Detection of Infectious Cryptosporidium parvum Oocysts from Lamb’s Lettuce: CC–qPCR’s Intake
Abstract
1. Introduction
2. Materials and Methods
2.1. Cryptosporidium parvum Oocysts and Purification
2.2. Cell Line and Routine Cell Culture
2.3. Infection of Cell Monolayers
2.4. DNA Extraction and Real-Time qPCR
2.5. Lamb’s Lettuce: Cryptosporidium Oocysts Spiking, Recovery, and Cell Infection
2.6. Statistical Analysis
3. Results
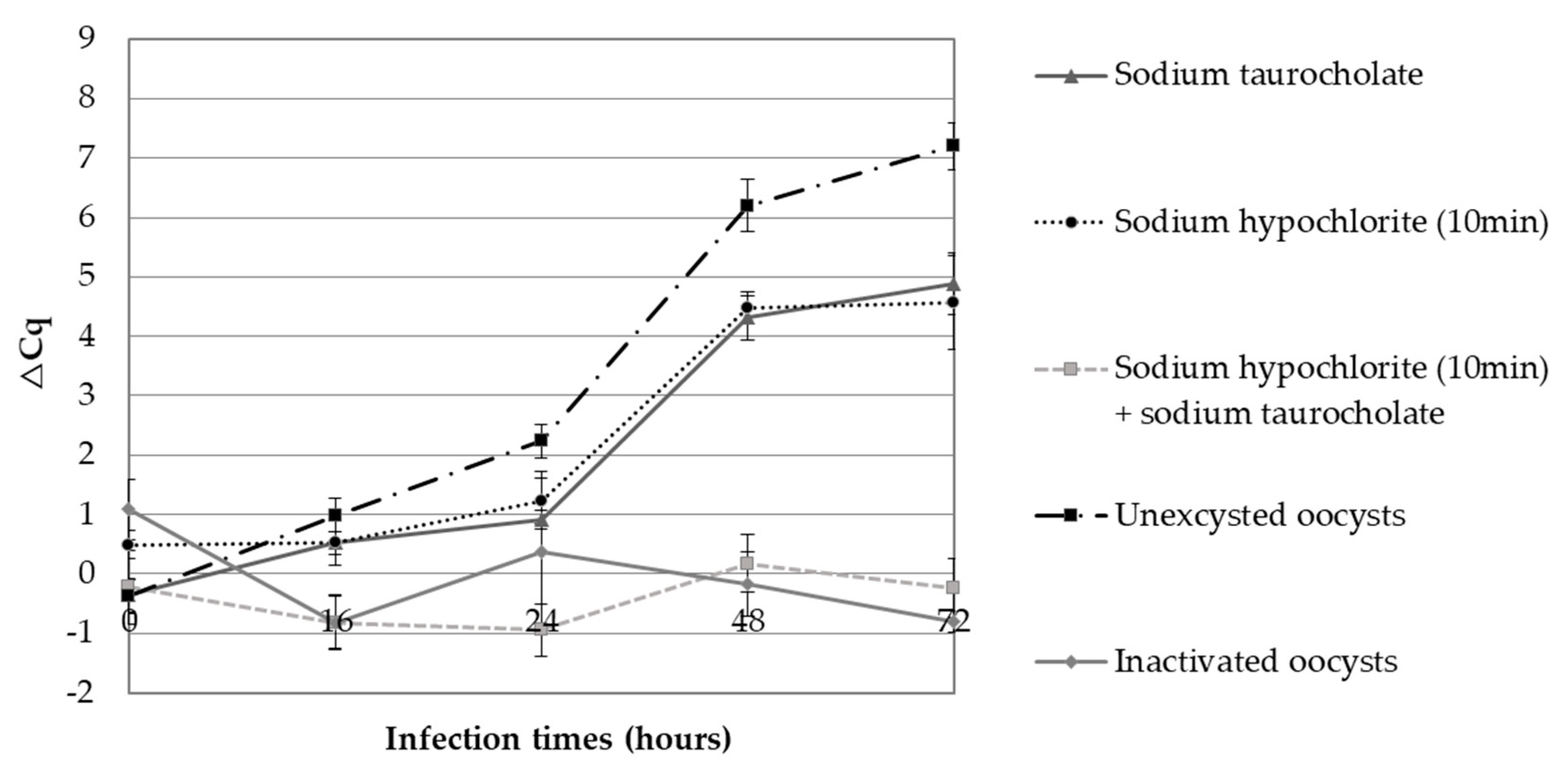
3.1. Parasitic Proliferation Kinetics Using Excysted and Unexcysted Oocysts Assessed by CC–qPCR
3.2. The Detection Limit of CC–qPCR Method Using Purified Unexcysted Oocysts
3.3. The Detection Limit of CC–qPCR Method Using Unexcysted Oocysts Recovered from Lamb’s Lettuce
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, F.A. The threat posed by the global emergence of livestock, food-borne, and zoonotic pathogens. Ann. N. Y. Acad. Sci. 1999, 894, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Macarisin, D.; Bauchan, G.; Fayer, R. Spinacia Oleracea L. leaf stomata harboring Cryptosporidium parvum oocysts: A potential threat to food safety. Appl. Environ. Microbiol. 2010, 76, 555–559. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J. 2012, 10, 2597. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 2013, 11, 3129. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014, 12, 3547. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, e04634. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, e05077. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union One Health 2018 zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.D.; Herman, L.; Hilbert, F.; et al. Public health risks associated with food-borne parasites. EFSA J. 2018, 16, e05495. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in Humans and animals—A One Health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef] [PubMed]
- NF EN ISO 18744-Juin. 2016. Available online: https://www.boutique.afnor.org/norme/nf-en-iso-18744/microbiologie-de-la-chaine-alimentaire-recherche-et-denombrement-de-cryptosporidium-et-giardia-dans-les-legumes-verts-frais-a-fe/article/818177/fa181274 (accessed on 15 December 2020).
- Ryan, U.; Hijjawi, N.; Xiao, L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Slifko, T.R.; Smith, H.V.; Rose, J.B. Emerging parasite zoonoses associated with water and food. Int. J. Parasitol. 2000, 30, 1379–1393. [Google Scholar] [CrossRef]
- Hohweyer, J.; Cazeaux, C.; Travaillé, E.; Languet, E.; Dumètre, A.; Aubert, D.; Terryn, C.; Dubey, J.P.; Azas, N.; Houssin, M.; et al. Simultaneous detection of the protozoan parasites Toxoplasma, Cryptosporidium and Giardia in food matrices and their persistence on basil leaves. Food Microbiol. 2016, 57, 36–44. [Google Scholar] [CrossRef]
- Cook, N.; Paton, C.A.; Wilkinson, N.; Nichols, R.A.B.; Barker, K.; Smith, H.V. Towards standard methods for the detection of Cryptosporidium parvum on lettuce and raspberries. Part 1: Development and optimization of methods. Int. J. Food Microbiol. 2006, 109, 215–221. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Karanis, P. An overview of methods/techniques for the detection of Cryptosporidium in food samples. Parasitol. Res. 2018, 117, 629–653. [Google Scholar] [CrossRef]
- Rousseau, A.; La Carbona, S.; Dumètre, A.; Robertson, L.J.; Gargala, G.; Escotte-Binet, S.; Favennec, L.; Villena, I.; Gérard, C.; Aubert, D. Assessing viability and infectivity of foodborne and waterborne stages (cysts/oocysts) of Giardia duodenalis, Cryptosporidium spp. and Toxoplasma gondii: A review of methods. Parasite 2018, 25, 14. [Google Scholar] [CrossRef]
- Pecková, R.; Stuart, P.D.; Sak, B.; Květoňová, D.; Kváč, M.; Foitová, I. Statistical comparison of excystation methods in Cryptosporidium parvum oocysts. Vet. Parasitol. 2016, 230, 1–5. [Google Scholar] [CrossRef]
- Di Giovanni, G.D.; Hashemi, F.H.; Shaw, N.J.; Abrams, F.A.; LeChevallier, M.W.; Abbaszadegan, M. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl. Environ. Microbiol. 1999, 65, 3427–3432. [Google Scholar] [CrossRef]
- Tyzzer, E.E. Cryptosporidium parvum (sp. nov.), a coccidium found in the small intestine of the common mouse. Arch. Protistenkd. 1912, 26, 394–412. [Google Scholar]
- Fontaine, M.; Guillot, E. Development of a taqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol. Lett. 2002, 214, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus, C. Valeriana locusta var. olitoria Linnaeus, 1753, var. nov. Zenodo 1753. [Google Scholar] [CrossRef]
- Robertson, L.J.; Gjerde, B. Factors affecting recovery efficiency in isolation of Cryptosporidium oocysts and Giardia cysts from vegetables for standard method development. J. Food Protect. 2001, 64, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Razakandrainibe, R.; Kubina, S.; Costa, D.; Robinson, G.; La Carbona, S.; Aubert, D.; David, A.; Gargala, G.; Villena, I.; Favennec, L.; et al. Evaluation of a modified method for the detection of Cryptosporidium oocysts on spinach leaves. Food Waterborne Parasitol. 2020, 21, e00097. [Google Scholar] [CrossRef] [PubMed]
- Dixon, B.; Parrington, L.; Cook, A.; Pollari, F.; Farber, J. Detection of Cyclospora, Cryptosporidium, and Giardia in ready-to-eat packaged leafy greens in Ontario, Canada. J. Food. Prot. 2013, 76, 307–313. [Google Scholar] [CrossRef]
- Robertson, L.J. Cryptosporidium as a Foodborne Pathogen; Springer Briefs in Food, Health, and Nutrition; Springer: New York, NY, USA, 2014; ISBN 978-1-4614-9377-8. [Google Scholar]
- Rochelle, P.A.; Marshall, M.M.; Mead, J.R.; Johnson, A.M.; Korich, D.G.; Rosen, J.S.; De Leon, R. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 2002, 68, 3809–3817. [Google Scholar] [CrossRef]
- Meloni, B.P.; Thompson, R.C. Simplified methods for obtaining purified oocysts from mice and for growing Cryptosporidium parvum in vitro. J. Parasitol. 1996, 82, 757–762. [Google Scholar] [CrossRef]
- Upton, S.J.; Tilley, M.; Brillhart, D.B. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol. Lett. 1994, 118, 233–236. [Google Scholar] [CrossRef]
- Slifko, T.R.; Huffman, D.E.; Dussert, B.; Owens, J.H.; Jakubowski, W.; Haas, C.N.; Rose, J.B. Comparison of tissue culture and animal models for assessment of Cryptospridium parvum infection. Exp. Parasitol. 2002, 101, 97–106. [Google Scholar] [CrossRef]
- Aboytes, R.; Giovanni, G.; Abrams, F.; Rheinecker, C.; Mcelroy, W.; Shaw, N.; Lechevallier, M. Detection of infectious Cryptosporidium in filtered drinking water. J. Am. Water Work Assoc. 2004, 96, 88–98. [Google Scholar] [CrossRef]
- Arrowood, M.J. In vitro cultivation of Cryptosporidium species. Clin. Microbiol. Rev. 2002, 15, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Nie, W.; Sheoran, A.; Zhang, Q.; Tzipori, S. Bile acids enhance invasiveness of Cryptosporidium spp. into cultured cells. Infect. Immun. 2006, 74, 3342–3346. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Daugschies, A.; Cakmak, A.; Yilmazer, N.; Dittmar, K.; Bangoura, B. Cryptosporidium parvum oocyst viability and behaviour of the residual body during the excystation process. Parasitol. Res. 2011, 109, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Gold, D.; Stein, B.; Tzipori, S. The utilization of sodium taurocholate in excystation of Cryptosporidium parvum and infection of tissue culture. J. Parasitol. 2001, 87, 997–1000. [Google Scholar] [CrossRef]
- Siripanth, C.; Punpoowong, B.; Amarapal, P.; Thima, N.; Eampokalap, B.; Kaewkungwal, J. Comparison of Cryptosporidium parvum development in various cell lines for screening in vitro drug testing. Southeast Asian J. Trop. Med. Public Health 2004, 35, 540–546. [Google Scholar]
- Delling, C.; Lendner, M.; Müller, U.; Daugschies, A. Improvement of in vitro evaluation of chemical disinfectants for efficacy on Cryptosporidium parvum oocysts. Vet. Parasitol. 2017, 245, 5–13. [Google Scholar] [CrossRef]

| Number of Inoculated Purified Oocysts | 10,000 | 1000 | 100 | 50 | 25 | 10 | 5 | 1 | |
|---|---|---|---|---|---|---|---|---|---|
| Oocysts batch | 2 | 4 | 4 | 3 | 3 | 4 | 3 | 3 | |
| Cell culture replicates | 9 | 15 | 15 | 12 | 12 | 15 | 12 | 12 | |
| qPCR repeats | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Number of positive wells detected | with cells | 18/18 | 30/30 | 30/30 | 22/24 | 22/24 | 23/30 | 5/24 | 8/30 |
| w/out cells | 18/18 | 29/30 | 14/30 | 6/24 | 7/24 | 3/30 | 3/24 | 2/30 | |
| Cq with cells | 25.55 | 29.03 | 31.90 | 33.16 | 33.63 | 34.92 | 35.18 | 34.35 | |
| SD with cells | 1.09 | 1.72 | 1.57 | 1.55 | 1.34 | 1.58 | 1.37 | 0.78 | |
| Cq w/out cells | 30.47 | 34.54 | 36.21 | 36.28 | 36.24 | 37.68 | 36.69 | 35.63 | |
| SD w/out cells | 0.82 | 1.36 | 0.73 | 0.57 | 0.98 | 1.24 | 0.28 | 0.00 | |
| Number of Inoculated Oocysts Per 30 g of Lamb’s Lettuce | 10,000 | 1000 | 100 | 10 | 1 | |
|---|---|---|---|---|---|---|
| Oocysts batch | 2 | 2 | 2 | 2 | 2 | |
| Cell culture replicates | 6 | 6 | 6 | 6 | 6 | |
| qPCR repeats | 2 | 2 | 2 | 2 | 2 | |
| Number of positive wells detected | with cells | 18/18 | 12/12 | 18/18 | 8/12 | 3/12 |
| w/out cells | 18/18 | 12/12 | 9/18 | 0/12 | 0/12 | |
| Cq with cells | 25.79 | 29.16 | 31.91 | 34.18 | 35.40 | |
| SD with cells | 2.38 | 1.74 | 2.52 | 1.58 | 0.87 | |
| Cq w/out cells | 30.82 | 33.94 | 34.86 | N/A | N/A | |
| SD w/out cells | 1.40 | 1.26 | 1.77 | N/A | N/A | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubina, S.; Costa, D.; Favennec, L.; Gargala, G.; Rousseau, A.; Villena, I.; La Carbona, S.; Razakandrainibe, R. Detection of Infectious Cryptosporidium parvum Oocysts from Lamb’s Lettuce: CC–qPCR’s Intake. Microorganisms 2021, 9, 215. https://doi.org/10.3390/microorganisms9020215
Kubina S, Costa D, Favennec L, Gargala G, Rousseau A, Villena I, La Carbona S, Razakandrainibe R. Detection of Infectious Cryptosporidium parvum Oocysts from Lamb’s Lettuce: CC–qPCR’s Intake. Microorganisms. 2021; 9(2):215. https://doi.org/10.3390/microorganisms9020215
Chicago/Turabian StyleKubina, Sophie, Damien Costa, Loïc Favennec, Gilles Gargala, Angélique Rousseau, Isabelle Villena, Stéphanie La Carbona, and Romy Razakandrainibe. 2021. "Detection of Infectious Cryptosporidium parvum Oocysts from Lamb’s Lettuce: CC–qPCR’s Intake" Microorganisms 9, no. 2: 215. https://doi.org/10.3390/microorganisms9020215
APA StyleKubina, S., Costa, D., Favennec, L., Gargala, G., Rousseau, A., Villena, I., La Carbona, S., & Razakandrainibe, R. (2021). Detection of Infectious Cryptosporidium parvum Oocysts from Lamb’s Lettuce: CC–qPCR’s Intake. Microorganisms, 9(2), 215. https://doi.org/10.3390/microorganisms9020215







