) (Figure 8A–E). In the nymph, plenty of Ophiocordyceps were widely distributed in hemolymph (Figure 8A). In the egg and new nymph, a few Ophiocordyceps were also found (Figure 8B,C), proving that Ophiocordyceps was vertical transmission from mother to offspring in soft scales.
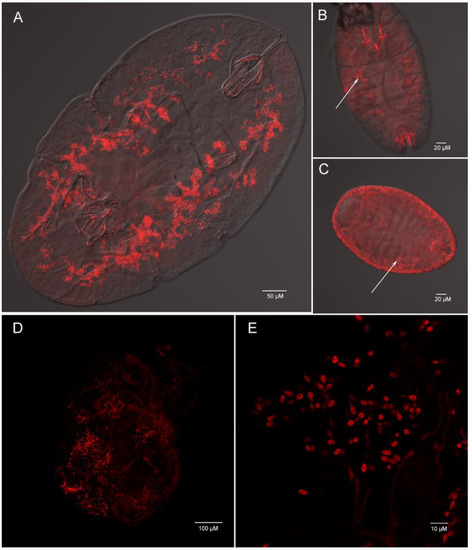
Figure 8.
Whole mount fluorescence in situ hybridization of Ophiocordyceps fungi (red) in Ceroplastes rubens (A,B,C) and Ceroplastes japonicus (D,E). Nymph (A), new nymph (B) and egg (C) of Ceroplastes rubens; (D) nymph of Ceroplastes japonicus; (E) high magnification of Ophiocordyceps cells in Ceroplastes japonicus. The arrows show the location of Ophiocordyceps fungi.
4. Discussion
Ophiocordyceps was clearly observed in the egg of C. japonicus, based on the methods of FISH and NGS, indicating the vertical transmission of Ophiocordyceps occurred from mother to offspring. A similar result has been confirmed in the egg of Parasaissetia nigra by Gomez-Polo et al. (2017) [14]. The vertical transmission of yeast-like symbionts (Ophiocordyceps species located within specialized cells in the fat body) have been observed in some cicadas [4]. These evidences in different species confirm that COF are vertical transmission. Strict vertical transmission of symbionts is typically (characteristically) observed for the primary symbiont [51]. However, many facultative symbionts are also vertically transmitted [52], for example, all species of the genus Rickettsia are vertically transmitted in invertebrates [53]. More evidence is needed to support the hypothesis that the COF are primary symbionts for soft scales.
Generally, a primary symbiont should be the presence of a symbiont in the majority of species in a single phylogenetic clade. Although the primary symbiont might be lost or replaced by other symbionts in some species of an insect group [9,54], they should still occupy the majority of that clade, indicating stable and ancient symbiosis. Gomez-Polo et al. (2017) [14] have confirmed that COF, widely known as entomopathogenic fungi, were prevalent in six tested species from soft scales. Here, 28 different Coccidae species from 14 localities were collected and analyzed in our study. Our result confirmed that COF were presented in all 28 tested species with high relative abundance, indicating that the symbiosis was stable.
The obligate and vertically transmitted symbionts have reduced genomes or lost some genes considered essential compared to their free-living relatives [52,55], and play functional roles at different developmental stages of their hosts. Genome sequencing of the Coccidae-associated Ophiocordyceps fungi are a key way for illustrating their biological and ecological roles in scale insects. However, the specific contribution and role of Ophiocordyceps in soft scales are still unknown. In our study, a very important and noteworthy work is to analyze the composition change of the fungal community over different developmental stages of C. japonicus. The COF appeared in all developmental stages, especially in the first and second instar, with high relative abundance, showing an intimate association with their hosts’ growth and metabolism. The time during the first and second instar is important for feeding and growth, the length of the body increases from about 300 um at the newly-hatched stage to 700 um at the second instar, close to the size of the female adult. At the second instar male, third instar female, prepupa, pupa, and male adult stage, low relative abundances of COF indicate that the contribution to pupa, mating, and reproduction of COF might not be remarkable. The fungal symbionts have large genome and provide a much more complete metabolic potential and synthesize all protein amino acids relative to bacterial symbionts [56]. Some examples have shown substitutive processes, from a bacterial symbiosis to a fungal one in some hemipteran groups [4,25]. Fungal symbiosis with larger genome revealed its metabolic versatility, including synthesis of almost all amino acids, vitamins, and other metabolites, which is sufficient to compensate for bacterial symbiosis loss [4,56,57]. Based on the above evidences in our study, we finally considered that Ophiocordyceps fungi are primary symbionts for soft scales.
In general, obligate symbionts have a long history of codiversification with their hosts, due to mutualism and indispensability for each other [14,54]. Many obligate bacterial associations of insects from 40 to 280 MYA [52]. Interestingly, the oldest evidence of animal parasitism by fungi (Ophiocordyceps) is from a fossil of a male scale insect estimated in the Upper Albian of the Lower Cretaceous (99–105 Mya) [18], indicating there is a long history between the Ophiocordyceps species and scale insects. In the phylogenetic tree, most hosts of Ophiocordyceps fungi were insects, indicating a strong relationship between pathogenic fungi and COF. The Ophiocordyceps of Saissetia clade is close to the zombie-ant fungus O. unilateralis in the phylogenetic tree, indicating that the Ophiocordyceps clade associated with the Saissetia clade might be related to ants’ pathogenic fungi. Ant attendance of scale insects is a well-known phenomenon [58,59]. Host shifts can be promoted by the overlapping ecological niche of the unrelated hosts [60]. Interestingly, Ophiocordyceps of Saissetia and Parasaissetia species collected from Israel and Cyprus [14] clustered with the Ceroplastes clade in our study. The COF in the same insect hosts from different countries have different lineages in the phylogenetic tree, providing evidence to support a hypothesis that soft scales might be occasionally and independently affected by Ophiocordyceps pathogens from other insects, then Ophiocordyceps species gradually reduced and lost their virulence with a long evolutionary history. Finally, some Ophiocordyceps species became obligate fungal symbionts in soft scales, which played functionally equivalent roles.
In conclusion, Ophiocordyceps fungi are widespread in soft scales, and show intimate association with the growth and development of soft scales. Our study further confirms that COF should be primary symbionts for soft scales, which might be acquired, newly and independently, from pathogenic relatives in the genus Ophiocordyceps. Our analysis may indicate a long-term evolutionary switch from insect pathogens to mutualistic symbionts in different hosts. Further works, including genomic and histological research, are needed to investigate the biological function, transmission mechanism, and evolutionary history of Ophiocordyceps in soft scales.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/2/404/s1, Table S1: collection detail of soft scales used in this study. Table S2: primers used for PCR amplification of genes of fungal symbiont and host insects, Table S3: alpha diversity of 28 soft scales through sequencing analysis of ITS gene amplicons and relative abundance.
Author Contributions
Conceptualization, J.D.; methodology, Y.Y. and X.W.; software, Y.Y. and Q.L.; data collection, curation and visualization, Y.Y. and X.W.; writing—original draft preparation, J.D. and Y.Y.; writing—review and editing, J.D. and X.H.; supervision, X.H.; project administration, X.H.; funding acquisition, J.D. and X.H. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (grant numbers: 31601860 and 31772504). The National Key R&D Program of China (2016YFE0203100), and Fujian Provincial Department of Science & Technology (2015J06005).
Data Availability Statement
The sequences reported in this paper have been deposited in the GenBank database www.ncbi.nlm.nih.gov/genbank/ (accessed on 24 January 2021) (accession nos. MW380751-MW380786, MW450918-MW450959, and SAMN171362219-SAMN17136304; BioProject nos. PRJNA687140).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Brumin, M.; Levy, M.; Ghanim, M. Transovarial transmission of Rickettsia spp. and organ-specific infection of the Whitefly Bemisia tabaci. Appl. Environ. Microbiol. 2012, 78, 5565–5574. [Google Scholar] [CrossRef]
- Hansen, A.; Moran, N. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2013, 23, 1473–1496. [Google Scholar] [CrossRef]
- Wilson, A.; Ashton, P.; Calevro, F.; Charles, H.; Colella, S.; Febvay, G.; Jander, G.; Kushlan, P.; Macdonald, S.; Coombs, J.; et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 2010, 19 (Suppl. 2), 249–258. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Moriyama, M.; Łukasik, P.; Vanderpool, D.; Tanahashi, M.; Meng, X.-Y.; McCutcheon, J.P.; Fukatsu, T. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. USA 2018, 115, E5970–E5979. [Google Scholar] [CrossRef]
- Husnik, F.; McCutcheon, J.P. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc. Natl. Acad. Sci. USA 2016, 113, E5416–E5424. [Google Scholar] [CrossRef] [PubMed]
- Sabree, Z.L.; Huang, C.Y.; Okusu, A.; Moran, N.A.; Normark, B.B. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ. Microbiol. 2013, 15, 1988–1999. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera Sp APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef]
- Baumann, P.; Baumann, L.; Lai, C.Y.; Rouhbakhsh, D.; Moran, N.A.; Clark, M.A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu. Rev. Microbiol. 1995, 49, 55–94. [Google Scholar] [CrossRef]
- Bustamante-Brito, R. Endosymbiotic microorganisms of scale insects. TIP Rev. Espec. Cienc. Químico-Biológicas 2018, 21, 51–69. [Google Scholar]
- Gill, R.J. The Scale Insects of California: The Soft Scales (Homoptera: Coccoidea: Coccidae); Analysis and Identification Branch, Division of Plant Industry: Sacramento, CA, USA, 1988. [Google Scholar]
- Mayrolin, G.M.; Denno, B.D.; Miller, D.R.; Miller, G.L.; Yair, B.D.; Hardy, N.B. ScaleNet: A literature-based model of scale insect biology and systematics. Database 2016, 2016, bav118. [Google Scholar]
- Hamon, A.B.; Williams, M.L. The Soft Scale Insects of Florida (Homoptera: Coccoidea: Coccidae); Arthropods of Florida and Neighboring Land Areas, Florida Department of Agriculture & Consumer Services Division Plant Industry: Gainesville, FL, USA, 1984; 194p. [Google Scholar]
- Rosenblueth, M.; Sayavedra, L.; Sámano-Sánchez, H.; Roth, A.; Martínez-Romero, E. Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J. Evol. Biol. 2012, 25, 2357. [Google Scholar] [CrossRef]
- Gomez-Polo, P.; Ballinger, M.J.; Lalzar, M.; Malik, A.; Chiel, E. An exceptional family: Ophiocordyceps -allied fungus dominates the microbiome of soft scale insects (Hemiptera: Sternorrhyncha: Coccidae). Mol. Ecol. 2017, 26, 5855–5868. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Roskov, Y.; Abucay, L.; Orrell, T.; Nicolson, D.; Bailly, N.; Kirk, P.; Bourgoin, T.; DeWalt, R.; Decock, W.; De Wever, A. Species 2000 & ITIS Catalogue of Life, 2017 Annual Checklist; Naturalis: Leiden, The Netherlands, 2018; ISSN 2405-884X. Available online: www.catalogueoflife.org/annual-checklist/2017 (accessed on 24 January 2021).
- Sanjuan, T.I.; Franco-Molano, A.E.; Kepler, R.M.; Spatafora, J.W.; Tabima, J.; Vasco-Palacios, A.M.; Restrepo, S. Five new species of entomopathogenic fungi from the Amazon and evolution of neotropical Ophiocordyceps. Fungal Biol. 2015, 119, 901–916. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.H.; Poinar, G.O.; Spatafora, J.W. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 2008, 49, 495–502. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.D.; Yang, R.H.; Hsiang, T.; Wang, K.; Liang, D.Q.; Liang, F.; Cao, D.M.; Zhou, F.; Wen, G.; et al. Complete mitochondrial genome of the medicinal fungus Ophiocordyceps sinensis. Sci. Rep. 2015, 5, 13892. [Google Scholar] [CrossRef] [PubMed]
- Evans, H.C.; Elliot, S.L.; Hughes, D.P.; Corrie, M. Hidden Diversity behind the Zombie-Ant Fungus Ophiocordyceps unilateralis: Four new species described from Carpenter Ants in Minas Gerais, Brazil. PLoS ONE 2011, 6, e17024. [Google Scholar] [CrossRef]
- Kobmoo, N.; Mongkolsamrit, S.; Tasanathai, K.; Thanakitpipattana, D.; Luangsa-Ard, J.J. Molecular phylogenies reveal host-specific divergence of Ophiocordyceps unilateralis sensu lato following its host ants. Mol. Ecol. 2012, 21, 3022–3031. [Google Scholar] [CrossRef]
- Chen, S.; Liu, X. Control of the soybean cyst nematode by the fungi Hirsutella rhossiliensis and Hirsutella minnesotensis in greenhouse studies. Biol. Control 2005, 32, 208–219. [Google Scholar] [CrossRef]
- Podsiadło, E.B.; Michalik, K.; Michalik, A.; Szklarzewicz, T. Yeast-like microorganisms in the scale insect Kermes quercus (Insecta, Hemiptera, Coccomorpha: Kermesidae). Newly acquired symbionts? Arthropod Struct. Dev. 2018, 47, 56–63. [Google Scholar] [CrossRef]
- Vashishtha, A.; Sharama, K.K.; Lakhanpaul, S. Co-Existence, Phylogeny and putative role of Wolbachia and Yeast-Like Symbiont (YLS) in Kerria lacca (Kerr). Curr. Microbiol. 2011, 63, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Tanahashi, M.; Lin, C.P.; Koga, R.; Fukatsu, T. Fungal and bacterial endosymbionts of eared leafhoppers of the subfamily Ledrinae (Hemiptera: Cicadellidae). Appl. Entomol. Zool. 2016, 51, 465–477. [Google Scholar] [CrossRef]
- Kobiałka, M.; Michalik, A.; Walczak, M.; Szklarzewicz, T. Dual “Bacterial-Fungal” symbiosis in Deltocephalinae Leafhoppers (Insecta, Hemiptera, Cicadomorpha: Cicadellidae). Microb. Ecol. 2018, 75, 771–782. [Google Scholar] [CrossRef]
- Haynes, S.; Darby, A.C.; Daniell, T.J.; Webster, G.; van Veen, F.J.F. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 2003, 69, 7216–7223. [Google Scholar] [CrossRef] [PubMed]
- Sepúlveda, D.A.; Zepeda-Paulo, F.; Ramírez, C.C.; Lavandero, B.; Figueroa, C.C. Diversity, frequency, and geographic distribution of facultative bacterial endosymbionts in introduced aphid pests. Insect Sci. 2017, 24, 511–521. [Google Scholar] [CrossRef]
- Baumann, L.; Baumann, P. Cospeciation Between the primary endosymbionts of Mealybugs and their hosts. Curr. Microbiol. 2005, 50, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Gruwell, M.E.; Hardy, N.B.; Gullan, P.J.; Dittmar, K. Evolutionary relationships among primary endosymbionts of the Mealybug subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl. Environ. Microbiol. 2010, 76, 7521–7525. [Google Scholar] [CrossRef]
- Ligi, T.; Oopkaup, K.; Truu, M.; Preem, J.K.; Nõlvak, H.; Mitsch, W.J.; Mander, ü.; Truu, J. Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol. Eng. 2014, 72, 56–66. [Google Scholar] [CrossRef]
- Vierheilig, J.; Savio, D.; Ley, R.E.; Mach, R.L.; Reischer, G.H. Potential applications of next generation DNA sequencing of 16S rRNA gene amplicons in microbial water quality monitoring. Water Sci. Technol. 2015, 72, 1962–1972. [Google Scholar] [CrossRef]
- Ivens, A.B.F.; Gadau, A.; Kiers, E.T.; Kronauer, D.J.C. Can social partnerships influence the microbiome? Insights from ant farmers and their trophobiont mutualists. Mol. Ecol. 2018, 27, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Deng, J.; Zhang, J.T.; Zhou, Q.S.; Zhang, Y.Z.; Wu, S.A. DNA barcoding of common soft scales (Hemiptera: Coccoidea: Coccidae) in China. Bull. Entomol. Res. 2015, 105, 545–554. [Google Scholar] [CrossRef]
- Tang, F.T. The Coccidae of China; Shanxi United University Press: Taiyuan, China, 1991; 377p. (In Chinese) [Google Scholar]
- Deng, J.; Yu, F.; Zhang, T.X.; Hu, H.Y.; Zhu, C.D.; Wu, S.A.; Zhang, Y.Z. DNA barcoding of six Ceroplastes species (Hemiptera: Coccoidea: Coccidae) from China. Mol. Ecol. Resour. 2012, 12, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.G.; Freedman, E.Z.; White, T.J.; Taylor, J.W. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J. Clin. Microbiol. 1994, 32, 253–255. [Google Scholar] [CrossRef]
- Bolger, A.M.; Marc, L.; Bjoern, U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 15, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5264–5267. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.; Bahram, M.; Bates, S.; Bruns, T.; Bengtsson-Palme, J.; Callaghan, T.; et al. Towards a unified paradigm for sequence-based identification of Fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Hhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.; von Haeseler, A.; Minh, B. IQ-TREE: A fast and effective Stochastic Algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; Haeseler, A.V. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef]
- Gibson, C.M.; Hunter, M.S. Extraordinarily widespread and fantastically complex: Comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 2010, 13, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.J.; Zchori-Fein, H.E. The emerging diversity of Rickettsia. Proc. Biol. Sci. 2006, 273, 2097–2106. [Google Scholar] [CrossRef]
- Koga, R.; Bennett, G.M.; Cryan, J.R.; Moran, N.A. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 2013, 15, 2073–2081. [Google Scholar] [CrossRef]
- Moran, N.A.; Bennett, G.M. The tiniest tiny genomes. Annu. Rev. Microbiol. 2014, 68, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Vogel, K.J.; Moran, N.A. Functional and evolutionary analysis of the genome of an obligate fungal symbiont. Genome Biol. Evol. 2013, 5, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.W.; Noda, H.; Xie, H.Q.; Suetsugu, Y.; Zhu, Q.H.; Zhang, C.X. Genomic analysis of an ascomycete fungus from the rice planthopper reveals how it adapts to an endosymbiotic lifestyle. Genome Biol. Evol. 2015, 7, 2623–2634. [Google Scholar] [CrossRef] [PubMed]
- Bendov, Y.; Hodgson, C.J. Soft Scale Insects: Their Biology, Natural Enemies and Control; World Crop Pests; Elsevier: Amsterdam, The Netherlands, 1997; Volume 7, p. 452. [Google Scholar]
- Ueda, S.; Quek, S.P.; Itioka, T.; Inamori, K.; Sato, Y.; Murase, K.; Itino, T. An ancient tripartite symbiosis of plants, ants and scale insects. Proc. Biol. Sci. 2008, 275, 2319–2326. [Google Scholar] [CrossRef]
- Nikoh, N.; Fukatsu, T. Interkingdom host jumping underground: Phylogenetic analysis of entomoparasitic fungi of the genus cordyceps. Mol. Biol. Evol. 2000, 17, 629–638. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).