Intracellular Bacteria in Plants: Elucidation of Abundant and Diverse Cytoplasmic Bacteria in Healthy Plant Cells Using In Vitro Cell and Callus Cultures
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Tissue Cultures
2.2. Cultivation Based Testing of Plant Tissue Cultures for Bacterial Association
2.3. Protoplast Preparation from Cell Cultures
2.4. Bright-Field Microscopy on Cell, Callus, and Protoplasts
2.5. Epi-Fluorescence Microscopy
2.6. Confocal Laser Scanning Microscopy (CSLM)
2.7. Electron Microscopy
2.8. Distinguishing Intracellular Bacteria from Plant Micro-Organelles
2.9. Fluorescent In Situ Hybridization (FISH)
2.10. 16S rRNA Gene V3 Taxonomic Profiling on Grape Cell Culture
2.11. 16S rRNA Gene V3–V4 Taxonomic Profiling on Grape Callus Tissue
2.12. Bacterial Activation and Identification
2.13. Functional Profiling of Bacterial Communities in Grape Callus
2.14. Microscopic Observations on Field Plant Tissues
2.15. Accession Numbers
3. Results
3.1. Callus and Cell Cultures
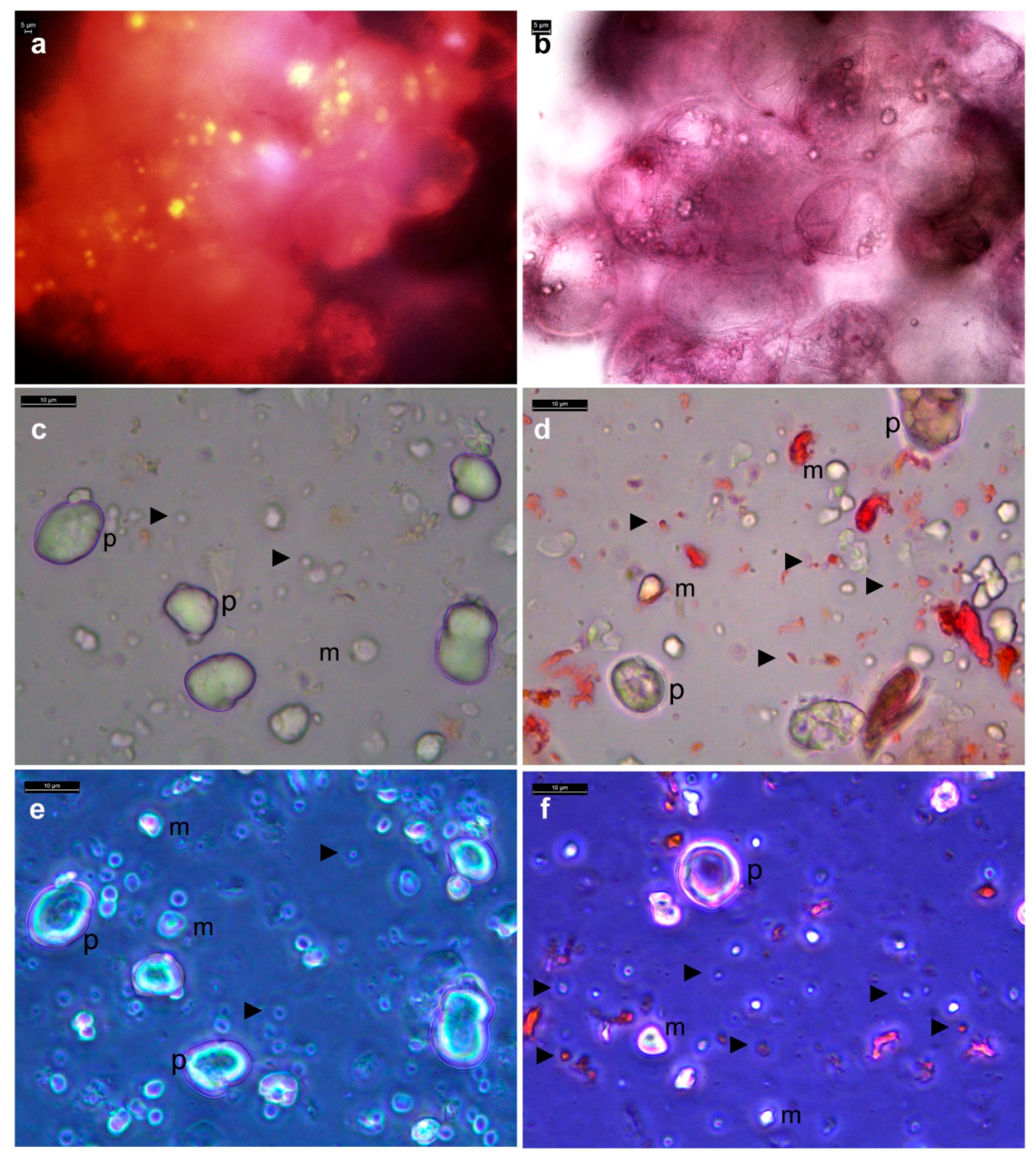
3.2. Live Imaging of Grape Cells under Bright-Field Microscopy
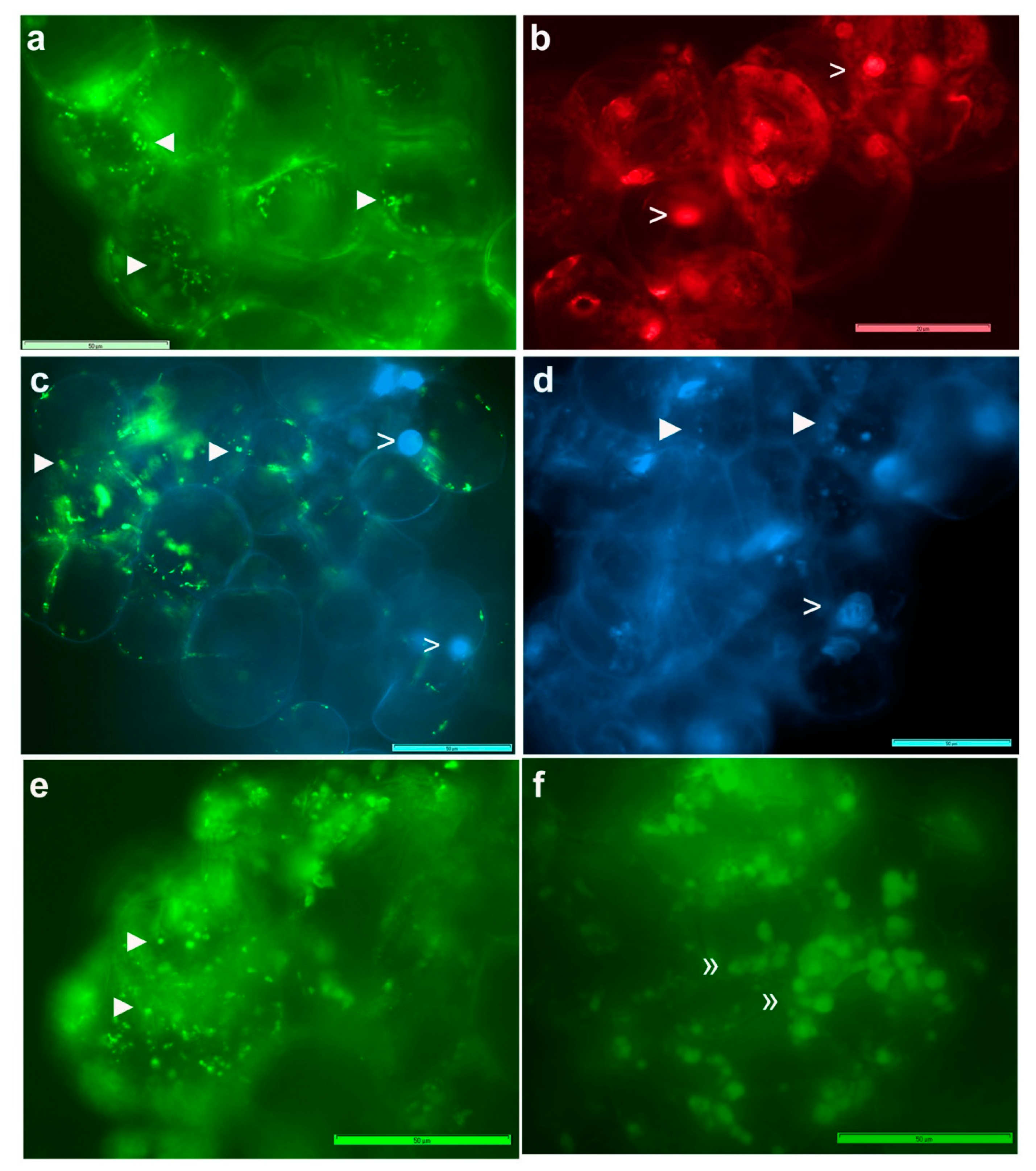
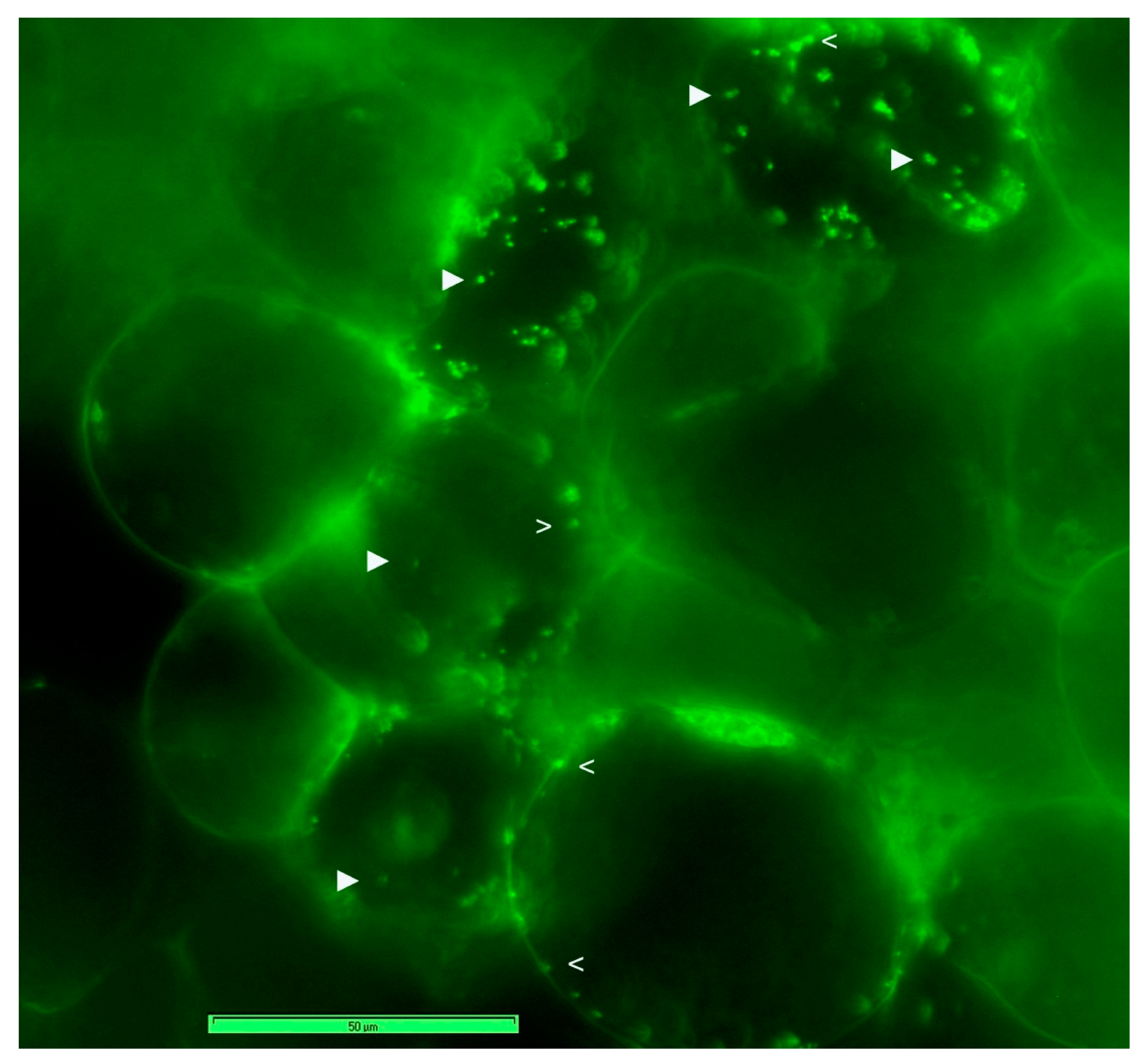
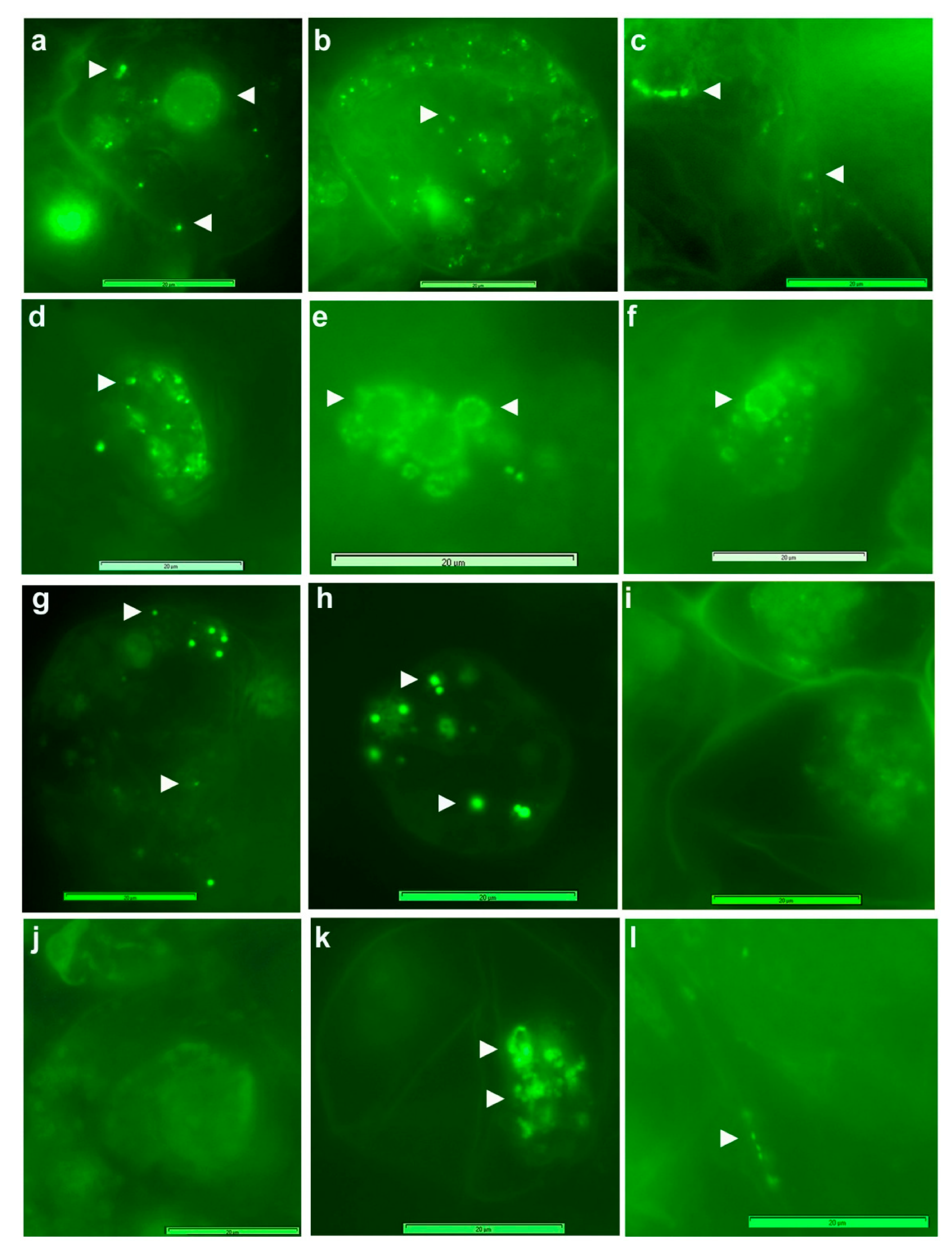
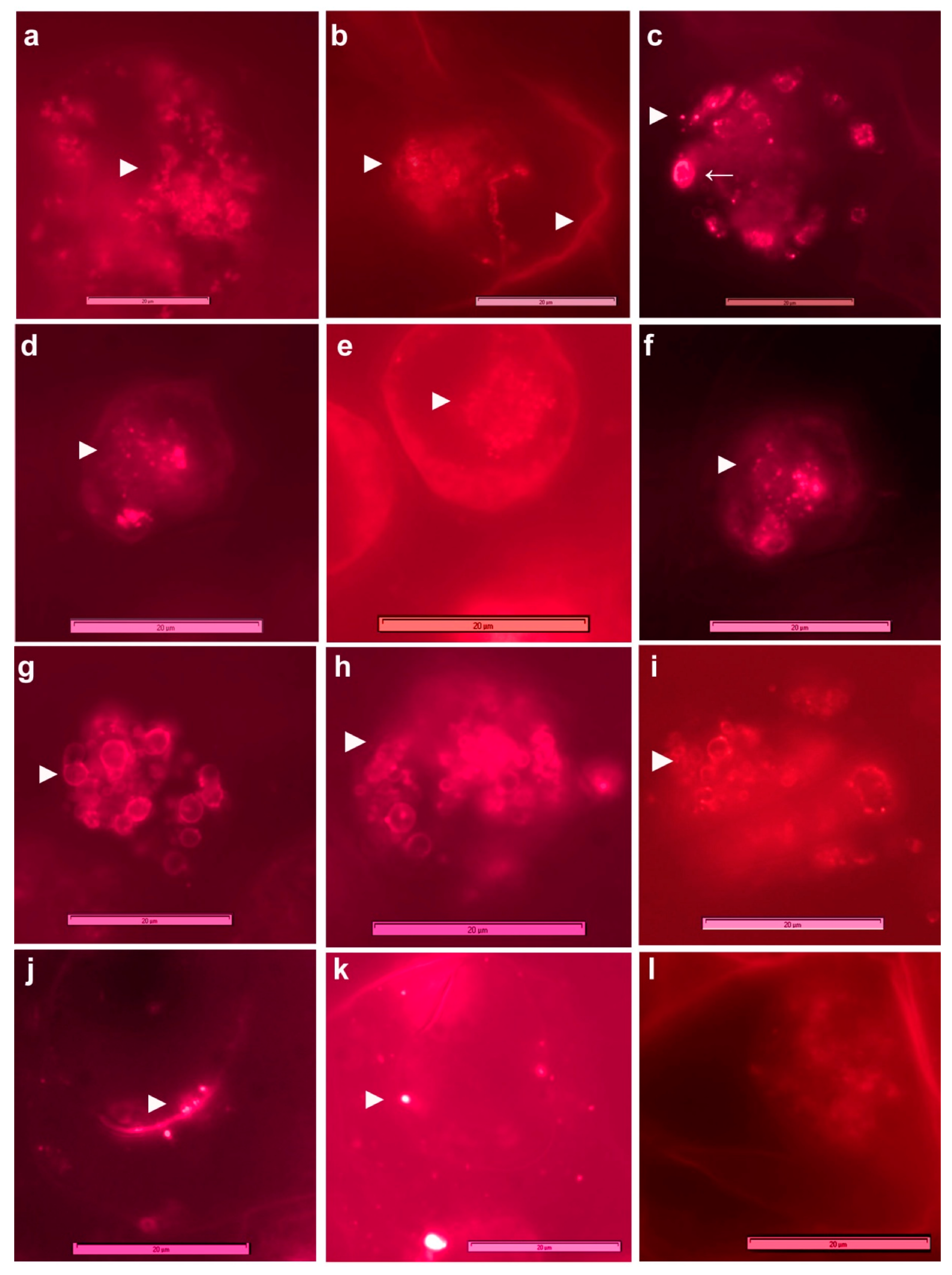
3.3. Live imaging of Grape Cells under Epi-Fluorescence Microscopy
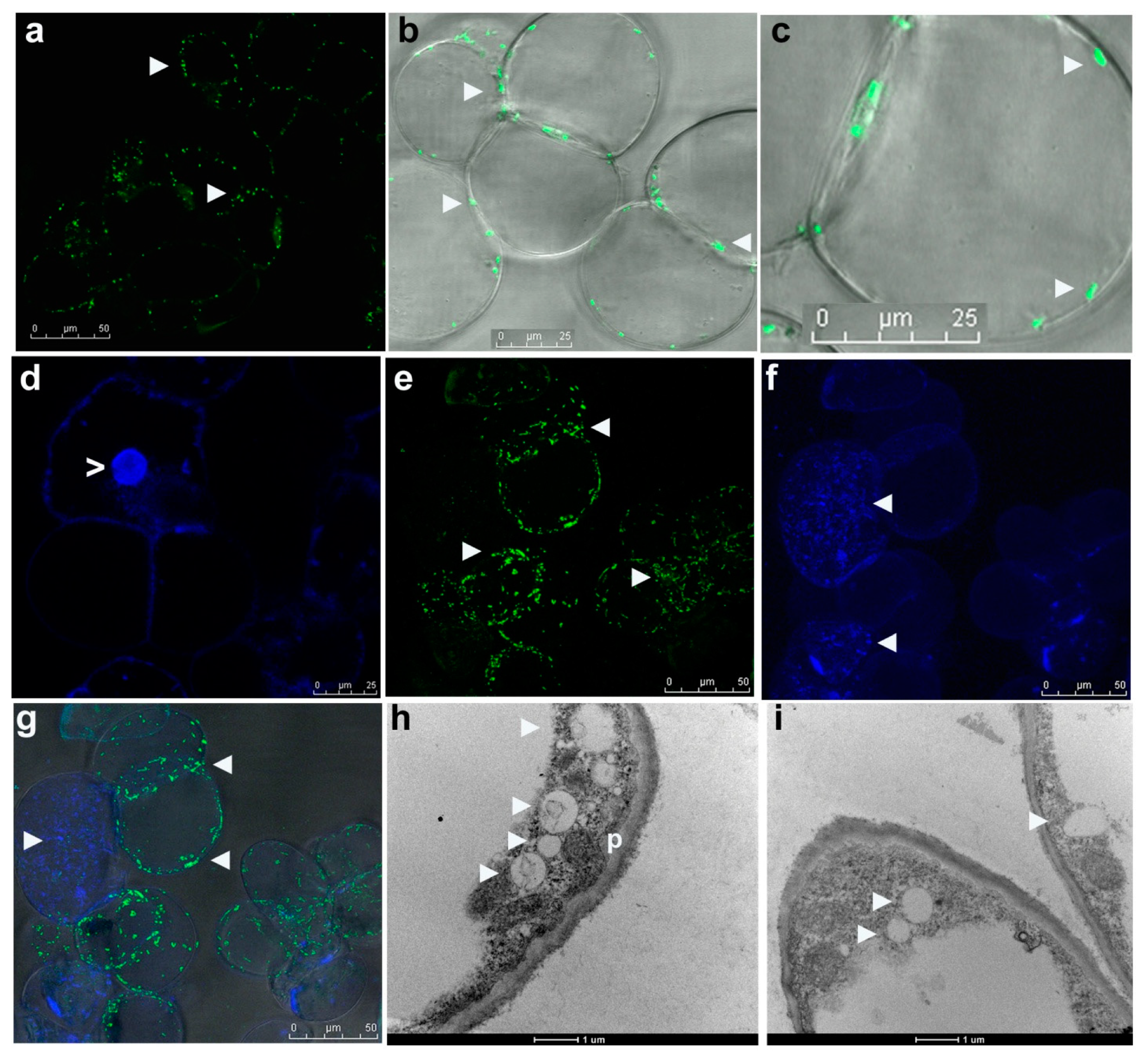
3.4. Live Imaging of Grape Cells through CLSM
3.5. Electron Microscopy
3.6. Distinguishing Intracellular Bacteria and Plant Cell Organelles
3.7. FISH on Grape Cell Cultures Using Eubacterial Probes
3.8. Observations on Callus/Protoplast from Other Sources and Plant Species
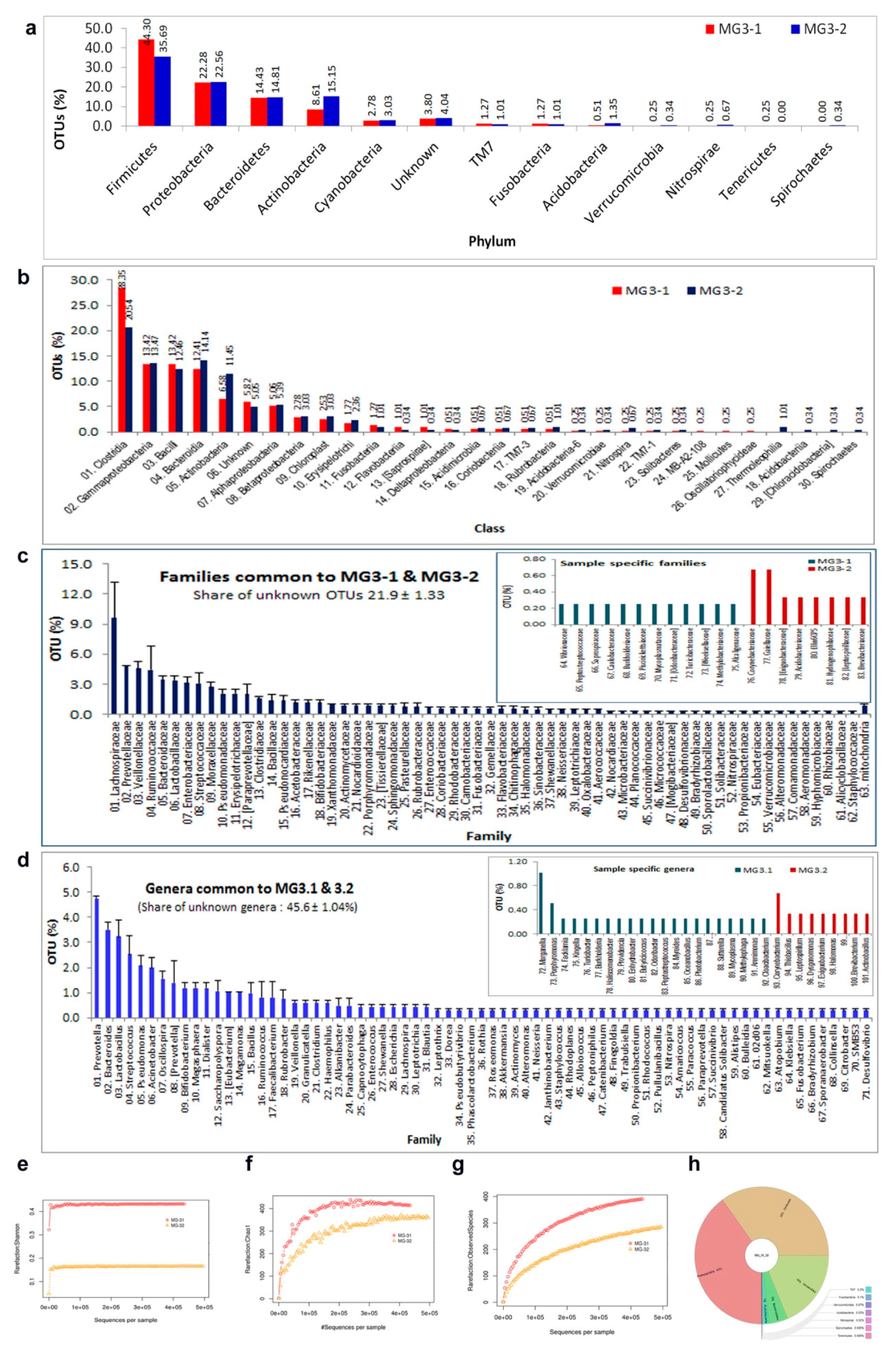
3.9. 16S rRNA Gene V3 Amplicon Profiling of Grape Cell Culture
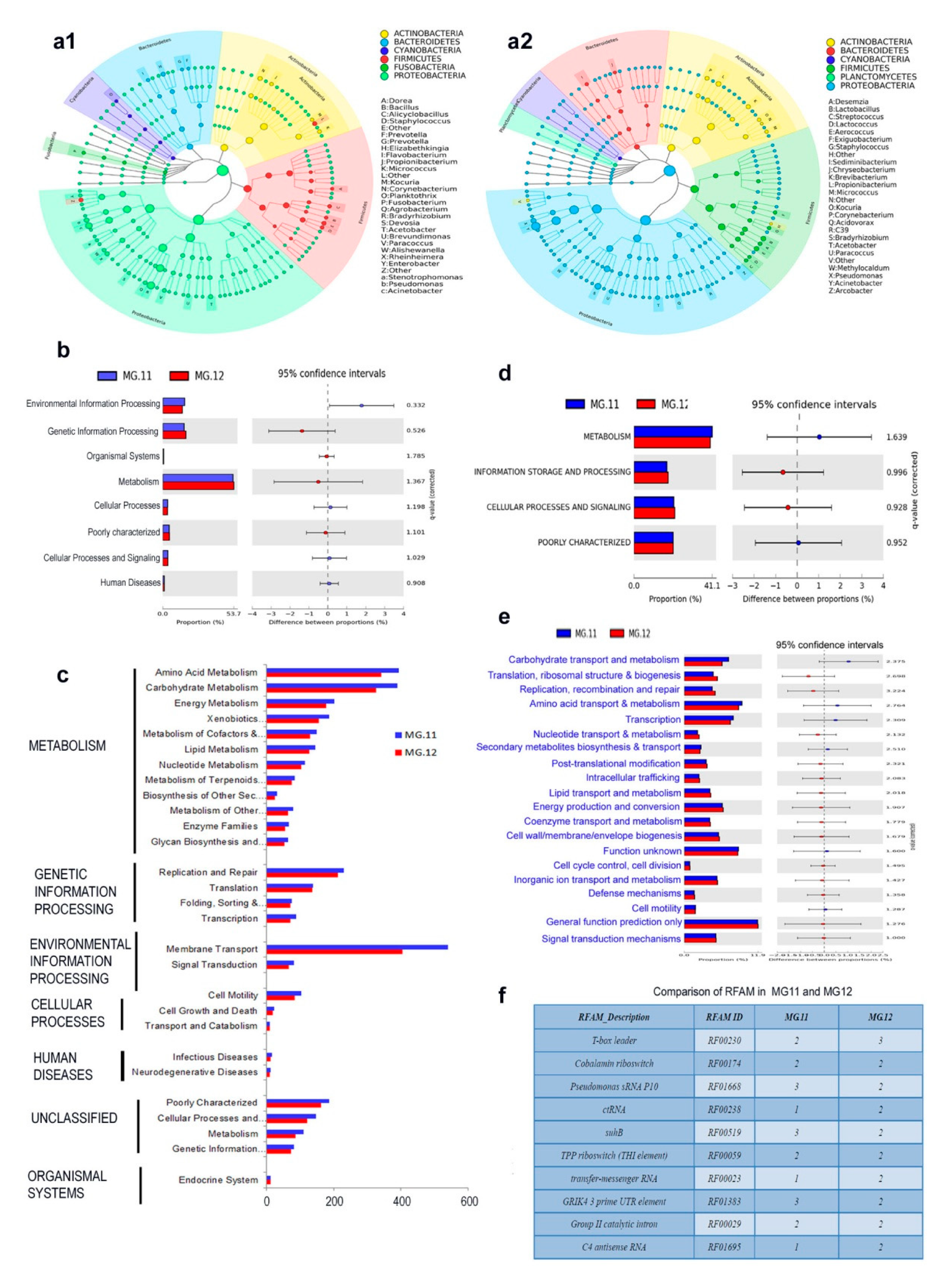
3.10. 16S rRNA Gene V3–V4 Profiling on Grape Callus
3.11. Activation of Uncultured Bacteria from “FU01” Cells to Cultivation
3.12. PICRUSt Functional Analysis on 16S rRNA Gene V3–V4 Data
3.13. Microscopic Observations on Field Plant Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLSM | confocal laser scanning microscopy |
| FDW | filter sterilized distilled water post-autoclaving |
| NA | nutrient agar |
| TEM | transmission electron microscopy |
| HTH | host tissue homogenate |
| TSA | tryptone soy agar |
References
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of grapevine flowers, berries and seeds: Identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef]
- Conn, V.M.; Franco, C.M.M. Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl. Environ. Microbiol. 2004, 70, 1787–1794. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microb. Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef]
- Dos-Santos, C.M.; de Souza, D.G.; Balsanelli, E.; Cruz, L.M.; de Souza, E.M.; Baldani, J.I.; Schwab, S. A culture-independent approach to enrich endophytic bacterial cells from sugarcane stems for community characterization. Microb. Ecol. 2017, 74, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Sekhar, A.C. Cultivation versus molecular analysis of banana (Musa sp.) shoot-tip tissue reveals enormous diversity of normally uncultivable endophytic bacteria. Microb. Ecol. 2017, 73, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.C.; Saldierna Guzmán, J.P.; Shay, J.E. Transmission of bacterial endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Shaik, S.P.; Thomas, P. In vitro activation of seed-transmitted cultivation- recalcitrant endophytic bacteria in tomato and host-endophyte mutualism. Microorganisms 2019, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Butterworth, S.; Brindisi, L.; Gatei, J.W.; Elmore, M.T.; Verma, S.K.; Yao, X.; Kowalski, K.P. Seed-vectored microbes: Their roles in improving seedling fitness and competitor plant suppression. In Seed Endophytes; Verma, S.K., White, J.F., Jr., Eds.; Springer: Cham, Switzerland, 2019; pp. 3–20. [Google Scholar]
- Schaffer, W.I. Terminology associated with cell, tissue and organ culture, molecular biology and molecular genetics. Vitro Cell. Dev. Biol. Plant. 1990, 26, 97–101. [Google Scholar] [CrossRef]
- Smith, R.H. Plant Tissue Culture. Techniques and Experiments; Elsevier Inc.: London, UK, 2013. [Google Scholar]
- Thomas, P. A three-step screening procedure for detection of covert and endophytic bacteria in plant tissue cultures. Curr. Sci. 2004, 87, 67–72. [Google Scholar]
- Orlikowska, T.; Nowak, K.; Reed, B. Bacteria in the plant tissue culture environment. Plant Cell Tiss. Org. Cult. 2017, 128, 487–508. [Google Scholar] [CrossRef]
- Thomas, P.; Swarna, G.K.; Patil, P.; Rawal, R.D. Ubiquitous presence of normally non-culturable endophytic bacteria in field shoot-tips of banana and their gradual activation to quiescent cultivable form in tissue cultures. Plant Cell Tiss. Org. Cult. 2008, 93, 39–54. [Google Scholar] [CrossRef]
- Thomas, P.; Sekhar, A.C.; Pasha, S.S. High taxonomic diversity of cultivation-recalcitrant endophytic bacteria in grapevine field shoots, their in vitro introduction and unsuspected persistence. Planta 2017, 246, 879–898. [Google Scholar] [CrossRef]
- Pirttilä, A.M.; Laukkanen, H.; Pospiech, H.; Myllyla, R.; Hohtola, A. Detection of intracellular bacteria in the buds of scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl. Environ. Microbiol. 2000, 66, 3073–3077. [Google Scholar] [CrossRef] [PubMed]
- Esposito-Polesi, N.P.; de Abreu-Tarazi, M.F.; de Almeida, C.V.; Tsai, S.M.; de Almeida, M. Investigation of endophytic bacterial community in supposedly axenic cultures of pineapple and orchids with evidence on abundant intracellular bacteria. Curr. Microbiol. 2017, 74, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Sekhar, A.C. Live cell imaging reveals extensive intracellular cytoplasmic colonization of banana genotypes by normally non-cultivable endophytic bacteria. AoB Plants 2014, 6, plu002. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Agrawal, M.; Bharathkumar, C.B. Diverse cellular colonizing endophytic bacteria in field shoots and in vitro cultured papaya with physiological and functional implications. Physiol. Plant. 2019, 166, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Quambusch, M.; Winkelmann, T. Bacterial endophytes in plant tissue culture: Mode of action, detection, and control. In Plant Cell Culture Protocols; Loyola-Vargas, V.M., Ochoa-Alejo, N., Eds.; Humana Press: New York, NY, USA, 2018; pp. 69–88. [Google Scholar]
- Hambeck, M.; Senula, A.; Kodym, A. Occurrence of latent bacteria during cryopreservation of long-term in vitro cultures of coltsfoot, Tussilago Farfara. Cryoletters 2019, 40, 333–340. [Google Scholar]
- Wilson, S.A.; Roberts, S.C. Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotech. J. 2012, 10, 249–268. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.; Jang, M.O.; Jin, Y.W.; Lee, E.K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149. [Google Scholar] [CrossRef]
- Pham, N.T.; Meier-Dinkel, A.; Höltken, A.M.; Quambusch, M.; Mahnkopp, F.; Winkelmann, T. Endophytic bacterial communities in in vitro shoot cultures derived from embryonic tissue of hybrid walnut (Juglans × intermedia). Plant Cell Tiss. Org. Cult. 2017, 130, 153–165. [Google Scholar] [CrossRef]
- Conn, S.; Franco, C.; Zhang, W. Characterization of anthocyanic vacuolar inclusions in Vitis vinifera L. cell suspension cultures. Planta 2010, 231, 1343–1360. [Google Scholar] [CrossRef]
- Vuong, T.V.; Franco, C.; Zhang, W. Treatment strategies for high resveratrol induction in Vitis vinifera L. cell suspension culture. Biotech. Rep. 2014, 1, 15–21. [Google Scholar] [CrossRef]
- Cormier, F.; Crevier, H.A.; Do, C.B. Effects of sucrose concentration on the accumulation of anthocyanins in grape (Vitis vinifera) cell suspension. Can. J. Bot. 1990, 68, 1822–1825. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Thomas, P.; Reddy, M.K. Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall–plasma membrane peri-space in the shoot-tip tissue of banana. AoB Plants 2013, 5, plt011. [Google Scholar] [CrossRef]
- Jedd, G.; Chua, N.H. Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 2002, 43, 384–392. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mathur, J.; Mathur, N.; Hulskamp, M. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule based peroxisome motility in plants. Plant Physiol. 2002, 128, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Stahl, D.A.; Amann, R. Development and application of nucleic acid probes. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons Ltd.: Chichester, UK, 1991; pp. 205–248. [Google Scholar]
- Daims, H.; Brühl, A.; Amann, R.; Schleier, K.H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef]
- Meier, H.; Amann, R.; Ludwig, W.; Schleifer, K.H. Specific oligo-nucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 1999, 22, 186–196. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Thomas, P.; Soly, T.A. Endophytic bacteria associated with growing shoot tips of banana (Musa sp.) cv. Grand Naine and the affinity of endophytes to the host. Microb. Ecol. 2009, 58, 952–964. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Koskimäki, J.J.; Pirttilä, A.M.; Ihantola, E.L.; Halonen, O.; Frank, A.C. The intracellular scots pine shoot symbiont Methylobacterium extorquens DSM13060 aggregates around the host nucleus and encodes eukaryote-like proteins. MBio 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Prieto, P.; Schilirò, E.; Maldonado-González, M.M.; Valderrama, R.; Barroso-Albarracín, J.B.; Mercado-Blanco, J. Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 2011, 62, 435–445. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Torres, M.S.; Somu, M.P.; Johnson, H.; Irizarry, I.; Chen, Q.; Zhang, N.; Walsh, E.; Tadych, M.; Bergen, M. Hydrogen peroxide staining to visualize intracellular bacterial infections of seedling root cells. Microsc. Res. Tech. 2014, 77, 566–573. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Thomas, P.; Rajendran, T.P.; Franco, C.M.M. Cytobacts: Diverse, vertically transmissible, cultivation-recalcitrant intracellular bacteria ubiquitous to vascular plants. Manuscript in preparation. 2021. [Google Scholar]
- Brown, R. A brief account of microscopical observations made in the months of June, July, and August, 1827, on the particles contained in the pollen of plants; and on the general existence of active molecules in organic and inorganic bodies. Edinb. New Philos. J. 1828, 5, 358–371. [Google Scholar] [CrossRef]
- Mathur, J.; Mammone, A.; Barton, K.A. Organelle extensions in plant cells. J. Integr. Plant Biol. 2012, 54, 851–867. [Google Scholar] [CrossRef]
- Evert, R.F. Esau’s Plant Anatomy, 3rd ed.; John Wiley and Sons, Inc.: New Jersey, NY, USA, 2006. [Google Scholar]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Kantor, R.S.; Wrighton, K.C.; Handley, K.M.; Sharon, I.; Hug, L.A.; Castelle, C.J.; Thomas, B.C.; Banfield, J.F. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 2013, 4, e00708-13. [Google Scholar] [CrossRef]
- Sun, L.; Qiu, F.; Zhang, X.; Dai, X.; Dong, X.; Song, W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb. Ecol. 2008, 55, 415–424. [Google Scholar] [CrossRef]
- Müller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 2015, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Shaik, S.P. Molecular profiling on surface-disinfected tomato seeds reveals high diversity of cultivation recalcitrant endophytic bacteria with low share of spore forming Firmicutes. Microb. Ecol. 2020, 79, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; Brugère, J.F.; Gribaldo, S.; Schmitz, R.A.; Moissl-Eichinger, C. The host-associated archaeome. Nat. Rev. Microbiol. 2020, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P. Intense association of non-culturable endophytic bacteria with antibiotic-cleansed in vitro watermelon and their activation in degenerating cultures. Plant Cell Rep. 2011, 30, 2313–2325. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Mitter, B.; Trognitz, F.; Sessitsch, A. Metabolic potential of endophytic bacteria. Curr Opin. Biotech. 2014, 27, 30–37. [Google Scholar] [CrossRef]
- Holland, M.A.; Polacco, J.C. PPFMs and other covert contaminants: Is there more to plant physiology than just plant? Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 197–209. [Google Scholar] [CrossRef]
- Dale, C.; Moran, N.A. Molecular interactions between bacterial symbionts and their hosts. Cell 2006, 126, 453–465. [Google Scholar] [CrossRef]
- Bordenstein, S.R. Genomic and cellular complexity from symbiotic simplicity. Cell 2014, 158, 1236–1237. [Google Scholar] [CrossRef][Green Version]
- Van Leuven, J.T.; Meister, R.C.; Simon, C.; McCutcheon, J.P. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 2014, 158, 1270–1280. [Google Scholar] [CrossRef]
- Nelson, E.B. The seed microbiome: Origins, interactions and impacts. Plant Soil 2018, 422, 7–34. [Google Scholar] [CrossRef]
- Thomas, P. Vertical transmission of diverse cultivation-recalcitrant endophytic bacteria elucidated using watermelon seed embryos. Manuscript in preparation. 2021. [Google Scholar]
- Margulis, L.; Bermudes, D. Symbiosis as a mechanism of evolution: Status of cell symbiosis theory. Symbiosis 1985, 1, 101–124. [Google Scholar] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, P.; Franco, C.M.M. Intracellular Bacteria in Plants: Elucidation of Abundant and Diverse Cytoplasmic Bacteria in Healthy Plant Cells Using In Vitro Cell and Callus Cultures. Microorganisms 2021, 9, 269. https://doi.org/10.3390/microorganisms9020269
Thomas P, Franco CMM. Intracellular Bacteria in Plants: Elucidation of Abundant and Diverse Cytoplasmic Bacteria in Healthy Plant Cells Using In Vitro Cell and Callus Cultures. Microorganisms. 2021; 9(2):269. https://doi.org/10.3390/microorganisms9020269
Chicago/Turabian StyleThomas, Pious, and Christopher M. M. Franco. 2021. "Intracellular Bacteria in Plants: Elucidation of Abundant and Diverse Cytoplasmic Bacteria in Healthy Plant Cells Using In Vitro Cell and Callus Cultures" Microorganisms 9, no. 2: 269. https://doi.org/10.3390/microorganisms9020269
APA StyleThomas, P., & Franco, C. M. M. (2021). Intracellular Bacteria in Plants: Elucidation of Abundant and Diverse Cytoplasmic Bacteria in Healthy Plant Cells Using In Vitro Cell and Callus Cultures. Microorganisms, 9(2), 269. https://doi.org/10.3390/microorganisms9020269






