Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Media, and Cell Growth
2.2. Generation of Gene Deletion Mutants in C. diphtheriae
2.3. Recombinant Plasmids
2.4. Protein Purification
2.5. RT-FeDEx Assay
2.6. RNA-Seq Analysis
2.7. Reverse Transcription Polymerase Chain Reactions
2.8. Detection of Diphtheria Toxin
2.9. Electron Microscopy
2.10. Chrome Azurol S (CAS) Assay
2.11. Caenorhabditis Elegans Killing Assay
2.12. Statistical Analysis
3. Results
3.1. C. Diphtheriae Encodes Ribonuclease J
3.2. Genetic Disruption of rnj Alters Cell Growth and Morphology
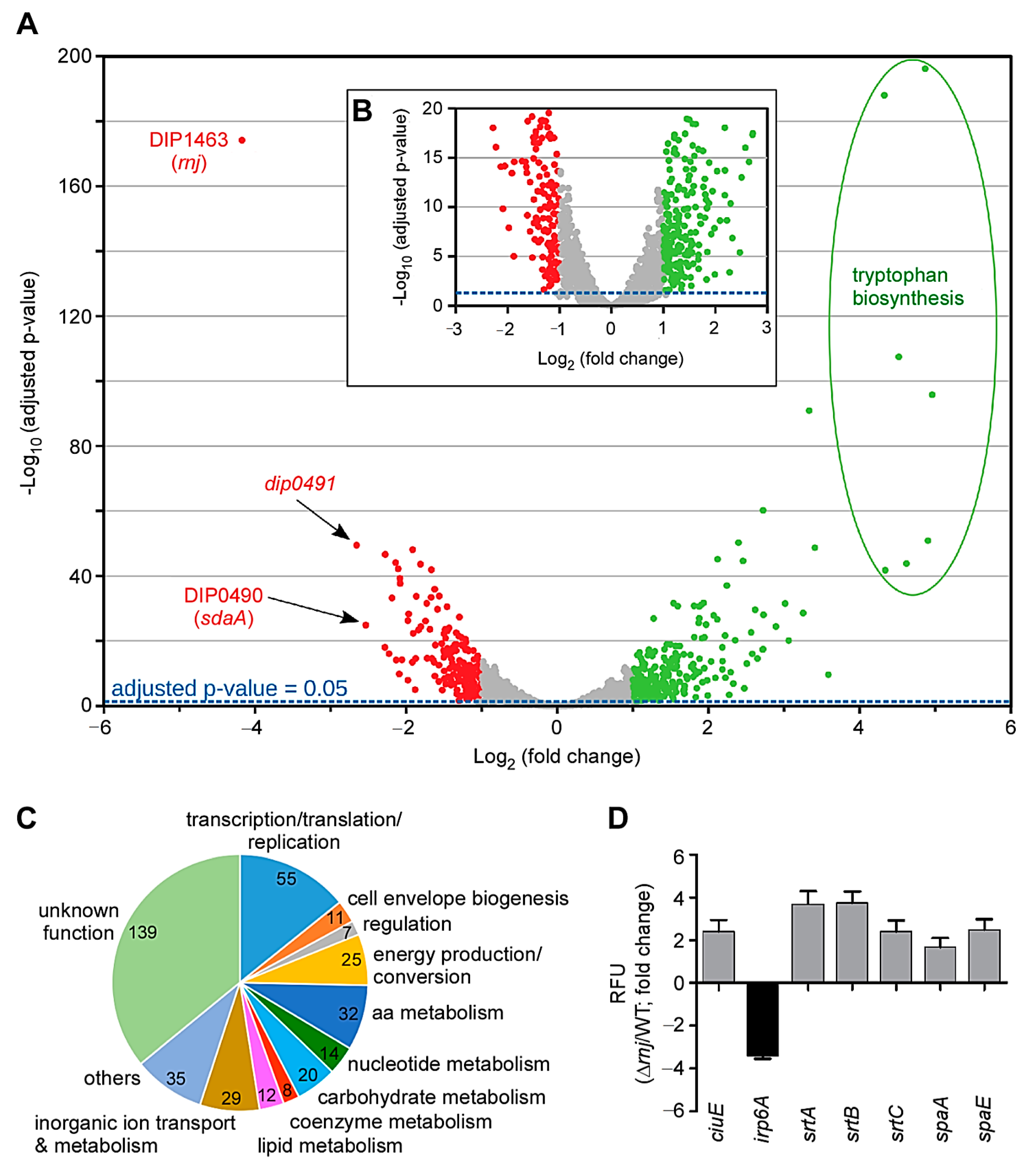
3.3. Transcriptome Analysis of the rnj Mutant
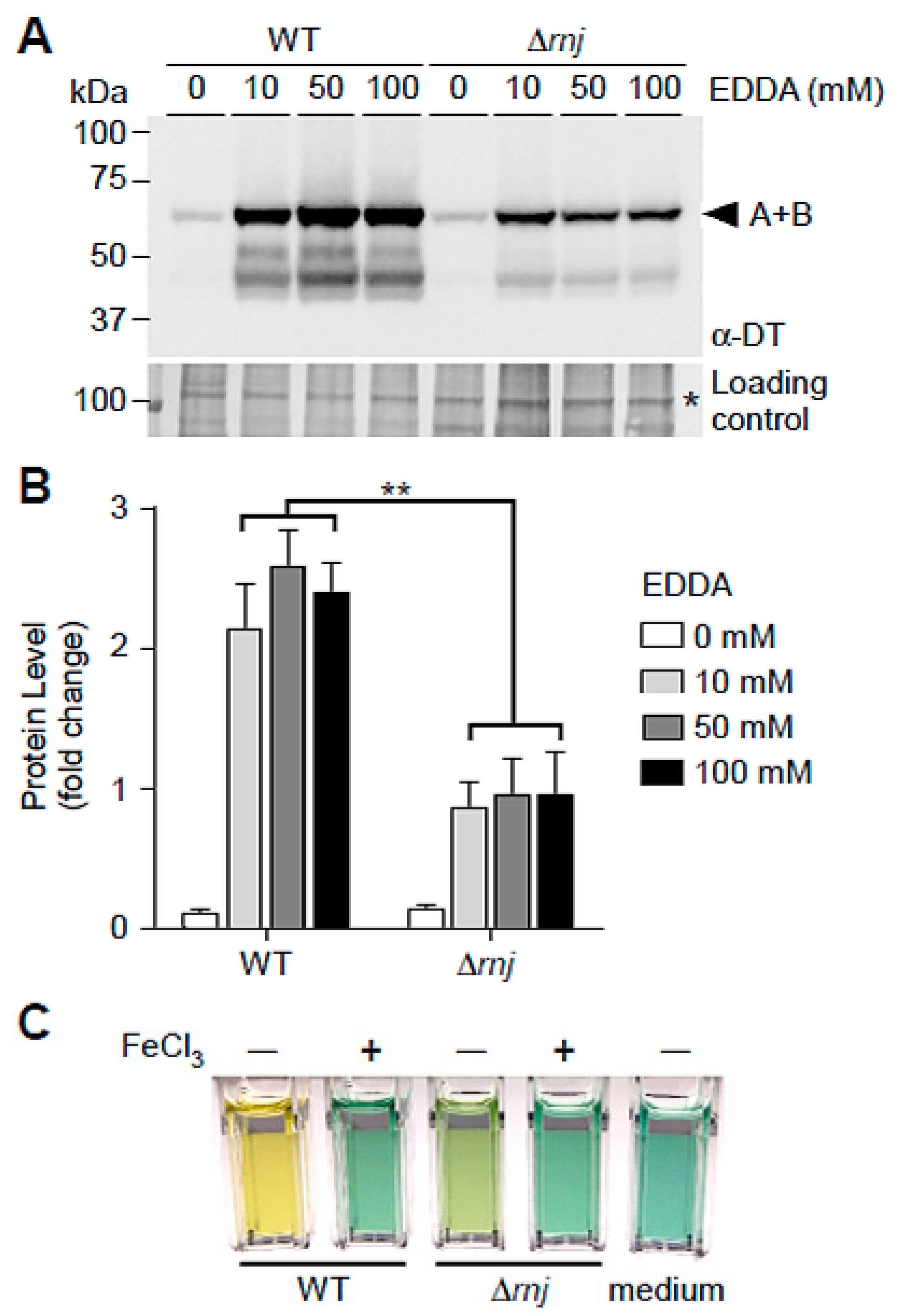
3.4. Reduction of Secreted Diphtheria Toxin in the ∆rnJ Mutant
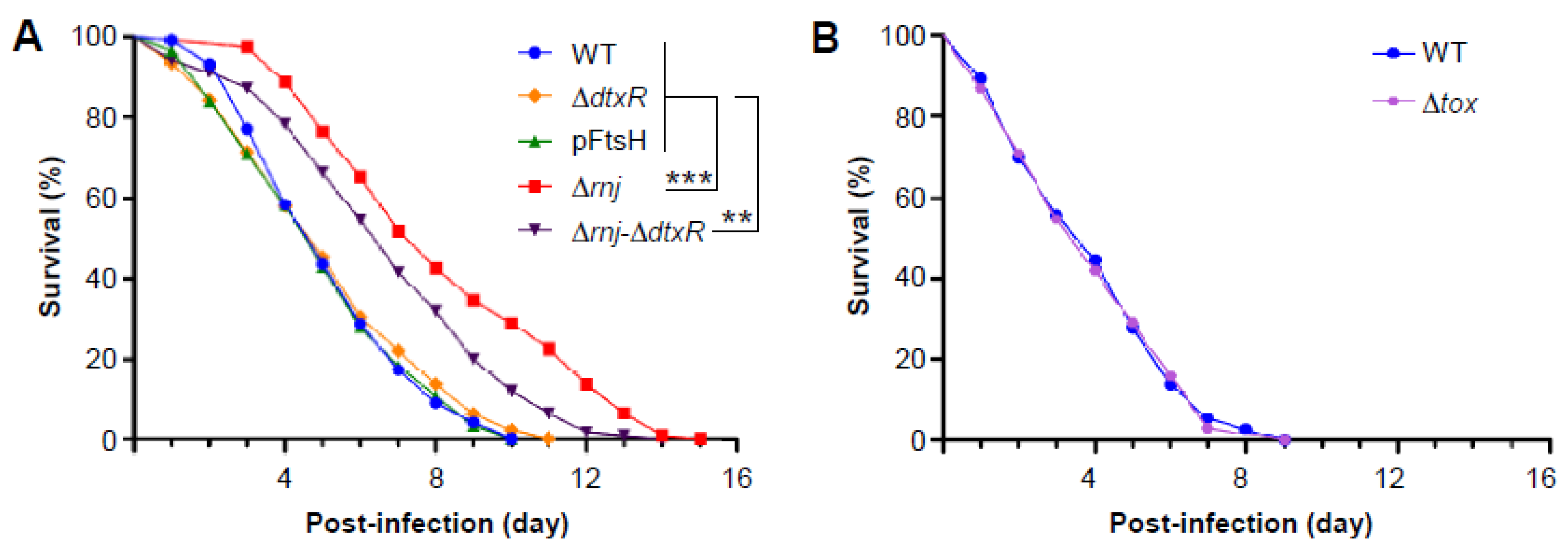
3.5. Virulence Attenuation of the ∆rnj Mutant in a Caenorhabditis Elegans Model of Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef]
- Hoe, C.-H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.-H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Repoila, F.; Majdalani, N.; Gottesman, S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: The RpoS paradigm. Mol. Microbiol. 2003, 48, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Bechhofer, D.H. Nucleotide specificity in bacterial mRNA recycling. Proc. Natl. Acad. Sci. USA 2013, 110, 8765–8766. [Google Scholar] [CrossRef] [PubMed]
- Hui, M.P.; Foley, P.L.; Belasco, J.G. Messenger RNA Degradation in Bacterial Cells. Annu. Rev. Genet. 2014, 48, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Apirion, D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics 1978, 90, 659–671. [Google Scholar]
- Grunberg-Manago, M. Messenger RNA Stability and Its Role in Control of Gene Expression in Bacteria and Phages. Annu. Rev. Genet. 1999, 33, 193–227. [Google Scholar] [CrossRef]
- Carpousis, A.J. The RNA Degradosome of Escherichia coli: An mRNA-Degrading Machine Assembled on RNase E. Annu. Rev. Microbiol. 2007, 61, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Condon, C.; Putzer, H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002, 30, 5339–5346. [Google Scholar] [CrossRef]
- Lehnik-Habrink, M.; Newman, J.; Rothe, F.M.; Solovyova, A.S.; Rodrigues, C.; Herzberg, C.; Commichau, F.M.; Lewis, R.J.; Stülke, J. RNase Y in Bacillus subtilis: A Natively Disordered Protein That is the Functional Equivalent of RNase E from Escherichia coli. J. Bacteriol. 2011, 193, 5431–5441. [Google Scholar] [CrossRef]
- Even, S. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005, 33, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Condon, C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010, 7, 316–321. [Google Scholar] [CrossRef]
- Mathy, N.; Hébert, A.; Mervelet, P.; Bénard, L.; Dorléans, A.; De La Sierra-Gallay, I.L.; Noirot, P.; Putzer, H.; Condon, C. Bacillus subtilisribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol. Microbiol. 2010, 75, 489–498. [Google Scholar] [CrossRef]
- Bugrysheva, J.V.; Scott, J.R. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol. Microbiol. 2009, 75, 731–743. [Google Scholar] [CrossRef]
- Gao, P.; Pinkston, K.L.; Nallapareddy, S.R.; Van Hoof, A.; Murray, B.E.; Harvey, B.R. Enterococcus faecalis rnjB is Required for Pilin Gene Expression and Biofilm Formation. J. Bacteriol. 2010, 192, 5489–5498. [Google Scholar] [CrossRef]
- Gao, P.; Pinkston, K.L.; Bourgogne, A.; Murray, B.E.; Van Hoof, A.; Harvey, B.R. Functional studies of E. faecalis RNase J2 and its role in virulence and fitness. PLoS ONE 2017, 12, e0175212. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Merritt, J.; Qi, F.; Khajotia, S.; Liu, N. RNases J1 and J2 are critical pleiotropic regulators in Streptococcus mutans. Microbiology 2015, 161, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.; Shinde, P.; Zou, Z.; Kreth, J.; Merritt, J. Examining the Protein Interactome and Subcellular Localization of RNase J2 Complexes in Streptococcus mutans. Front. Microbiol. 2019, 10, 2150. [Google Scholar] [CrossRef] [PubMed]
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2013, 71, 1799–1828. [Google Scholar] [CrossRef]
- Pei, X.-Y.; Bralley, P.; Jones, G.H.; Luisi, B.F. Linkage of catalysis and 5’ end recognition in ribonuclease RNase J. Nucleic Acids Res. 2015, 43, 8066–8076. [Google Scholar] [CrossRef]
- Jones, S.E.; Leong, V.; Ortega, J.; Elliot, M.A. Development, Antibiotic Production, and Ribosome Assembly in Streptomyces venezuelae Are Impacted by RNase J and RNase III Deletion. J. Bacteriol. 2014, 196, 4253–4267. [Google Scholar] [CrossRef]
- Bralley, P.; Aseem, M.; Jones, G.H. SCO5745, a Bifunctional RNase J Ortholog, Affects Antibiotic Production in Streptomyces coelicolor. J. Bacteriol. 2014, 196, 1197–1205. [Google Scholar] [CrossRef]
- Rogers, E.A.; Das, A.; Ton-That, H. Adhesion by Pathogenic Corynebacteria. Adv. Exp. Med. Biol. 2011, 715, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Kandel, J. Structure and activity of diphtheria toxin. I. Thiol-dependent dissociation of a fraction of toxin into enzymically active and inactive fragments. J. Biol. Chem. 1971, 246, 24. [Google Scholar]
- Uchida, T.; Gill, D.M.; Pappenheimer, A.M. Mutation in the Structural Gene for Diphtheria Toxin carried by Temperate Phage β. Nat. New Biol. 1971, 233, 8–11. [Google Scholar] [CrossRef]
- Boyd, J.; Oza, M.N.; Murphy, J.R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 1990, 87, 5968–5972. [Google Scholar] [CrossRef] [PubMed]
- Fourel, G.; Phalipon, A.; Kaczorek, M. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtoxR, in Corynebacterium diphtheriae. Infect. Immun. 1989, 57, 3221–3225. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.K. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 2000, 181, 156–167. [Google Scholar] [CrossRef]
- Kunkle, C.A.; Schmitt, M.P. Analysis of a DtxR-Regulated Iron Transport and Siderophore Biosynthesis Gene Cluster in Corynebacterium diphtheriae. J. Bacteriol. 2005, 187, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Trost, E.; Blom, J.; Soares, S.D.C.; Huang, I.-H.; Al-Dilaimi, A.; Schröder, J.; Jaenicke, S.; Dorella, F.A.; Rocha, F.S.; Miyoshi, A.; et al. Pangenomic Study of Corynebacterium diphtheriae That Provides Insights into the Genomic Diversity of Pathogenic Isolates from Cases of Classical Diphtheria, Endocarditis, and Pneumonia. J. Bacteriol. 2012, 194, 3199–3215. [Google Scholar] [CrossRef]
- Ton-That, H.; Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003, 50, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Swierczynski, A.; Ton-That, H. Type III Pilus of Corynebacteria: Pilus Length is Determined by the Level of Its Major Pilin Subunit. J. Bacteriol. 2006, 188, 6318–6325. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.H.; Ton-That, H. Assembly of Distinct Pilus Structures on the Surface of Corynebacterium diphtheriae. J. Bacteriol. 2006, 188, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Reardon-Robinson, M.E.; Osipiuk, J.; Jooya, N.; Chang, C.; Joachimiak, A.; Das, A.; Ton-That, H. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriaeis essential for viability, pilus assembly, toxin production and virulence. Mol. Microbiol. 2015, 98, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Broadway, M.M.; Rogers, E.A.; Chang, C.; Huang, I.-H.; Dwivedi, P.; Yildirim, S.; Schmitt, M.P.; Das, A.; Ton-That, H. Pilus Gene Pool Variation and the Virulence of Corynebacterium diphtheriae Clinical Isolates during Infection of a Nematode. J. Bacteriol. 2013, 195, 3774–3783. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Nguyen, M.T.; Ton-That, H. Genetic Manipulation of Corynebacterium diphtheriae and Other Corynebacterium Species. Curr. Protoc. Microbiol. 2020, 58, 111. [Google Scholar] [CrossRef]
- Wittchen, M.; Busche, T.; Gaspar, A.H.; Lee, J.H.; Ton-That, H.; Kalinowski, J.; Tauch, A. Transcriptome sequencing of the human pathogen Corynebacterium diphtheriae NCTC 13129 provides detailed insights into its transcriptional landscape and into DtxR-mediated transcriptional regulation. BMC Genom. 2018, 19, 82. [Google Scholar] [CrossRef]
- Reyes, O.; Guyonvarch, A.; Bonainy, C.; Salti, V.; David, F.; LeBlon, G. ‘Integron’-bearing vectors: A method suitable for stable chromosomal integration in highly restrictive Corynebacteria. Gene 1991, 107, 61–68. [Google Scholar] [CrossRef]
- Luong, T.T.; Reardon-Robinson, M.E.; Siegel, S.D.; Ton-That, H. Reoxidation of the Thiol-Disulfide Oxidoreductase MdbA by a Bacterial Vitamin K Epoxide Reductase in the Biofilm-Forming Actinobacterium Actinomyces oris. J. Bacteriol. 2017, 199, e00817-16. [Google Scholar] [CrossRef]
- Siegel, S.D.; Amer, B.R.; Wu, C.; Sawaya, M.R.; Gosschalk, J.E.; Clubb, R.T.; Ton-That, H.; Otto, M.; Wu, H. Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein. mBio 2019, 10, e01580-18. [Google Scholar] [CrossRef]
- Sinturel, F.; Pellegrini, O.; Xiang, S.; Tong, L.; Condon, C.; Bénard, L. Real-time fluorescence detection of exoribonucleases. RNA 2009, 15, 2057–2062. [Google Scholar] [CrossRef]
- Louden, B.C.; Lynne, A.M.; Haarmann, D. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Chen, Y.; Ton-That, H. Corynebacterium diphtheriae Virulence Analyses Using a Caenorhabditis elegans Model. Curr. Protoc. Microbiol. 2020, 58, e109. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Sifri, C.D.; Mylonakis, E.; Qin, X.; Singh, K.V.; Murray, B.E.; Calderwood, S.B.; Ausubel, F.M. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 2001, 98, 10892–10897. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; De Almeida, D.F. Isolation and characterization of a new temperature-sensitive cell division mutant of Escherichia coli K-12. J. Bacteriol. 1975, 124, 1502–1507. [Google Scholar] [CrossRef]
- Zellmeier, S.; Zuber, U.; Schumann, W.; Wiegert, T. The absence of FtsH metalloprotease activity causes overexpression of the sigmaW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis. J. Bacteriol. 2003, 185, 973–982. [Google Scholar] [CrossRef]
- Tao, X.; Schiering, N.; Zeng, H.-Y.; Ringe, D.; Murphy, J.R. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 1994, 14, 191–197. [Google Scholar] [CrossRef]
- Zajdowicz, S.; Haller, J.C.; Krafft, A.E.; Hunsucker, S.W.; Mant, C.T.; Duncan, M.W.; Hodges, R.S.; Jones, D.N.M.; Holmes, R.K. Purification and Structural Characterization of Siderophore (Corynebactin) from Corynebacterium diphtheriae. PLoS ONE 2012, 7, e34591. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.M.; Cryz, S.J.; Holmes, R.K. Genetic and biochemical evidence for a siderophore-dependent iron transport system in Corynebacterium diphtheriae. Infect. Immun. 1984, 45, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.E.; Burgos, J.M.; Schmitt, M.P. Analysis of Novel Iron-Regulated, Surface-Anchored Hemin-Binding Proteins in Corynebacterium diphtheriae. J. Bacteriol. 2013, 195, 2852–2863. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lu, M.; Zhang, H.; Hu, J.; Zhou, C.; Xu, Q.; Shah, A.M.U.H.; Xu, H.; Wang, L.; Hua, Y. Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucleic Acids Res. 2015, 43, 5550–5559. [Google Scholar] [CrossRef] [PubMed]
- Deikus, G.; Condon, C.; Bechhofer, D.H. Role of Bacillus subtilis RNase J1 Endonuclease and 5′-Exonuclease Activities in trp Leader RNA Turnover. J. Biol. Chem. 2008, 283, 17158–17167. [Google Scholar] [CrossRef] [PubMed]
- Langklotz, S.; Baumann, U.; Narberhaus, F. Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta Bioenerg. 2012, 1823, 40–48. [Google Scholar] [CrossRef]
- Yanofsky, C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 2007, 13, 1141–1154. [Google Scholar] [CrossRef]
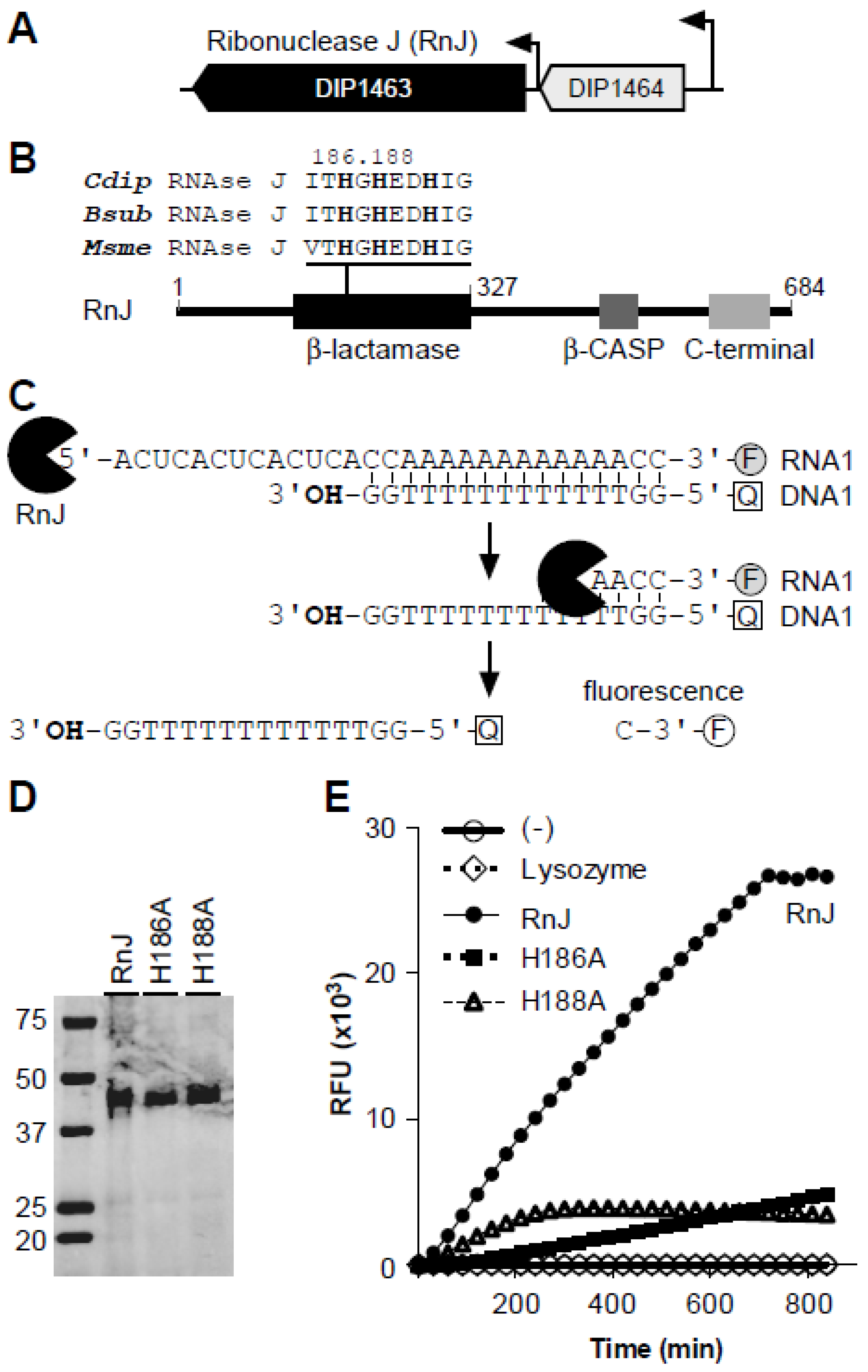
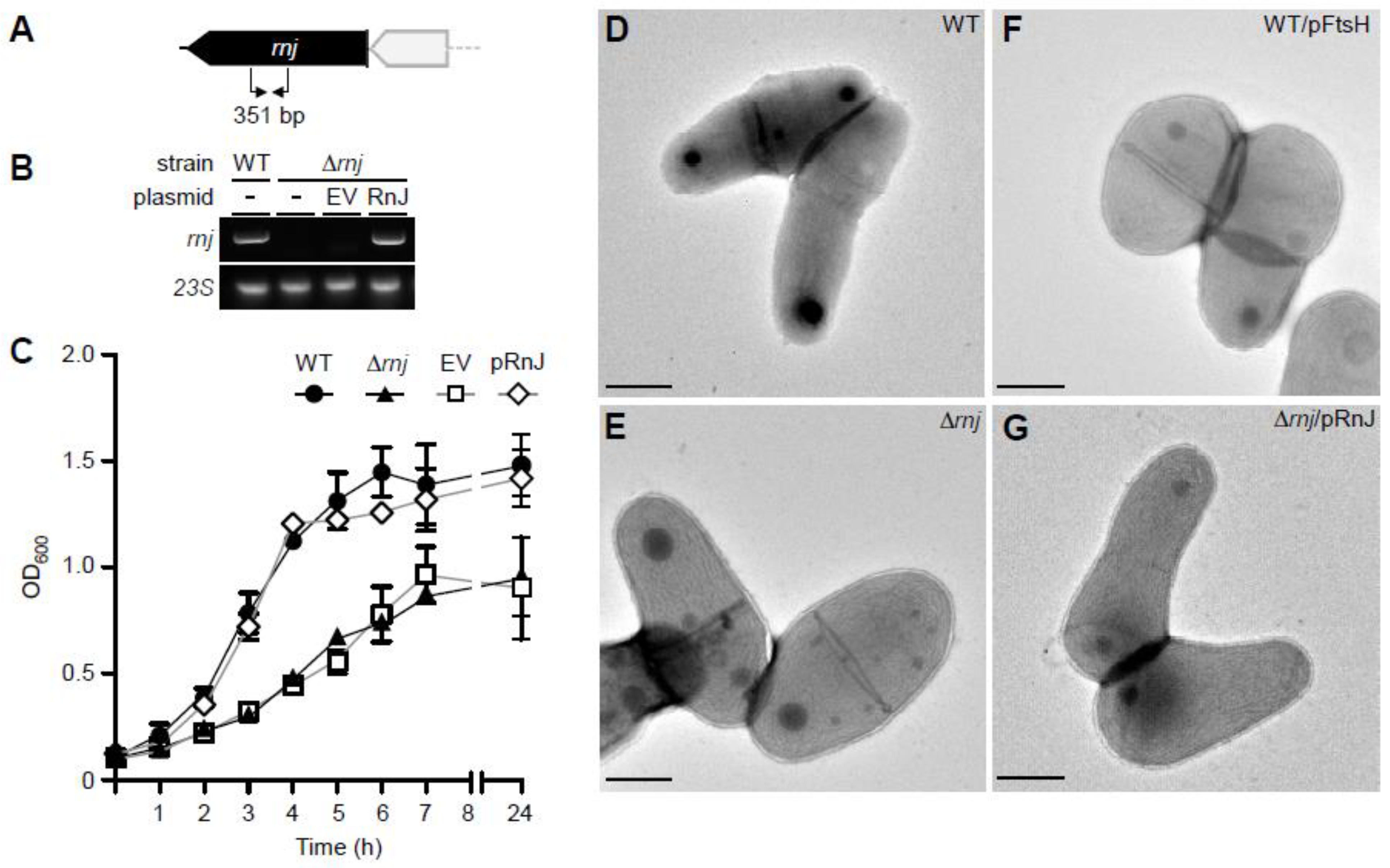
| Strains | Width (µm) | Length (µm) |
|---|---|---|
| WT | 0.64 ± 0.06 | 1.26 ± 0.40 |
| ∆rnj | 0.99 ± 0.13 | 1.39 ± 0.36 |
| WT/pFtsH | 0.90 ± 0.07 | 1.32 ± 0.33 |
| Gene Name | Function | Log2-Fold Change |
|---|---|---|
| iutD | Putative ABC-type iron protein | 1.03 |
| ciuC | Iron transport system membrane protein | 1.18 |
| ciuE | Corynebactin biosynthetic gene | 1.24 |
| sufB | Fe–S cluster assembly protein | 1.33 |
| sufR | Iron–sulfur cluster biosynthesis transcriptional regulator | 1.56 |
| piuB | Iron-uptake factor | 1.83 |
| irp4 | DtxR-dependent, iron-regulated promoter/operator | 2.65 |
| irp6A | Ferrisiderophore receptor (putative ABC transporter) | −1.30 |
| irp5 | DtxR-dependent, iron-regulated promoter/operator | −1.33 |
| Strains | +Fe3+ | −Fe3+ | ||
|---|---|---|---|---|
| OD600 | Siderophore Production (%) | OD600 | Siderophore Production (%) | |
| WT | 3.38 ± 0.08 | 19.30 ± 2.99 | 2.62 ± 0.29 | 67.77 ± 2.90 |
| ∆rnj | 2.92 ± 0.31 | 20.42 ± 1.97 | 2.38 ± 0.23 | 44.49 ± 3.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luong, T.T.; Nguyen, M.T.; Chen, Y.-W.; Chang, C.; Lee, J.H.; Wittchen, M.; Ton-That, H.; Cruz, M.; Garsin, D.A.; Das, A.; et al. Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms 2021, 9, 389. https://doi.org/10.3390/microorganisms9020389
Luong TT, Nguyen MT, Chen Y-W, Chang C, Lee JH, Wittchen M, Ton-That H, Cruz M, Garsin DA, Das A, et al. Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms. 2021; 9(2):389. https://doi.org/10.3390/microorganisms9020389
Chicago/Turabian StyleLuong, Truc Thanh, Minh Tan Nguyen, Yi-Wei Chen, Chungyu Chang, Ju Huck Lee, Manuel Wittchen, HyLam Ton-That, Melissa Cruz, Danielle A. Garsin, Asis Das, and et al. 2021. "Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae" Microorganisms 9, no. 2: 389. https://doi.org/10.3390/microorganisms9020389
APA StyleLuong, T. T., Nguyen, M. T., Chen, Y.-W., Chang, C., Lee, J. H., Wittchen, M., Ton-That, H., Cruz, M., Garsin, D. A., Das, A., Tauch, A., & Ton-That, H. (2021). Ribonuclease J-Mediated mRNA Turnover Modulates Cell Shape, Metabolism and Virulence in Corynebacterium diphtheriae. Microorganisms, 9(2), 389. https://doi.org/10.3390/microorganisms9020389






