The Impact of Pre-Slaughter Fasting on the Ruminal Microbial Population of Commercial Angus Steers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Selection and Handling
2.2. Rumen Content Collection and Storage
2.3. DNA Extraction and Sequencing
2.4. Volatile Fatty Acid Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tajima, K.; Aminov, R.I.; Nagamine, T.; Matsui, H.; Nakamura, M.; Benno, Y. Diet-Dependent Shifts in the Bacterial Population of the Rumen Revealed with Real-Time PCR. Appl. Environ. Microbiol. 2001, 67, 2766–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Menezes, A.B.; Lewis, E.; O’Donovan, M.; O’Neill, B.F.; Clipson, N.; Doyle, E.M. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 2011, 78, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Carberry, C.A.; Kenny, D.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of Phenotypic Residual Feed Intake and Dietary Forage Content on the Rumen Microbial Community of Beef Cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizrahi, I. Rumen Symbioses, the Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Springer: Berlin/Heidelberg, Germany, 2013; pp. 533–544. [Google Scholar]
- Flint, H.J.; Bayer, E.A. Plant Cell Wall Breakdown by Anaerobic Microorganisms from the Mammalian Digestive Tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef]
- Hobson, P.N.; Stewart, C.S. The Rumen Microbial Ecosystem; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Krause, D.O.; Denman, S.; Mackie, R.I.; Morrison, M.; Rae, A.L.; Attwood, G.; McSweeney, C. Opportunities to improve fiber degradation in the rumen: Microbiology, ecology, and genomics. FEMS Microbiol. Rev. 2003, 27, 663–693. [Google Scholar] [CrossRef] [Green Version]
- Wolin, M.J. Volatile Fatty Acids and the Inhibition of Escherichia coli Growth by Rumen Fluid. Appl. Microbiol. 1969, 17, 83–87. [Google Scholar] [CrossRef]
- Brownlie, L.E.; Grau, F.H. Effect of Food Intake on Growth and Survival of Salmonellas and Escherichia coli in the Bovine Rumen. J. Gen. Microbiol. 1967, 46, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Pointon, A.; Kiermeier, A.; Fegan, N. Review of the impact of pre-slaughter feed curfews of cattle, sheep and goats on food safety and carcase hygiene in Australia. Food Control. 2012, 26, 313–321. [Google Scholar] [CrossRef]
- Rasmussen, M.A.; Cray, W.C.; Casey, T.A.; Whipp, S.C. Rumen contents as a reservoir of enterohemorrhagic Escherichia coli. FEMS Microbiol. Lett. 1993, 114, 79–84. [Google Scholar] [CrossRef]
- Callaway, T.; Elder, R.; Keen, J.; Anderson, R.; Nisbet, D. Forage Feeding to Reduce Preharvest Escherichia coli Populations in Cattle, a Review. J. Dairy Sci. 2003, 86, 852–860. [Google Scholar] [CrossRef] [Green Version]
- Nastasijevic, I. STEC O157 in the beef chain—Risk assessment and management. CAB Rev. Perspect. Agric. Veter-Sci. Nutr. Nat. Resour. 2011, 6, 61–80. [Google Scholar] [CrossRef]
- Small, A.; Reid, C.-A.; Avery, S.M.; Karabasil, N.; Crowley, C.; Buncic, S. Potential for the Spread of Escherichia coli O157, Salmonella, and Campylobacter in the Lairage Environment at Abattoirs. J. Food Prot. 2002, 65, 931–936. [Google Scholar] [CrossRef]
- Arthur, J.P.; Herd, R. Residual feed intake in beef cattle. Rev. Bras. Zootec. 2008, 37, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Herd, R.M.; Archer, J.A.; Arthur, P.F. Reducing the cost of beef production through genetic improvement in residual feed intake: Opportunity and challenges to application. J. Anim. Sci. 2003, 81 (Suppl. S1), E9–E17. [Google Scholar] [CrossRef]
- Nkrumah, J.D.; Okine, E.K.; Mathison, G.W.; Schmid, K.; Li, C.; Basarab, J.A.; Price, M.A.; Wang, Z.; Moore, S.S. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle1. J. Anim. Sci. 2006, 84, 145–153. [Google Scholar] [CrossRef]
- Lourenco, J.; Callaway, T.; Kieran, T.; Glenn, T.; McCann, J.C.; Stewart, R.L.J. Analysis of the Rumen Microbiota of Beef Calves Supplemented During the Suckling Phase. Front. Microbiol. 2019, 10, 1131. [Google Scholar] [CrossRef] [Green Version]
- Rothrock, M.J.; Hiett, K.L.; Gamble, J.; Caudill, A.C.; Cicconi-Hogan, K.M.; Caporaso, J.G. A Hybrid DNA Extraction Method for the Qualitative and Quantitative Assessment of Bacterial Communities from Poultry Production Samples. J. Vis. Exp. 2014, 94, e52161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2009, 26, 266–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lourenco, J.M.; Kieran, T.; Seidel, D.S.; Glenn, T.C.; Da Silveira, M.F.; Callaway, T.R.; Stewart, R.L., Jr. Comparison of the ruminal and fecal microbiotas in beef calves supplemented or not with concentrate. PLoS ONE 2020, 15, e0231533. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef] [Green Version]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative Analysis of Microbial Profiles in Cow Rumen Fed with Different Dietary Fiber by Tagged 16S rRNA Gene Pyrosequencing. Curr. Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef]
- Dias, J.; Marcondes, M.; de Souza, S.M.; Silva, B.C.D.M.E.; Noronha, M.; Resende, R.T.; Machado, F.S.; Mantovani, H.C.; Dill-McFarland, K.A.; Suen, G. Bacterial Community Dynamics across the Gastrointestinal Tracts of Dairy Calves during Preweaning Development. Appl. Environ. Microbiol. 2018, 84, e02675-17. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Kim, J.; Kuehn, L.A.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J.E. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Zeineldin, M.; Aldridge, B.; Lowe, J. Dysbiosis of the fecal microbiota in feedlot cattle with hemorrhagic diarrhea. Microb. Pathog. 2018, 115, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Hoedt, E.; Parks, D.H.; Volmer, J.G.; Rosewarne, C.; Denman, S.; McSweeney, C.; Muir, J.G.; Gibson, P.R.; Cuív, P.Ó.; Hugenholtz, P.; et al. Culture- and metagenomics-enabled analyses of the Methanosphaera genus reveals their monophyletic origin and differentiation according to genome size. ISME J. 2018, 12, 2942–2953. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Hernandez-Sanabria, E.; Guan, L.L. Assessment of the Microbial Ecology of Ruminal Methanogens in Cattle with Different Feed Efficiencies. Appl. Environ. Microbiol. 2009, 75, 6524–6533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecours, P.B.; Marsolais, D.; Cormier, Y.; Berberi, M.; Haché, C.; Bourdages, R.; Duchaine, C. Increased Prevalence of Methanosphaera stadtmanae in Inflammatory Bowel Diseases. PLoS ONE 2014, 9, e87734. [Google Scholar] [CrossRef]
- Park, S.F. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 2002, 74, 177–188. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Scharff, R.L. Economic Burden from Health Losses Due to Foodborne Illness in the United States. J. Food Prot. 2012, 75, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Epiao, H.; Elachman, M.; Emalfatti, S.; Sczyrba, A.; Eknierim, B.; Eauer, M.; Tringe, S.; Mackie, R.I.; Yeoman, C.J.; Ehess, M. Temporal dynamics of fibrolytic and methanogenic rumen microorganisms during in situ incubation of switchgrass determined by 16S rRNA gene profiling. Front. Microbiol. 2014, 5, 307. [Google Scholar] [CrossRef] [Green Version]
- Kudo, H.; Cheng, K.-J.; Costerton, J.W. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can. J. Microbiol. 1987, 33, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Döpfer, D.; Anklam, K.; Mikheil, D.; Ladell, P. Growth curves and morphology of three Treponema subtypes isolated from digital dermatitis in cattle. Veter-J. 2012, 193, 685–693. [Google Scholar] [CrossRef]
- Sullivan, L.E.; Clegg, S.; Angell, J.W.; Newbrook, K.; Blowey, R.W.; Carter, S.D.; Bell, J.; Duncan, J.S.; Grove-White, D.H.; Murray, R.D.; et al. High-Level Association of Bovine Digital Dermatitis Treponema spp. with Contagious Ovine Digital Dermatitis Lesions and Presence of Fusobacterium necrophorum and Dichelobacter nodosus. J. Clin. Microbiol. 2015, 53, 1628–1638. [Google Scholar] [CrossRef] [Green Version]
- Trott, D.J.; Moeller, M.R.; Zuerner, R.L.; Goff, J.P.; Waters, W.R.; Alt, D.P.; Walker, R.L.; Wannemuehler, M.J. Characterization of Treponema phagedenis -Like Spirochetes Isolated from Papillomatous Digital Dermatitis Lesions in Dairy Cattle. J. Clin. Microbiol. 2003, 41, 2522–2529. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, L.E.; Carter, S.D.; Duncan, J.S.; Grove-White, D.H.; Angell, J.W.; Evans, N.J. The Gastrointestinal Tract as a Potential Infection Reservoir of Digital Dermatitis-Associated Treponemes in Beef Cattle and Sheep. Appl. Environ. Microbiol. 2015, 81, 7460–7469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, A.M. Variations in the pH and volatile fatty acid concentration within the bovine reticulo-rumen. New Zealand J. Agric. Res. 1964, 7, 694–706. [Google Scholar] [CrossRef]
- Stewart, W.E.; Stewart, D.G.; Schultz, L.H. Rates of Volatile Fatty Acid Production in the Bovine Rumen1. J. Anim. Sci. 1958, 17, 723–736. [Google Scholar] [CrossRef]
- Guan, L.L.; Nkrumah, J.D.; Basarab, J.A.; Moore, S. Linkage of microbial ecology to phenotype: Correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 2008, 288, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
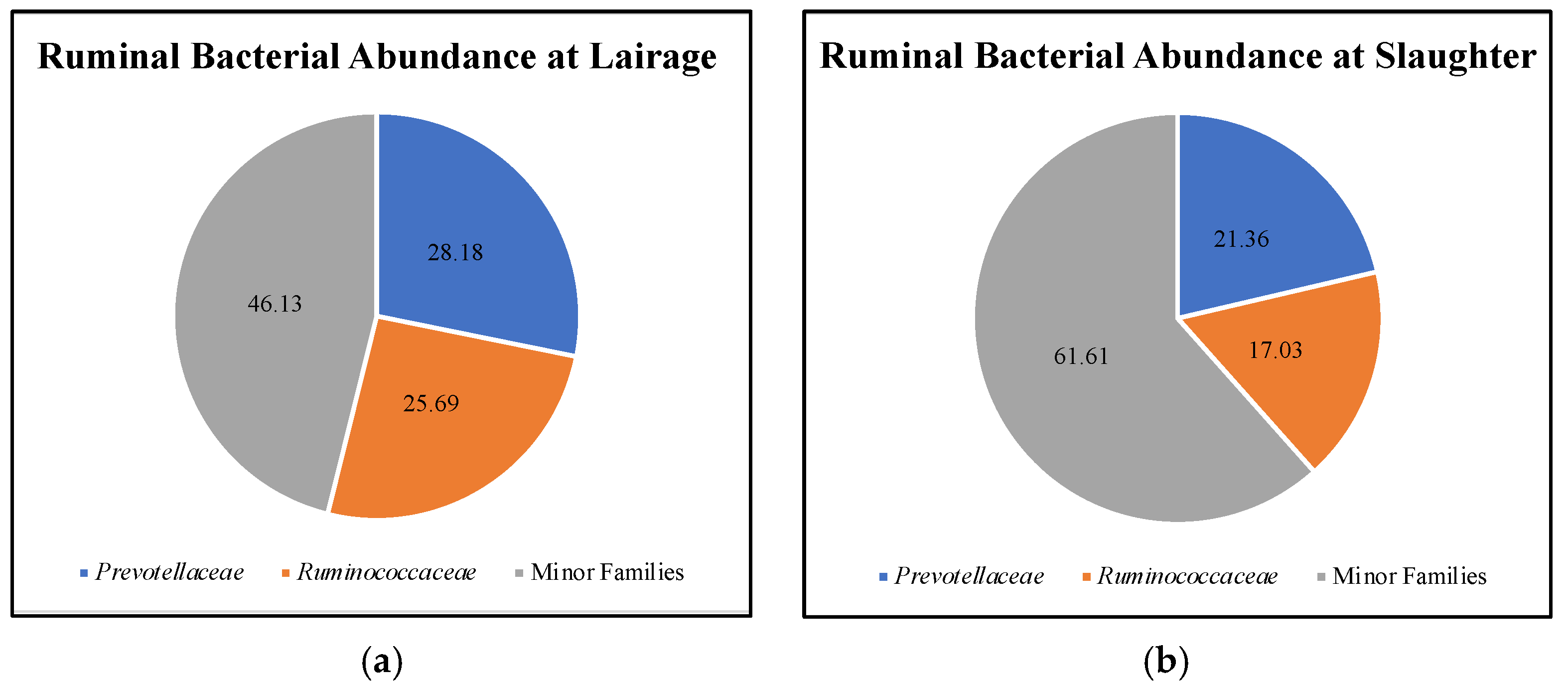

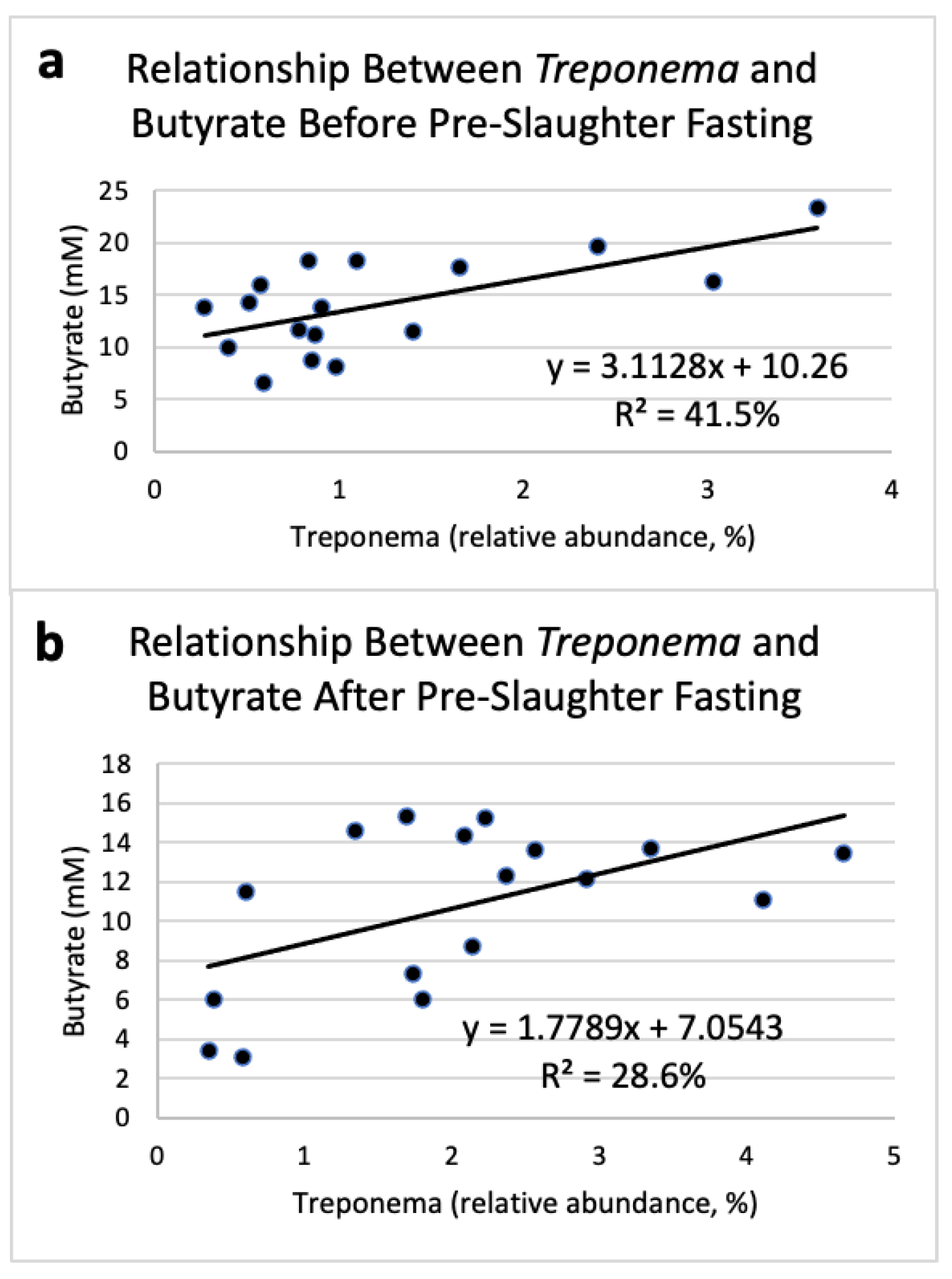
| Index | High-RFI Steers | |||
| Lairage | Slaughter | SEM | p-Value | |
| Chao 1 | 2791 | 3786 | 177 | 0.001 |
| Shannon Index | 8.18 | 8.59 | 0.072 | 0.001 |
| Evenness | 0.768 | 0.786 | 0.007 | 0.040 |
| Low-RFI Steers | ||||
| Lairage | Slaughter | SEM | p-Value | |
| Chao 1 | 2499 | 3051 | 204 | 0.027 |
| Shannon Index | 7.92 | 8.45 | 0.161 | 0.011 |
| Evenness | 0.753 | 0.788 | 0.014 | 0.030 |
| Volatile Fatty Acid | Lairage | Slaughter | SEM | p-Value |
|---|---|---|---|---|
| Acetate | 62.50 | 62.73 | 6.740 | 0.97 |
| Propionate | 26.13 | 22.50 | 4.520 | 0.43 |
| Isobutyrate | 1.29 | 1.27 | 0.081 | 0.88 |
| Butyrate | 14.06 | 10.71 | 1.710 | 0.07 |
| Isovalerate | 3.27 | 3.70 | 0.397 | 0.30 |
| Valerate | 1.71 | 1.76 | 0.467 | 0.91 |
| Total volatile fatty acid | 109.21 | 103.56 | 13.100 | 0.67 |
| Acetate: Propionate | 2.63 | 3.18 | 0.332 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welch, C.B.; Lourenco, J.M.; Seidel, D.S.; Krause, T.R.; Rothrock, M.J.; Pringle, T.D.; Callaway, T.R. The Impact of Pre-Slaughter Fasting on the Ruminal Microbial Population of Commercial Angus Steers. Microorganisms 2021, 9, 2625. https://doi.org/10.3390/microorganisms9122625
Welch CB, Lourenco JM, Seidel DS, Krause TR, Rothrock MJ, Pringle TD, Callaway TR. The Impact of Pre-Slaughter Fasting on the Ruminal Microbial Population of Commercial Angus Steers. Microorganisms. 2021; 9(12):2625. https://doi.org/10.3390/microorganisms9122625
Chicago/Turabian StyleWelch, Christina Breanne, Jeferson M. Lourenco, Darren S. Seidel, Taylor Rae Krause, Michael J. Rothrock, T. Dean Pringle, and Todd R. Callaway. 2021. "The Impact of Pre-Slaughter Fasting on the Ruminal Microbial Population of Commercial Angus Steers" Microorganisms 9, no. 12: 2625. https://doi.org/10.3390/microorganisms9122625
APA StyleWelch, C. B., Lourenco, J. M., Seidel, D. S., Krause, T. R., Rothrock, M. J., Pringle, T. D., & Callaway, T. R. (2021). The Impact of Pre-Slaughter Fasting on the Ruminal Microbial Population of Commercial Angus Steers. Microorganisms, 9(12), 2625. https://doi.org/10.3390/microorganisms9122625







