DNA Methylation Patterns Differ between Free-Living Rhizobium leguminosarum RCAM1026 and Bacteroids Formed in Symbiosis with Pea (Pisum sativum L.)
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genome Assembly and Comparison
3.2. Methylation Motif Search
3.3. Methylation System Genes in the RCAM1026 Genome
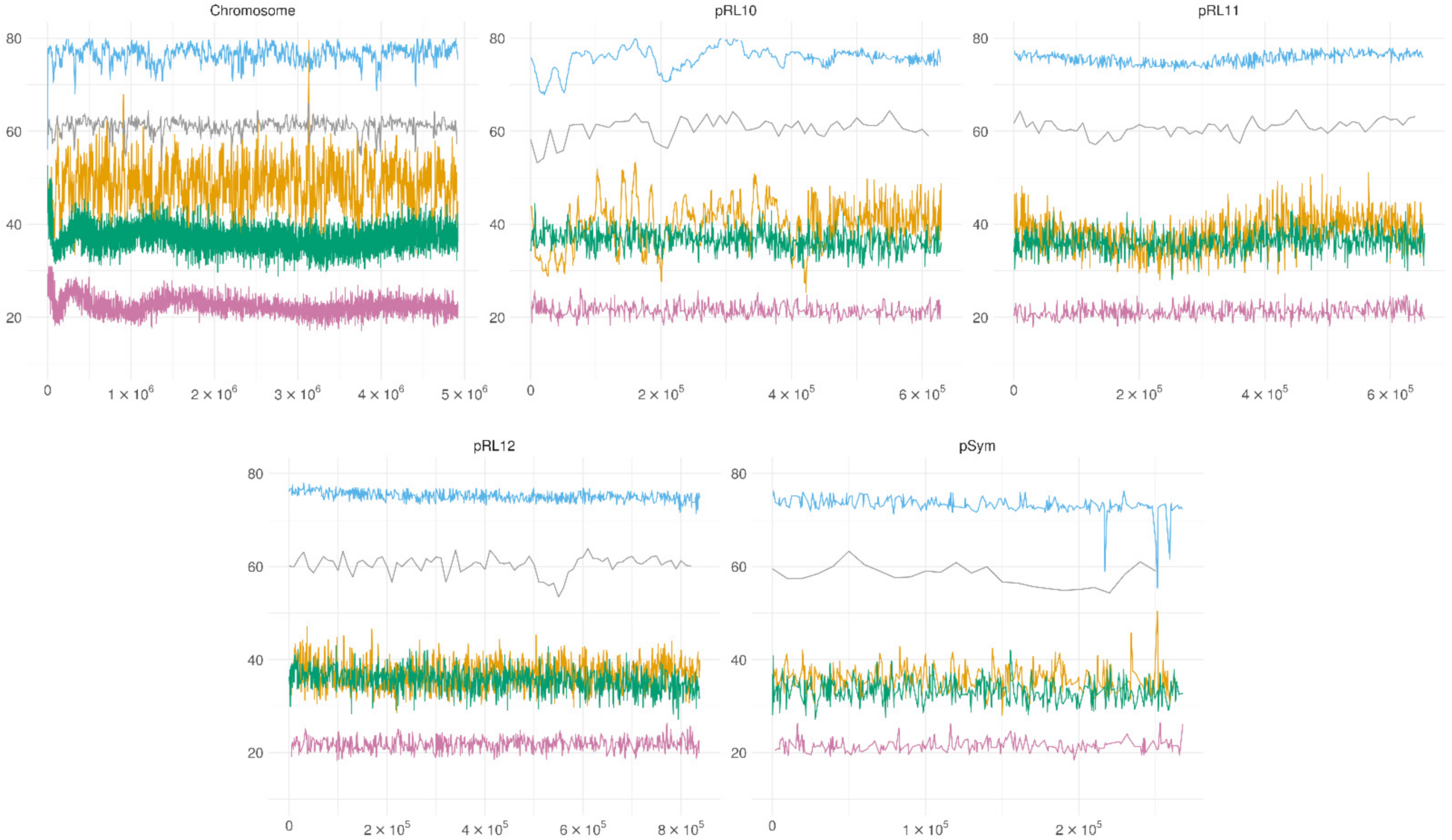
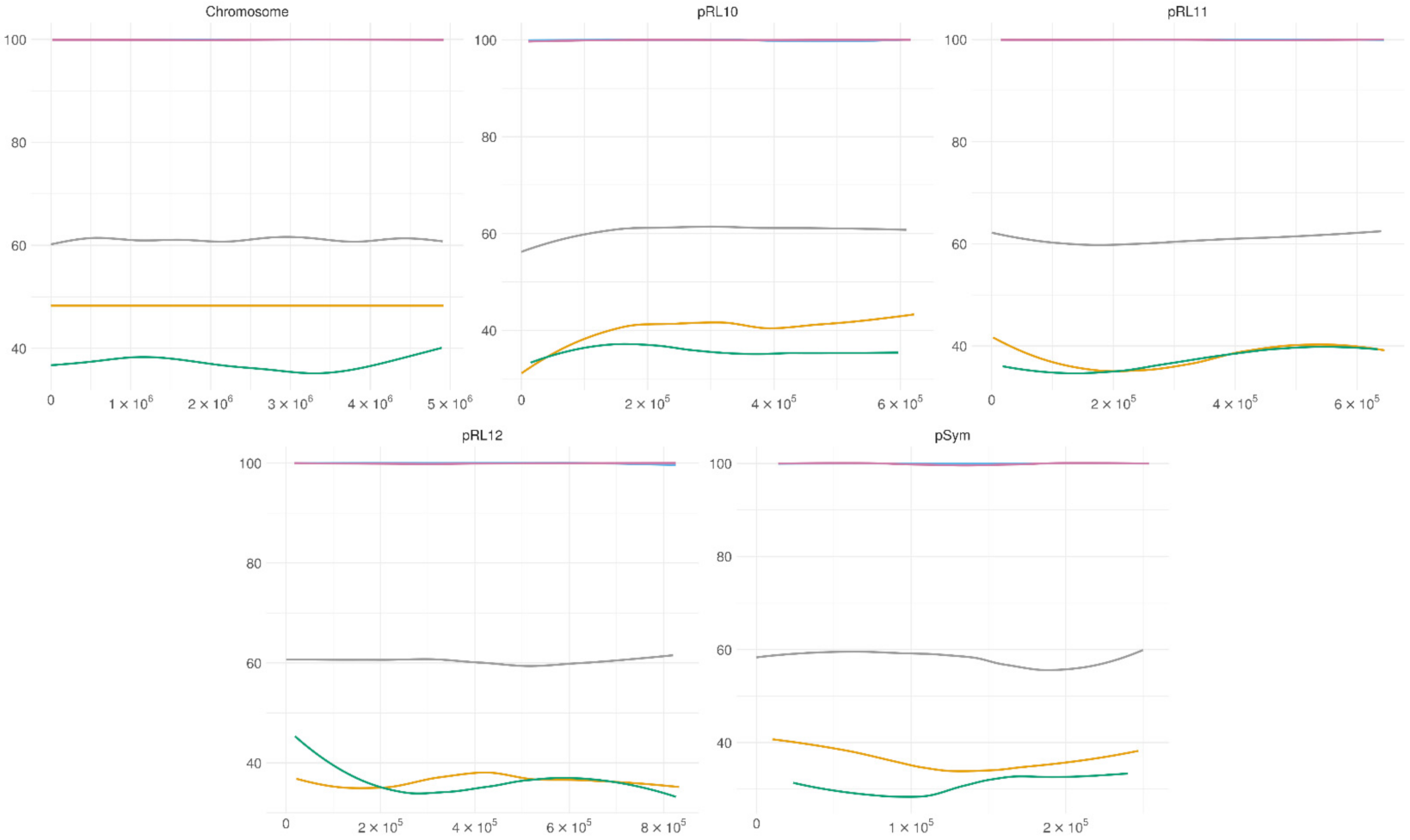
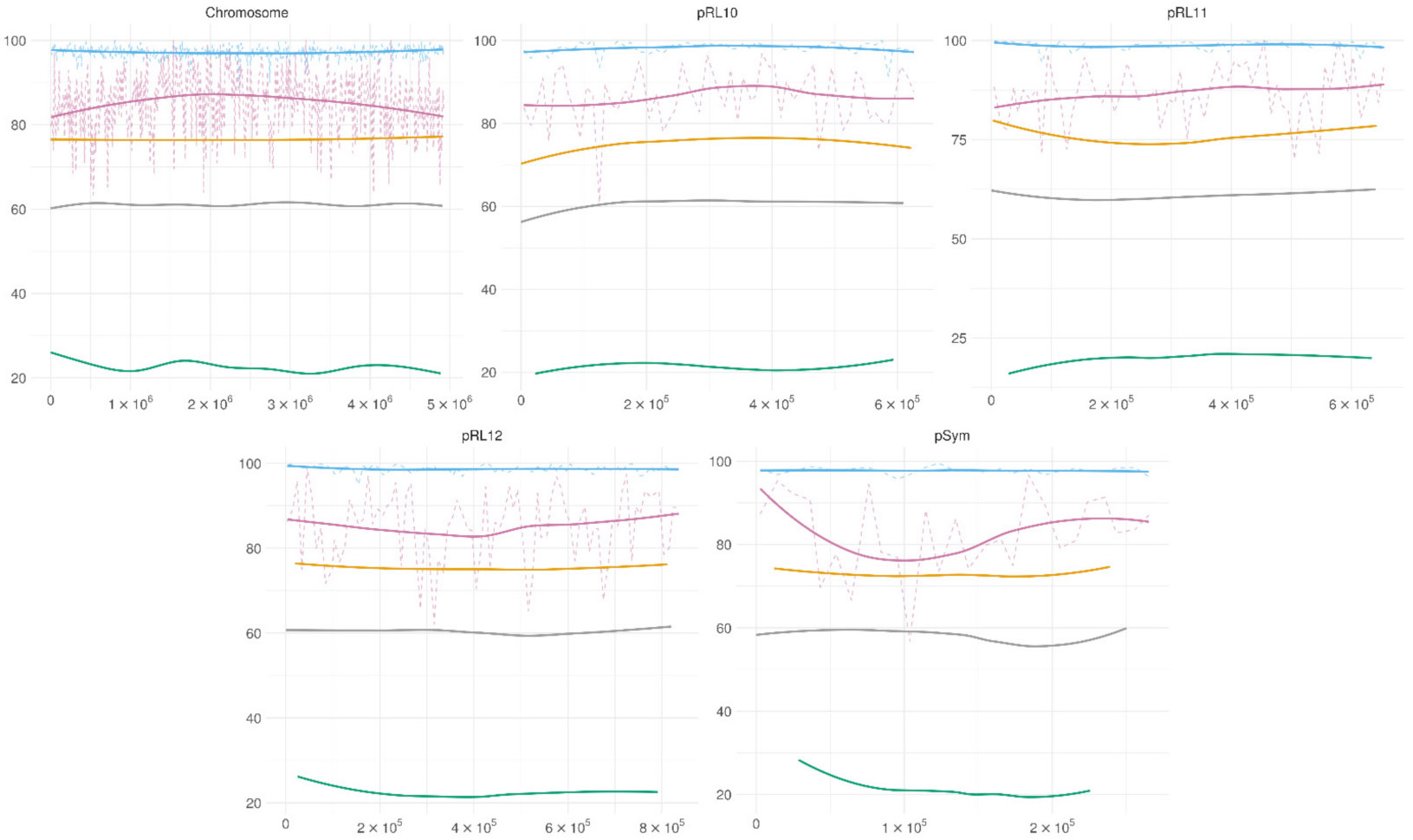
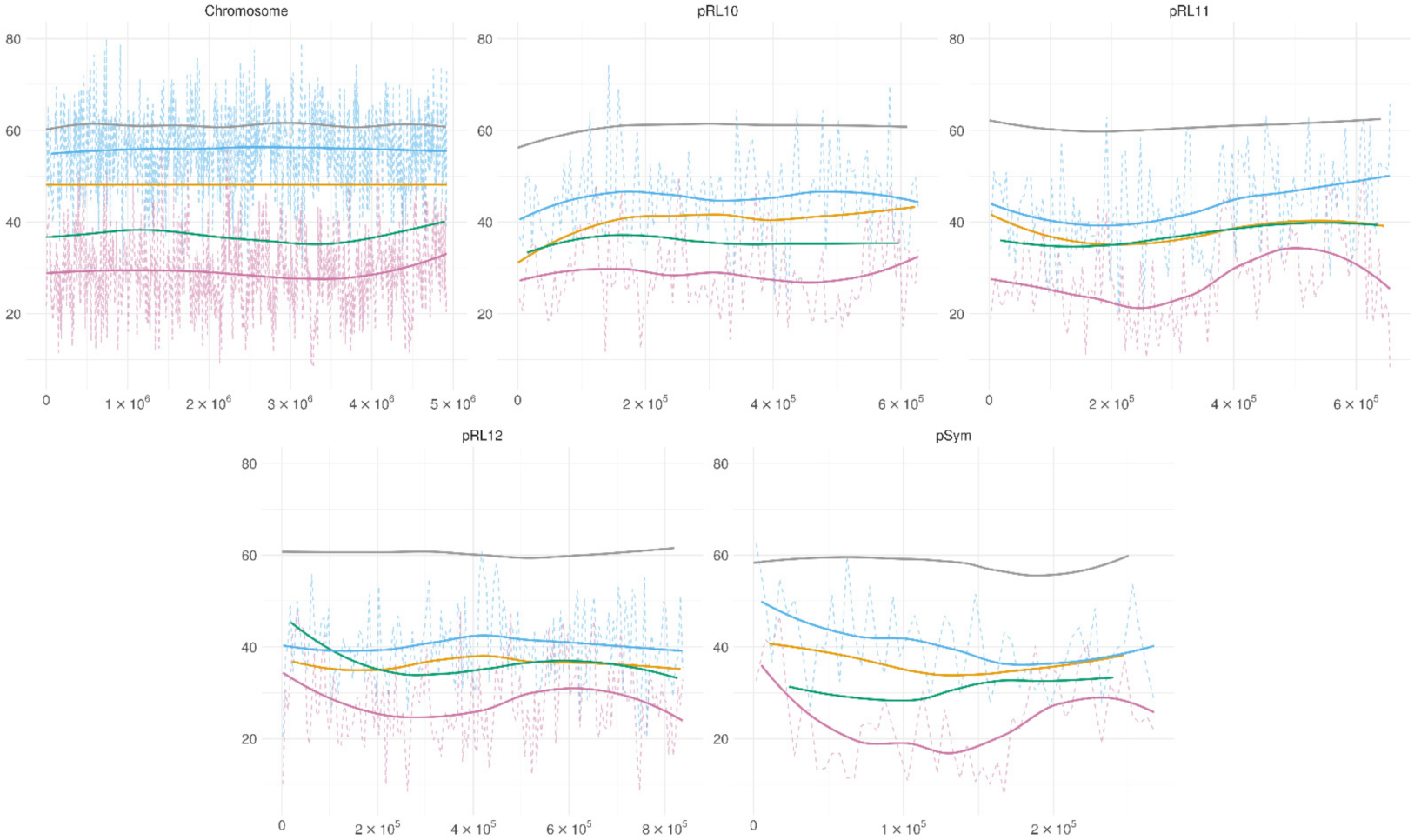
3.4. Genome-Wide Methylation Patterns
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bickle, T.A.; Krüger, D.H. Biology of DNA restriction. Microbiol. Rev. 1993, 57, 434–450. [Google Scholar] [CrossRef] [PubMed]
- Collier, J. Epigenetic regulation of the bacterial cell cycle. Curr. Opin. Microbiol. 2009, 12, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Beaulaurier, J.; Schadt, E.E.; Fang, G. Deciphering bacterial epigenomes using modern sequencing technologies. Nat. Rev. Genet. 2019, 20, 157–172. [Google Scholar] [CrossRef]
- Casadesús, J.; Low, D. Epigenetic Gene Regulation in the Bacterial World. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef] [PubMed]
- Boye, E.; Løbner-Olesen, A.; Skarstad, K. Limiting DNA replication to once and only once. EMBO Rep. 2000, 1, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Løbner-Olesen, A.; Marinus, M.G.; Hansen, F.G. Role of SeqA and Dam in Escherichia coli gene expression: A global/microarray analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 4672–4677. [Google Scholar] [CrossRef] [PubMed]
- Low, D.A.; Casadesús, J. Clocks and switches: Bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 2008, 11, 106–112. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Alexander, N.; Grigorev, K.; Bezdan, D.; Sichtig, H.; Chiu, C.Y.; Mason, C.E. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 2019, 10, 579. [Google Scholar] [CrossRef] [PubMed]
- Spadar, A.; Perdigão, J.; Phelan, J.; Charleston, J.; Modesto, A.; Elias, R.; de Sessions, P.F.; Hibberd, M.L.; Campino, S.; Duarte, A.; et al. Methylation analysis of Klebsiella pneumoniae from Portuguese hospitals. Sci. Rep. 2021, 11, 6491. [Google Scholar] [CrossRef]
- Downie, J.A. Legume nodulation. Curr. Biol. 2014, 24, R184–R190. [Google Scholar] [CrossRef]
- De La Peña, T.C.; Fedorova, E.; Pueyo, J.J.; Lucas, M.M. The Symbiosome: Legume and Rhizobia Co-evolution toward a Nitrogen-Fixing Organelle? Front. Plant Sci. 2018, 8, 2229. [Google Scholar] [CrossRef]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.; Ardley, J.; James, E. From North to South: A latitudinal look at legume nodulation processes. S. Afr. J. Bot. 2013, 89, 31–41. [Google Scholar] [CrossRef]
- Alunni, B.; Gourion, B. Terminal bacteroid differentiation in the legume−rhizobium symbiosis: Nodule-specific cysteine-rich peptides and beyond. New Phytol. 2016, 211, 411–417. [Google Scholar] [CrossRef]
- Ichida, H.; Matsuyama, T.; Abe, T.; Koba, T. DNA adenine methylation changes dramatically during establishment of symbiosis. FEBS J. 2007, 274, 951–962. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davis-Richardson, A.G.; Russell, J.T.; Dias, R.; McKinlay, A.J.; Canepa, R.; Fagen, J.R.; Rusoff, K.T.; Drew, J.C.; Kolaczkowski, B.; Emerich, D.W.; et al. Corrigendum: Integrating DNA Methylation and Gene Expression Data in the Development of the Soybean-Bradyrhizobium N2-Fixing Symbiosis. Front. Microbiol. 2016, 7, 952. [Google Scholar] [CrossRef]
- Dicenzo, G.; Cangioli, L.; Nicoud, Q.; Cheng, J.H.T.; Blow, M.; Shapiro, N.; Woyke, T.; Biondi, E.; Alunni, B.; Mengoni, A.; et al. DNA Methylation in Ensifer Species during Free-Living Growth and during Nitrogen-Fixing Symbiosis with Medicago spp. bioRxiv 2021. [Google Scholar] [CrossRef]
- Afonin, A.; Sulima, A.; Zhernakov, A.; Zhukov, V. Draft genome of the strain RCAM1026 Rhizobium leguminosarum bv. viciae. Genom. Data 2017, 11, 85–86. [Google Scholar] [CrossRef]
- Afonin, A.M.; Leppyanen, I.V.; Kulaeva, O.A.; Shtark, O.Y.; Tikhonovich, I.A.; Dolgikh, E.A.; Zhukov, V.A. A high coverage reference transcriptome assembly of pea (Pisum sativum L.) mycorrhizal roots. Vavilov J. Genet. Breed. 2020, 24, 331–339. [Google Scholar] [CrossRef]
- Catalano, C.M.; Lane, W.S.; Sherrier, D.J. Biochemical characterization of symbiosome membrane proteins fromMedicago truncatula root nodules. Electrophorrsis 2004, 25, 519–531. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology, 5th ed.; John Wiley & Sons: New York, NY, USA, 2002; ISBN 0471250929. [Google Scholar]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Afonin, A.M.; Gribchenko, E.S.; Sulima, A.S.; Zhukov, V.A. Complete Genome Sequence of an Efficient Rhizobium leguminosarum bv. viciae Strain, A1. Microbiol. Resour. Announc. 2020, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.; Coston, R.; Salzberg, S.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; Von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liu, Y.; Jiang, Y.; Li, J.; Gao, Y.; Cui, Z.; Liu, Y.; Liu, B.; Wang, Y. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020, 21, 1–24. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Tourancheau, A.; Mead, E.A.; Zhang, X.-S.; Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 2021, 18, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial Genome Analysis: The COG Approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- REBASE. The Restriction Enzyme Database. Available online: http://rebase.neb.com/rebase/rebase.htm (accessed on 30 August 2021).
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
- Lapidus, A.; Korobeynikov, A. Selected Abstracts of “Bioinformatics: From Algorithms to Applications 2021” Conference. BMC Bioinform. 2021, 21, 1–18. [Google Scholar] [CrossRef]
- Heuer, H.; Abdo, Z.; Smalla, K. Patchy distribution of flexible genetic elements in bacterial populations mediates robustness to environmental uncertainty. FEMS Microbiol. Ecol. 2008, 65, 361–371. [Google Scholar] [CrossRef]
- Wion, D.; Casadesús, J. N6-methyl-adenine: An epigenetic signal for DNA–protein interactions. Nat. Rev. Genet. 2006, 4, 183–192. [Google Scholar] [CrossRef]
- Ibryashkina, E.M.; Zakharova, M.V.; Baskunov, V.B.; Bogdanova, E.S.; Nagornykh, M.O.; Den’Mukhamedov, M.M.; Melnik, B.S.; Kolinski, A.; Gront, D.; Feder, M.; et al. Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct. Biol. 2007, 7, 48. [Google Scholar] [CrossRef]
- Naito, T.; Kusano, K.; Kobayashi, I. Selfish Behavior of Restriction-Modification Systems. Science 1995, 267, 897–899. [Google Scholar] [CrossRef] [PubMed]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
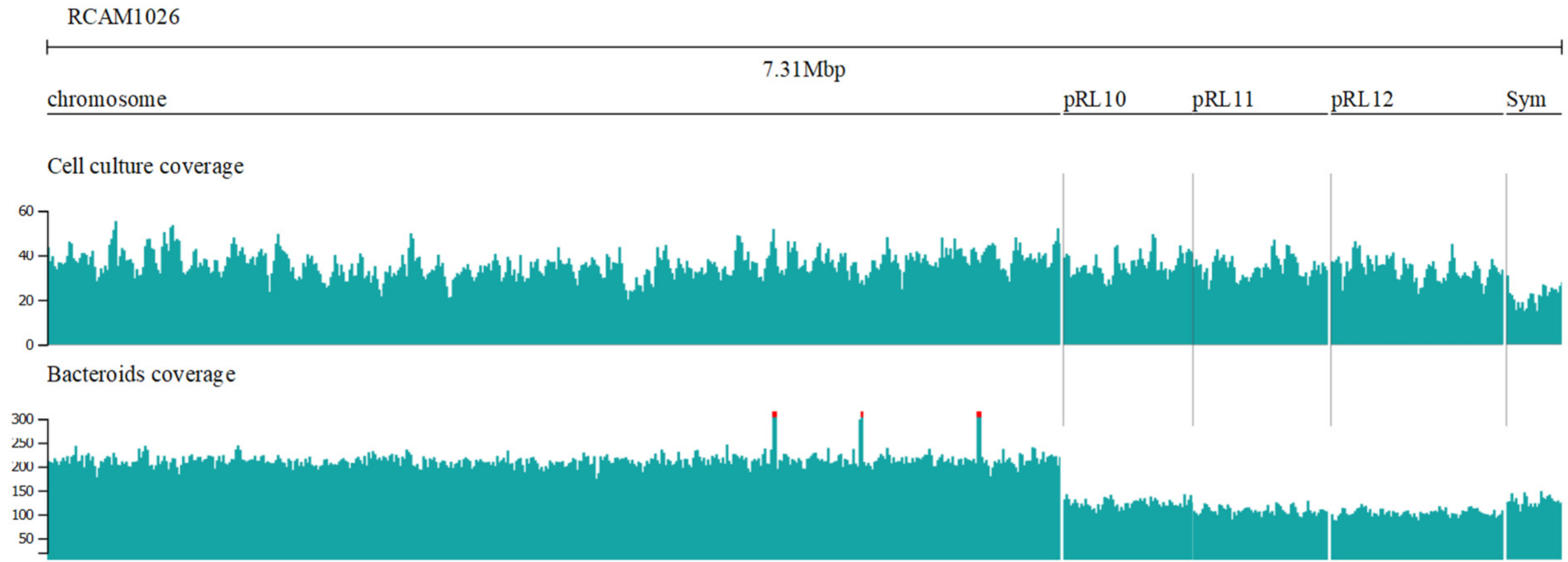
| Chromosome Name | Length | CDS | tRNA Genes | GC Composition | Coverage in Bacteroids | Coverage in Cell Culture 1 |
|---|---|---|---|---|---|---|
| Chromosome | 4,921,456 | 4736 | 51 | 61.09 | 1 | 1 |
| pRL10 | 629,474 | 575 | 2 | 60.61 | 0.91 | 0.58 |
| pRL11 | 655,637 | 625 | - | 60.89 | 0.94 | 0.5 |
| pRL12 | 838,366 | 756 | - | 60.40 | 0.97 | 0.48 |
| Symbiotic plasmid | 268,924 | 263 | - | 58.12 | 0.59 | 0.6 |
| Motif | Characterized Motif | Modified Base | Modified Position | Nanodisco Score |
|---|---|---|---|---|
| GANTC | G6mANTC | 6mA | 2 | 34.16 |
| GATC | GAT4mC | 4mC | 4 | 35.68 |
| GGCGCC | GG4mCGCC | 4mC | 3 | 41.49 |
| Motif | Chromosome | pRL10 | pRL11 | pRL12 | pSym |
|---|---|---|---|---|---|
| GANTC | 6873 (841) 1 | 770 (96) | 778 (136) | 1037 (149) | 406 (65) |
| GATC | 45,690 (6029) | 5963 (633) | 6042 (595) | 7889 (767) | 2264 (252) |
| GGCGCC | 6466 (812) | 756 (45) | 754 (57) | 888 (81) | 213 (23) |
| Gene | Replicon | Putative Enzyme Type | Motif | Homologue in REBASE | Similarity |
|---|---|---|---|---|---|
| 000897 | Chromosome | Methyltransferase | - | M.MspCH12ORF7910P | 54.422 |
| 000974 | Chromosome | Methyltransferase | - | M.Hhe1ORF5290P | 59.823 |
| 000982 | Chromosome | Methyltransferase | GATC | M.MspME121ORFAP | 51.515 |
| 001161 | Chromosome | Methyltransferase | GANTC | M.RleNORF744P | 100.000 |
| 001164 | Chromosome | Nicking endonuclease | - | N.Pec32ORF2247P | 56.267 |
| 001182 | Chromosome | Methyltransferase | GGCGCC | M.CspK31ORF2261P | 70.984 |
| 001183 | Chromosome | Restriction enzyme | - | Avi39ORF4780P | 64.912 |
| 001184 | Chromosome | Restriction enzyme | - | Sma240ORF2946P | 75.431 |
| 001185 | Chromosome | Nicking endonuclease | - | V.OspA1ORF4070P | 69.919 |
| 001185 | Chromosome | Helicase domain protein | - | H.AspSLV7ORF8235P | 82.676 |
| 001515 | Chromosome | Methyltransferase | - | M.CspCJ34ORFGP | 50.882 |
| 001573 | Chromosome | Methyltransferase | - | M.Hhe1ORF5290P | 58.850 |
| 002613 | Chromosome | Orphan methyltransferase | GATC | M.Sen6759Dam | 50.935 |
| 002688 | Chromosome | Methyltransferase | - | M.EcoF3113ORF24645P | 57.277 |
| 003957 | Chromosome | Methyltransferase | - | M.EcoF3113ORF24645P | 68.584 |
| 005751 | pRl11 | Restriction enzyme | GGCGCC | M.SfrNXT3ORF1642P | 89.922 |
| 005752 | pRl11 | Methyltransferase | GGCGCC | SfrNXT3ORF1642P | 92.014 |
| 005755 | pRl11 | Nicking endonuclease | - | V.OspA1ORF4070P | 67.424 |
| 006861 | pRl12 | Methyltransferase | - | M.Hhe1ORF5290P | 60.526 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afonin, A.M.; Gribchenko, E.S.; Zorin, E.A.; Sulima, A.S.; Zhukov, V.A. DNA Methylation Patterns Differ between Free-Living Rhizobium leguminosarum RCAM1026 and Bacteroids Formed in Symbiosis with Pea (Pisum sativum L.). Microorganisms 2021, 9, 2458. https://doi.org/10.3390/microorganisms9122458
Afonin AM, Gribchenko ES, Zorin EA, Sulima AS, Zhukov VA. DNA Methylation Patterns Differ between Free-Living Rhizobium leguminosarum RCAM1026 and Bacteroids Formed in Symbiosis with Pea (Pisum sativum L.). Microorganisms. 2021; 9(12):2458. https://doi.org/10.3390/microorganisms9122458
Chicago/Turabian StyleAfonin, Alexey M., Emma S. Gribchenko, Evgeny A. Zorin, Anton S. Sulima, and Vladimir A. Zhukov. 2021. "DNA Methylation Patterns Differ between Free-Living Rhizobium leguminosarum RCAM1026 and Bacteroids Formed in Symbiosis with Pea (Pisum sativum L.)" Microorganisms 9, no. 12: 2458. https://doi.org/10.3390/microorganisms9122458
APA StyleAfonin, A. M., Gribchenko, E. S., Zorin, E. A., Sulima, A. S., & Zhukov, V. A. (2021). DNA Methylation Patterns Differ between Free-Living Rhizobium leguminosarum RCAM1026 and Bacteroids Formed in Symbiosis with Pea (Pisum sativum L.). Microorganisms, 9(12), 2458. https://doi.org/10.3390/microorganisms9122458






