Diel Protein Regulation of Marine Picoplanktonic Communities Assessed by Metaproteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Sampling
2.2. Protein Isolation
2.3. Liquid Chromatography Tandem Mass Spectrometry Analysis
2.4. Ocean Sampling Day 2014 Metagenomic Data Set
2.5. Databases Creation and Protein Identification
2.6. Protein Annotation and Downstream Analyses
3. Results
3.1. Diel Structure of the Microbial Communities
3.2. Diel Functioning of the Microbial Communities
3.2.1. Protein Folding and Response to Stress
3.2.2. Replication, Transcription, and Translation
3.2.3. Energy Metabolism and Compounds Biosynthesis
3.2.4. Transport and Cell Division, Structure, and Mobility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedges, J.; Oades, J. Comparative organic geochemistries of soils and marine sediments. Org. Geochem. 1997, 27, 319–361. [Google Scholar] [CrossRef]
- Kujawinski, E.B. The Impact of Microbial Metabolism on Marine Dissolved Organic Matter. Annu. Rev. Mar. Sci. 2011, 3, 567–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, J.J.; Findlay, S.; Pace, M.L. Bacterial production in fresh and saltwater ecosystems: A cross-system overview. Mar. Ecol. Prog. Ser. 1988, 43, 1–10. [Google Scholar] [CrossRef]
- Turley, C.M.; Stutt, E.D. Depth-related cell-specific bacterial leucine incorporation rates on particles and its biogeochemical significance in the Northwest Mediterranean. Limnol. Oceanogr. 2000, 45, 419–425. [Google Scholar] [CrossRef]
- Simon, M.; Grossart, H.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef] [Green Version]
- DeLong, E.F.; Franks, D.G.; Alldredge, A.L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 1993, 38, 924–934. [Google Scholar] [CrossRef] [Green Version]
- Acinas, S.G.; Antón, J.; Rodríguez-Valera, F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1999, 65, 514–522. [Google Scholar] [CrossRef] [Green Version]
- Crump, B.C.; Armbrust, E.V.; Baross, J.A. Phylogenetic analysis of particle-attached and free-living bac-terial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 1999, 65, 3192–3204. [Google Scholar] [CrossRef] [Green Version]
- Hollibaugh, J.; Wong, P.; Murrell, M. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat. Microb. Ecol. 2000, 21, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Moeseneder, M.M.; Winter, C.; Herndl, G.J. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 2001, 46, 95–107. [Google Scholar] [CrossRef]
- Ghiglione, J.F.; Mevel, G.; Pujo-Pay, M.; Mousseau, L.; Lebaron, P.; Goutx, M. Diel and seasonal variations in abundance, activity, and community structure of particle-attached and free-living bacteria in NW Medi-terranean Sea. Microb. Ecol. 2007, 54, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Ditty, J.L.; Mackey, S.R.; Johnson, C.H. Bacterial Circadian Programs; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Galí, M.; Simó, R.; Vila-Costa, M.; Ruiz-González, C.; Gasol, J.M.; Matrai, P. Diel patterns of oceanic di-methylsulfide (DMS) cycling: Microbial and physical drivers. Glob. Biogeochem. Cycles 2013, 27, 620–636. [Google Scholar] [CrossRef]
- Kuipers, B.; Van Noort, G.; Vosjan, J.; Herndl, G. Diel periodicity of bacterioplankton in the euphotic zone of the subtropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 2000, 201, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Winter, C.; Herndl, G.; Weinbauer, M. Diel cycles in viral infection of bacterioplankton in the North Sea. Aquat. Microb. Ecol. 2004, 35, 207–216. [Google Scholar] [CrossRef]
- Gasol, J.; Doval, M.D.; Pinhassi, J.; Calderón-Paz, J.; Guixa-Boixareu, N.; Vaqué, D.; Pedrós-Alió, C. Diel variations in bacterial heterotrophic activity and growth in the northwestern Mediterranean Sea. Mar. Ecol. Prog. Ser. 1998, 164, 107–124. [Google Scholar] [CrossRef] [Green Version]
- Matallana-Surget, S.; Jagtap, P.D.; Griffin, T.J.; Beraud, M.; Wattiez, R. Comparative Metaproteomics to Study Environmental Changes. Metagenomics 2018, 327–363. [Google Scholar] [CrossRef]
- Poretsky, R.S.; Hewson, I.; Sun, S.; Allen, A.E.; Zehr, J.P.; Moran, M.A. Comparative day/night meta-transcriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 2009, 11, 1358–1375. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Field, D.; Swift, P.; Thomas, S.; Cummings, D.; Temperton, B.; Weynberg, K.; Huse, S.; Hughes, M.; Joint, I. The taxonomic and functional diversity of microbes at a temperate coastal site: A ‘multi-omic’ study of seasonal and diel temporal variation. PLoS ONE 2010, 5, e15545. [Google Scholar] [CrossRef]
- Ottesen, E.A.; Young, C.R.; Gifford, S.M.; Eppley, J.M.; Marin, R.; Schuster, S.C.; Scholin, C.A.; Delong, E.F. Multispecies diel tran-scriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 2014, 345, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Wilmes, P.; Bond, P. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 2004, 6, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.; Huttenhower, C. Sequencing and beyond: Integrating molecular ’omics’ for microbial community profiling. Nat. Rev. Genet. 2015, 13, 360–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kan, J.; Hanson, T.E.; Ginter, J.M.; Wang, K.; Chen, F. Metaproteomic analysis of Chesapeake Bay microbial communities. Saline Syst. 2005, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, M.A.; Bertrand, E.M.; Duffy, M.E.; Gaylord, D.A.; Held, N.A.; Hervey, W.J., IV; Hettich, R.L.; Jagtap, P.D.; Janech, M.G.; Kinkade, D.B.; et al. Progress and challenges in ocean metaproteomics and proposed best practices for data sharing. J. Proteome Res. 2019, 18, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Sowell, S.M.; Wilhelm, L.J.; Norbeck, A.D.; Lipton, M.S.; Nicora, C.D.; Barofsky, D.F.; Carlson, C.A.; Smith, R.; Giovanonni, S.J. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2008, 3, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.J.; Cavicchioli, R. Marine metaproteomics: Deciphering the microbial metabolic food web. Trends Microbiol. 2014, 22, 248–260. [Google Scholar] [CrossRef]
- Hawley, A.K.; Brewer, H.M.; Norbeck, A.D.; A-Toli, L.P.; Hallam, S.J. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc. Natl. Acad. Sci. USA 2014, 111, 11395–11400. [Google Scholar] [CrossRef] [Green Version]
- Bergauer, K.; Fernandez-Guerra, A.; Garcia, J.A.L.; Sprenger, R.; Stepanauskas, R.; Pachiadaki, M.; Jensen, O.N.; Herndl, G.J. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. Proc. Natl. Acad. Sci. USA 2017, 115, E400–E408. [Google Scholar] [CrossRef] [Green Version]
- Georges, A.A.; El-Swais, H.; Craig, S.E.; Li, W.K.; Walsh, D.A. Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton. ISME J. 2014, 8, 1301–1313. [Google Scholar] [CrossRef] [Green Version]
- Géron, A.; Werner, J.; Wattiez, R.; Lebaron, P.; Matallana Surget, S. Deciphering the functioning of mi-crobial communities: Shedding light on the critical steps in metaproteomics. Front. Microbiol. 2019, 10, 2395. [Google Scholar] [CrossRef] [Green Version]
- Heyer, R.; Schallert, K.; Zoun, R.; Becher, B.; Saake, G.; Benndorf, D. Challenges and perspectives of met-aproteomic data analysis. J. Biotechnol. 2017, 261, 24–36. [Google Scholar] [CrossRef]
- Werner, J.; Géron, A.; Kerssemakers, J.; Matallana-Surget, S. mPies: A novel metaproteomics tool for the creation of relevant protein databases and automatized protein annotation. Biol. Direct 2019, 14, 21. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2018, 47, D1211–D1217. [Google Scholar] [CrossRef] [Green Version]
- John, J.S. SeqPrep: Tool for Stripping Adaptors and/or Merging Paired Reads with Overlap into Single Reads. 2011. Available online: https://githubcom/jstjohn/SeqPrep (accessed on 8 November 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef] [Green Version]
- Rho, M.; Tang, H.; Ye, Y. FragGeneScan: Predicting genes in short and error-prone reads. Nucleic Acids Res. 2010, 38, e191. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matias Rodrigues, J.F.; Schmidt, T.S.; Tackmann, J.; von Mering, C. MAPseq: Highly efficient k-mer search with confidence estimates, for rRNA sequence analysis. Bioinformatics 2017, 33, 3808–3810. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN community edi-tion-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K. Glutamine signalling in bacteria. Front. Biosci. 2007, 12, 358–370. [Google Scholar] [CrossRef] [Green Version]
- Morris, R.M.; Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.J. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 2002, 420, 806–810. [Google Scholar] [CrossRef]
- Williams, T.J.; Wilkins, D.; Long, E.; Evans, F.; DeMaere, M.Z.; Raftery, M.J.; Cavicchioli, R. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 2013, 15, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Long, E.; Evans, F.; DeMaere, M.Z.; Lauro, F.M.; Raftery, M.J.; Ducklow, H.; Grzymski, J.J.; Murray, A.E.; Cavicchioli, R. A metaproteomic assess-ment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012, 6, 1883–1900. [Google Scholar] [CrossRef] [Green Version]
- Feingersch, R.; Suzuki, M.T.; Shmoish, M.; Sharon, I.; Sabehi, G.; Partensky, F.; Béjà, O. Microbial community genomics in eastern Mediterranean Sea surface waters. ISME J. 2009, 4, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, R.M.; Nunn, B.; Frazar, C.; Goodlett, D.R.; Ting, Y.; Rocap, G. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 2010, 4, 673–685. [Google Scholar] [CrossRef] [Green Version]
- Mella-Flores, D.; Mazard, S.; Humily, F.; Partensky, F.; Mahé, F.; Bariat, L.; Courties, C.; Marie, D.; Ras, J.; Mauriac, R.; et al. Is the distribution of Prochlorococcus and Synechococcus ecotypes in the Mediterranean Sea affected by global warming? Biogeosciences 2011, 8, 2785–2804. [Google Scholar] [CrossRef] [Green Version]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; Gonzalez, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- Mayot, N.; d’Ortenzio, F.; Taillandier, V.; Prieur, L.; De Fommervault, O.P.; Claustre, H.; Bosse, A.; Testor, P.; Conan, P. Physical and biogeochemical controls of the phytoplankton blooms in North Western Mediterranean Sea: A multiplat-form approach over a complete annual cycle (2012–2013 DEWEX experiment). J. Geophys. Res. Ocean. 2017, 122, 9999–10019. [Google Scholar] [CrossRef]
- Cohen, S.E.; Golden, S.S. Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 373–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sowell, S.M.; Abraham, P.E.; Shah, M.; Verberkmoes, N.C.; Smith, D.P.; Barofsky, D.F.; Giovannoni, S.J. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 2010, 5, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Matallana-Surget, S.; Cavicchioli, R.; Fauconnier, C.; Wattiez, R.; Leroy, B.; Joux, F.; Raftery, M.J.; LeBaron, P. Shotgun Redox Proteomics: Identification and Quantitation of Carbonylated Proteins in the UVB-Resistant Marine Bacterium, Photobacterium angustum S14. PLoS ONE 2013, 8, e68112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoch, M.P.; Snyder, R.A.; Jeffrey, W.H.; Dillon, K.S.; Coffin, R.B. Expression of glutamine synthetase and glutamate dehydrogenase by marine bacterioplankton: Assay optimizations and efficacy for assessing ni-trogen to carbon metabolic balance in situ. Limnol. Oceanogr. Methods 2006, 4, 308–328. [Google Scholar] [CrossRef] [Green Version]
- Waldbauer, J.R.; Rodrigue, S.; Coleman, M.L.; Chisholm, S.W. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS ONE 2012, 7, e43432. [Google Scholar] [CrossRef] [Green Version]
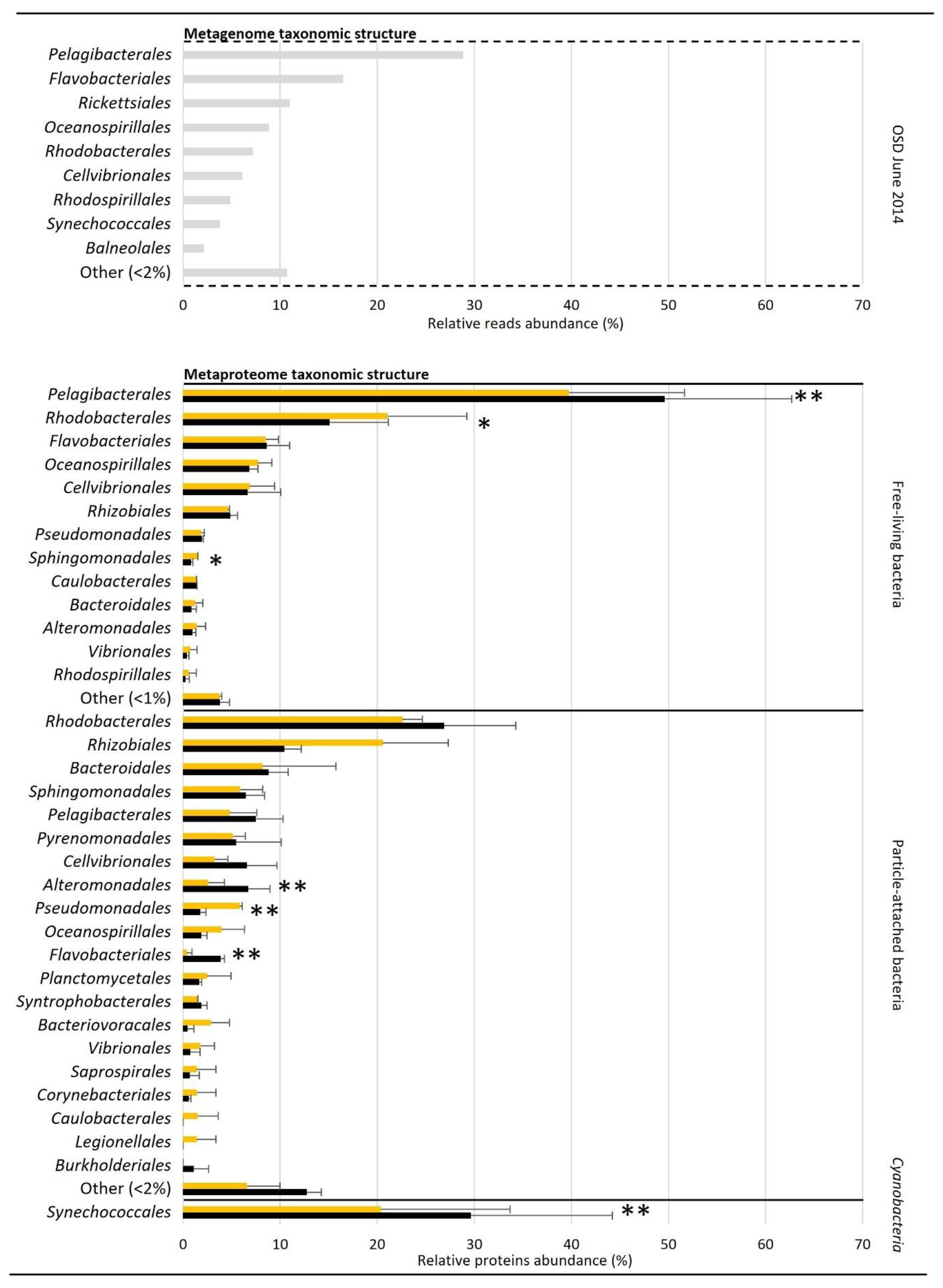


| Metagenome Taxonomic Structure | Metaproteome Taxonomic Structure | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.2 µm Size-Fraction | 0.8 µm Size-Fraction | ||||||||
| OSD June 2014 | Day | SD | Night | SD | Day | SD | Night | SD | |
| Total reads/proteins | 761 | 550 | ±49 | 452 | ±4 | 123 | ±28 | 170 | ±28 |
| Phylum | |||||||||
| Proteobacteria | 66.89 | ** 89.34 | ±1.62 | ** 92.43 | ±1.19 | 34.75 | ±7.92 | 33.19 | ±6.02 |
| Bacteroidetes | 15.51 | 6.48 | ±0.61 | 5.48 | ±0.40 | 2.71 | ±1.35 | 4.47 | ±0.84 |
| Cyanobacteria | 12.22 | 2.30 | ±1.83 | 0.43 | ±0.15 | 60.52 | ±8.06 | 60.83 | ±6.60 |
| Rhodothermaeota | 1.84 | 0.69 | ±0.22 | 0.56 | ±0.61 | ||||
| Planctomycetes | 0.13 | 0.03 | ±0.05 | 0.75 | ±0.79 | 0.46 | ±0.03 | ||
| Other (<1%) | 3.42 | 1.19 | 1 | 1.26 | 1.05 | ||||
| Class | |||||||||
| Alphaproteobacteria | 47.35 | * 67.06 | ±2.88 | * 71.54 | ±4.87 | 20.88 | ±2.04 | 20.86 | ±4.04 |
| Gammaproteobacteria | 17.77 | 23.29 | ±0.69 | 21.57 | ±4.01 | 12.79 | ±4.64 | 11.41 | ±0.97 |
| Flavobacteriia | 14.32 | 4.77 | ±0.75 | 4.94 | ±0.75 | ** 0.10 | ±0.14 | ** 1.46 | ±0.10 |
| Unclassified Cyanobacteria | 12.33 | 0.54 | ±0.77 | 0.45 | ±0.16 | 60.25 | ±7.55 | 60.26 | ±5.63 |
| Bacteroidia | 0.13 | 0.68 | ±0.47 | 0.51 | ±0.32 | 2.27 | ±1.95 | 2.65 | ±0.86 |
| Deltaproteobacteria | 0.06 | ±0.08 | * 0.43 | ±0.03 | * 0.78 | ±0.21 | |||
| Oligoflexia | 0.06 | ±0.09 | 0.07 | ±0.09 | 0.87 | ±0.66 | 0.44 | ±0.29 | |
| Planctomycetia | 0.03 | ±0.05 | 0.77 | ±0.80 | 0.49 | ±0.03 | |||
| Other (<1%) | 8.09 | 3.54 | 0.9 | 1.64 | 1.64 | ||||
| Free-Living Bacteria | Particle-Attached Bacteria | Cyanobacteria | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
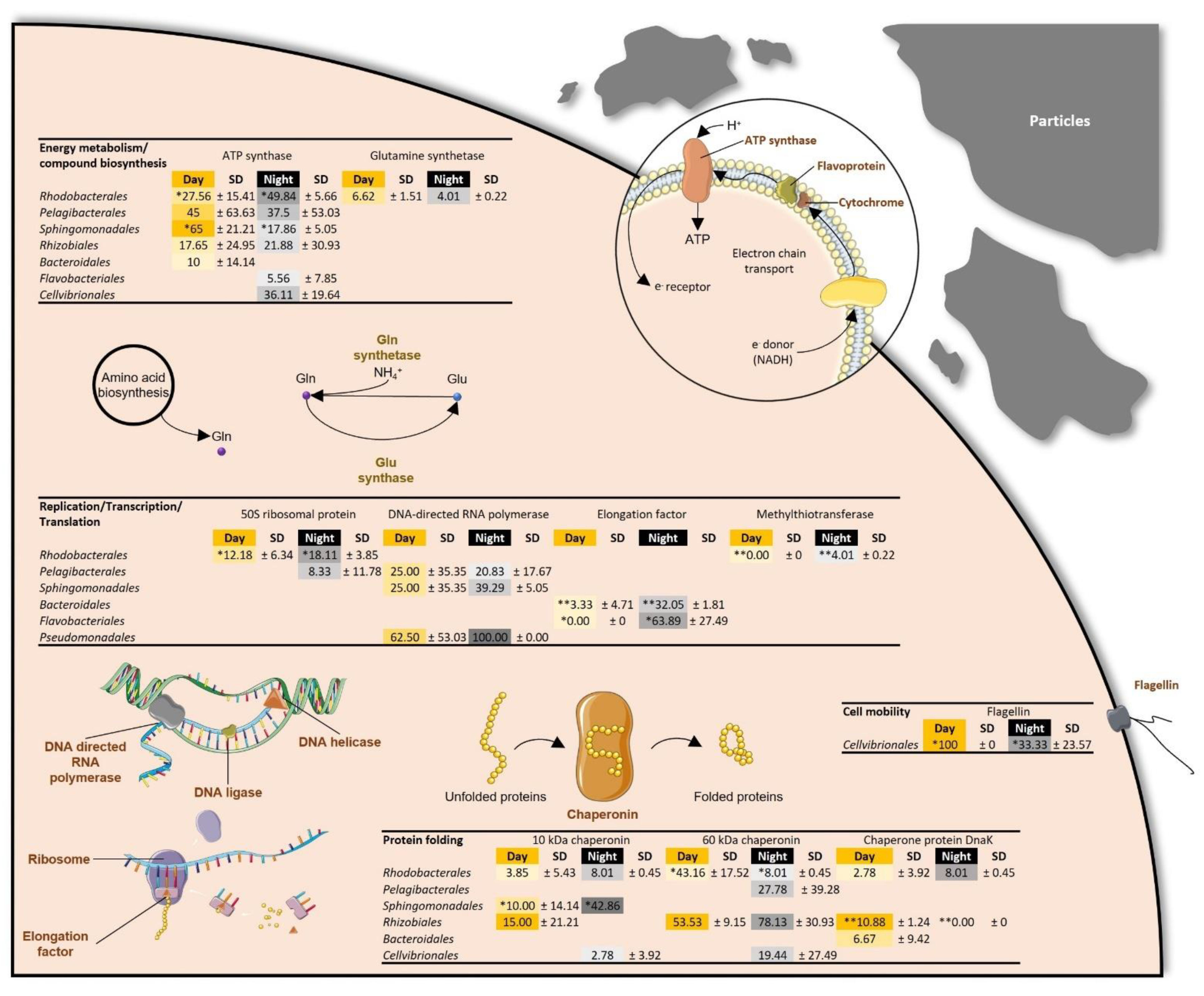
| Protein folding/ Stress response | Day | SD | Night | SD | Day | SD | Night | SD | Day | SD | Night | SD |
| 10 kDa chaperonin | 5.36 | ±0.22 | 5.52 | ±0.74 | 3.48 | ±1.66 | 4.64 | ±1.32 | 0.94 | ±1.33 | 5.91 | ±3.97 |
| 60 kDa chaperonin | 32.46 | ±1.19 | 30.29 | ±5.42 | ** 20.54 | ±6.29 | ** 25.52 | ±6.96 | 13.02 | ±6.48 | 17.02 | ±2.24 |
| ATP-dependent Clp protease proteolytic subunit | 1.11 | ±0.62 | ||||||||||
| Chaperone protein DnaK | 3.29 | ±0.04 | 3.72 | ±0.62 | 5.77 | ±3.22 | 4.01 | ±1.01 | 1.20 | ±1.70 | 1.68 | ±2.37 |
| Cold shock protein | 0.66 | ±0.05 | 0.97 | ±0.31 | ||||||||
| Rubrerythrin | 1.35 | ±0.17 | 1.98 | ±0.84 | ||||||||
| Energy metabolism/Compounds biosynthesis | ||||||||||||
| ATP synthase | 3.43 | ±0.23 | 2.97 | ±0.01 | ** 18.23 | ±6.28 | ** 11.68 | ±5.59 | * 7.75 | ±4.15 | * 16.55 | ±6.96 |
| Aconitate hydratase B | 0.12 | ±0.16 | 1.74 | ±2.46 | ||||||||
| Cysteine synthase | * 0.31 | ±0.44 | * 1.39 | ±0.87 | ||||||||
| Fructose-1,6-bisphosphatase | 1.72 | ±2.42 | ||||||||||
| Glutamine synthetase | 2.56 | ±0.21 | 2.17 | ±0.44 | ** 1.44 | ±1.21 | ** 1.27 | ±1.22 | *2.14 | ±0.36 | * 1.44 | ±0.14 |
| Glyceraldehyde-3-phosphate dehydrogenase | * 0.20 | ±0.04 | * 0.09 | ±0.00 | 3.77 | ±3.70 | 4.35 | ±4.32 | *2.46 | ±0.07 | * 1.78 | ±0.32 |
| Isocitrate dehydrogenase [NADP] | 0.87 | ±1.23 | 2.49 | ±2.30 | ||||||||
| Molybdopterin molybdenumtransferase | 1.73 | ±0.80 | 0.41 | ±0.58 | ||||||||
| Phycoerythrin | * 9.30 | ±4.63 | * 0.33 | ±0.47 | ||||||||
| Allophycocyanin | * 1.54 | ±0.48 | * 0 | |||||||||
| Carbon dioxide-concentrating mechanism protein CcmK | 0.04 | ±0.05 | 0.05 | ±0.07 | 0.94 | ±1.33 | 0.34 | ±0.47 | ||||
| Formate dehydrogenase | 0.72 | ±0.40 | 0.74 | ±0.91 | ||||||||
| Glucose-1-phosphate adenylyltransferase | 0.31 | ±0.44 | 0.72 | ±0.07 | ||||||||
| Replication/Transcription/Translation | ||||||||||||
| 30S ribosomal protein | 2.91 | ±0.39 | 2.66 | ±0.81 | 0.31 | ±0.44 | 1.11 | ±0.62 | ||||
| 50S ribosomal protein | 13.47 | ±1.37 | 12.86 | ±4.44 | 2.32 | ±1.65 | 10.40 | ±7.14 | 2.52 | ±3.55 | 7.23 | ±0.73 |
| DNA-binding protein HU | 7.16 | ±0.09 | 7.69 | ±0.63 | ||||||||
| DNA-directed RNA polymerase | 0.54 | ±0.10 | 0.76 | ±0.23 | 7.77 | ±8.51 | 9.52 | ±4.73 | 0.31 | ±0.44 | 0.67 | ±0.94 |
| Elongation factor | 5.62 | ±0.47 | 6.77 | ±2.98 | 6.94 | ±0.05 | 9.40 | ±3.58 | 7.36 | ±1.51 | 17.43 | ±3.82 |
| Histone-like protein | 0.16 | ±0.10 | 0.15 | ±0.07 | * 11.25 | ±5.22 | * 3.80 | ±1.89 | ||||
| Glycine-tRNA ligase | 1.44 | ±1.21 | 1.25 | ±0.55 | ||||||||
| Ribosomal protein S12 methylthiotransferase RimO | 1.07 | ±1.51 | ||||||||||
| Transport | ||||||||||||
| Amino-acid ABC transporter-binding protein | 5.61 | ±0.74 | 6.31 | ±0.36 | ||||||||
| Fructose import binding protein FrcB | 1.13 | ±0.50 | 1.26 | ±0.60 | ||||||||
| Phosphate-binding protein | 0.28 | ±0.15 | 0.29 | ±0.28 | 44.56 | ±23.88 | 23.09 | ±15.58 | ||||
| Cell motility, structure, and division | ||||||||||||
| Cell division protein FtsZ | 0.94 | ±1.33 | 0.78 | ±1.09 | ||||||||
| Actin-like protein | * 0.70 | ±0.23 | * 0.90 | ±0.15 | ||||||||
| Tubulin | 3.77 | ±2.07 | 2.57 | ±3.04 | ||||||||
| Peptidoglycan-associated lipoprotein | 0.37 | ±0.02 | 0.75 | ±0.63 | ||||||||
| Flagellin | 4.26 | ±0.41 | 5.37 | ±0.40 | 4.91 | ±1.18 | 3.58 | ±0.40 | ||||
| Other (<1%) | 7.60 | 5.72 | 4.03 | 2.30 | 4.03 | 1.40 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Géron, A.; Werner, J.; Lebaron, P.; Wattiez, R.; Matallana-Surget, S. Diel Protein Regulation of Marine Picoplanktonic Communities Assessed by Metaproteomics. Microorganisms 2021, 9, 2621. https://doi.org/10.3390/microorganisms9122621
Géron A, Werner J, Lebaron P, Wattiez R, Matallana-Surget S. Diel Protein Regulation of Marine Picoplanktonic Communities Assessed by Metaproteomics. Microorganisms. 2021; 9(12):2621. https://doi.org/10.3390/microorganisms9122621
Chicago/Turabian StyleGéron, Augustin, Johannes Werner, Philippe Lebaron, Ruddy Wattiez, and Sabine Matallana-Surget. 2021. "Diel Protein Regulation of Marine Picoplanktonic Communities Assessed by Metaproteomics" Microorganisms 9, no. 12: 2621. https://doi.org/10.3390/microorganisms9122621
APA StyleGéron, A., Werner, J., Lebaron, P., Wattiez, R., & Matallana-Surget, S. (2021). Diel Protein Regulation of Marine Picoplanktonic Communities Assessed by Metaproteomics. Microorganisms, 9(12), 2621. https://doi.org/10.3390/microorganisms9122621







