Subgingival Microbiota and Cytokines Profile Changes in Patients with Periodontitis: A Pilot Study Comparing Healthy and Diseased Sites in the Same Oral Cavities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Inclusion/Exclusion Criteria
2.3. Clinical Examination
2.4. Gingival Crevicular Fluid (GCF) and Subgingival Plaque (SP) Samples Collection
2.5. Cytokine Quantification by Multiplexed Bead Immunoassay
2.6. Microbial Community Analysis
2.6.1. Bacterial 16S rRNA Gene Amplification and Sequencing
2.6.2. 16S Sequencing Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Cytokine Profile Is Altered in Diseased Sites Compared to Healthy Sites
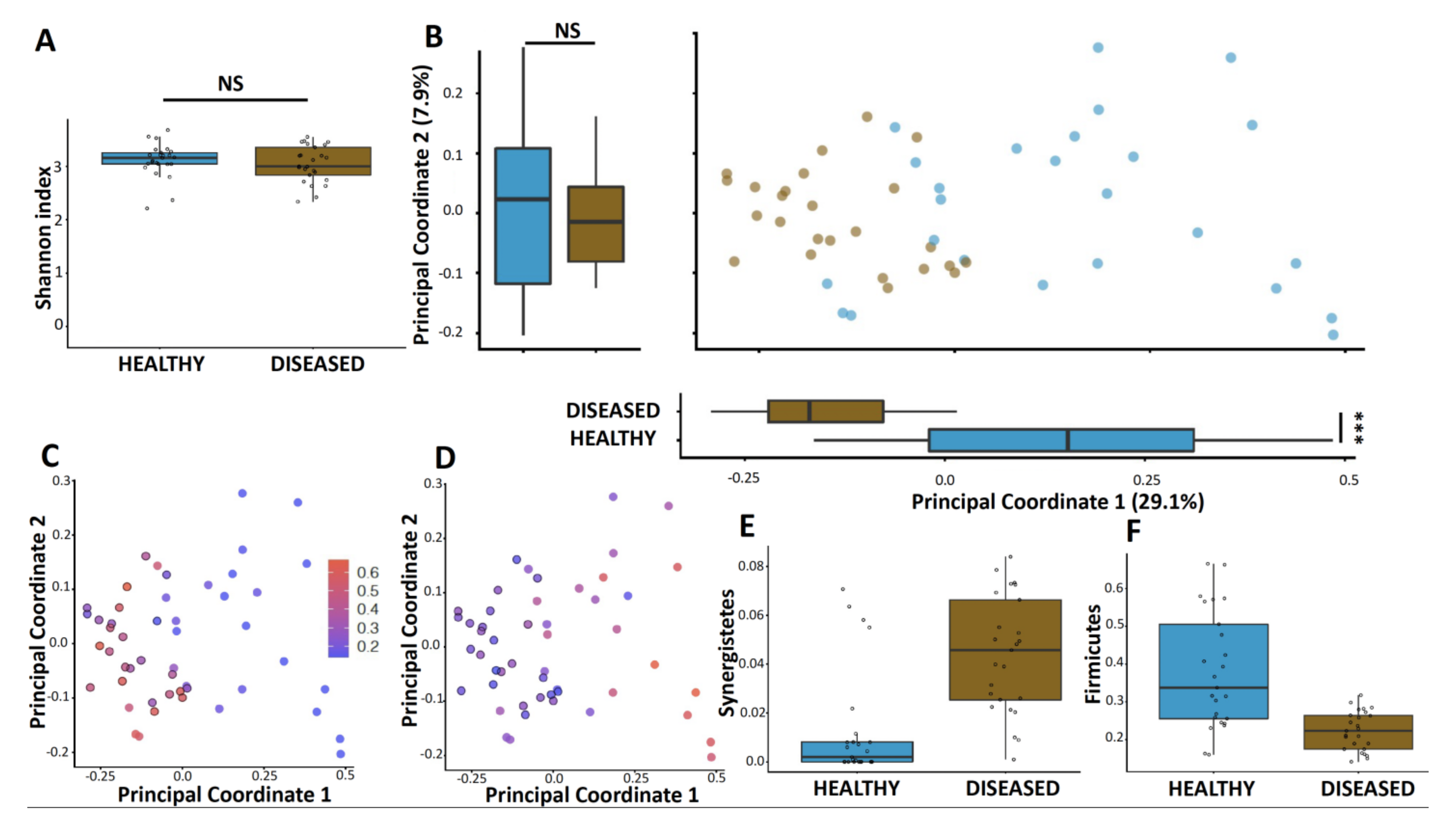
3.2. Microbiome Diversity Is Altered in Diseased Sites Compared to Healthy Sites
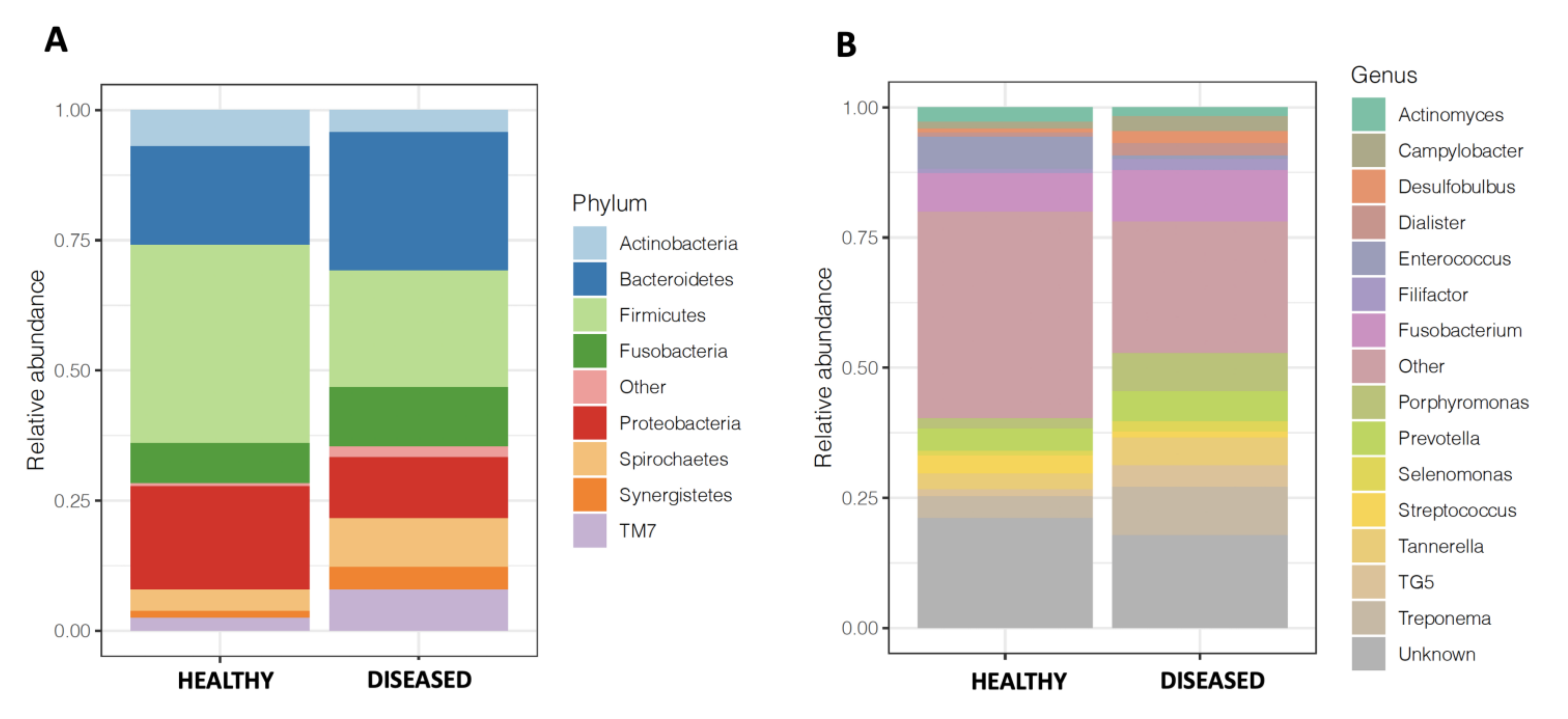
3.3. Microbiome of Diseased Sites Was Enriched in Synergistetes and Bacteroidetes and Depleted in Firmicutes and Proteobacteria
3.4. Prediction of Functional Profiles Using 16S rRNA Data in Diseased Sites Compared to Healthy Sites
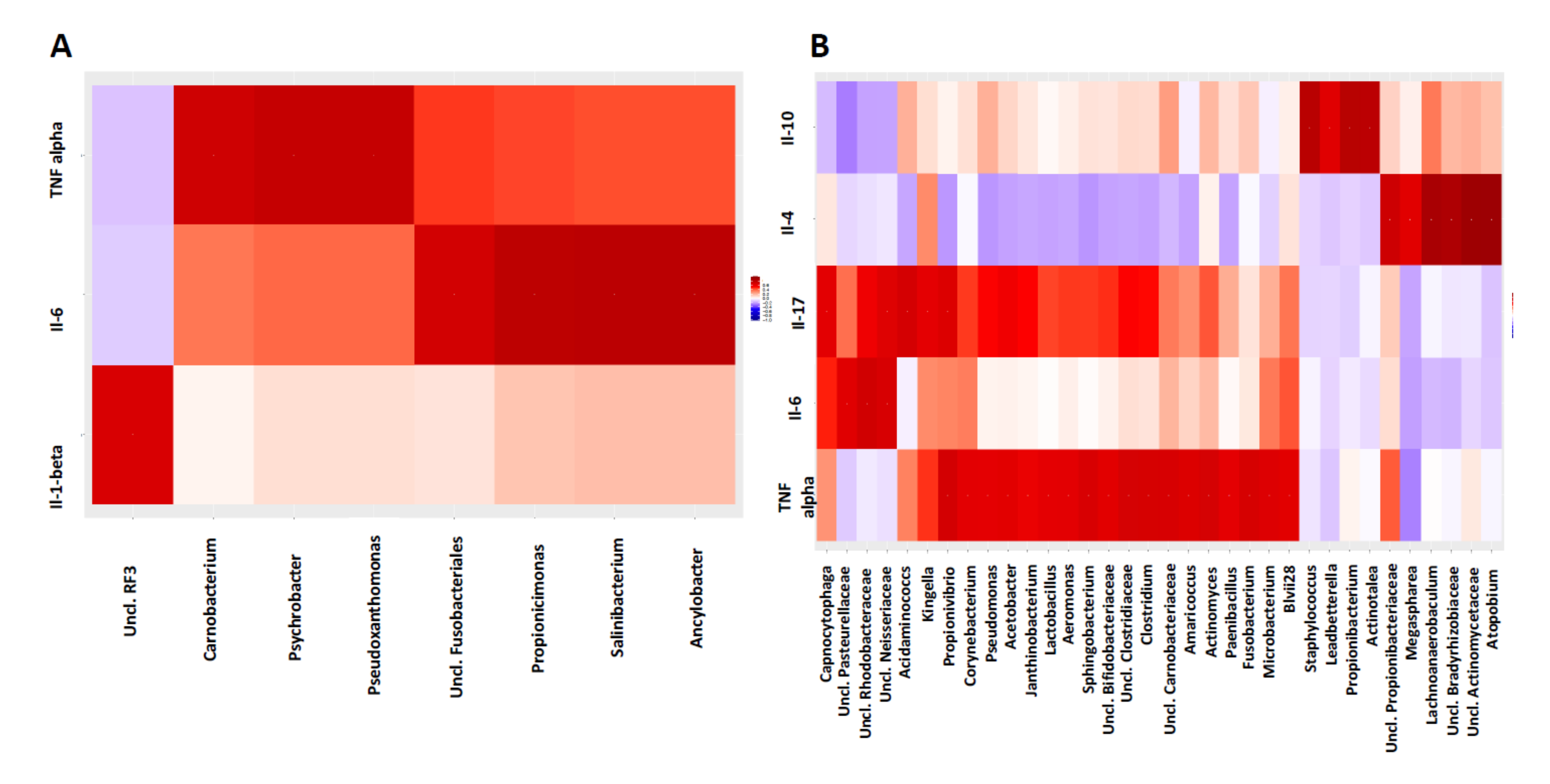
3.5. Host-Microbiome Interaction Is Altered in Diseased Sites Compared to Healthy Sites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petersen, P.E.; Ogawa, H. The global burden of periodontal disease: Towards integration with chronic disease prevention and control. Periodontology 2000 2012, 60, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Armitage, G.C. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 1999, 4, 1–6. [Google Scholar] [CrossRef]
- Armitage, G.C. Classifying periodontal diseases—A long-standing dilemma. Periodontology 2002, 30, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemming, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef] [Green Version]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Hemme, C.; Beleno, J.; Shi, Z.J.; Ning, D.; Qin, Y.; Tu, Q.; Jorgensen, M.; He, Z.; Wu, L. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018, 12, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
- Meuric, V.; Le Gall-David, S.; Boyer, E.; Acuña-Amador, L.; Martin, B.; Fong, S.B.; Barloy-Hubler, F.; Bonnaure-Mallet, M. Signature of microbial dysbiosis in periodontitis. Appl. Environ. Microbiol. 2017, 83, e00462–e00517. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Yao, Y.; Jiao, K.; Zhang, J.; Zheng, X.; Wu, F.; Hu, X.; Li, J.; Yu, Z.; Zhang, G. Relationship between gingival crevicular fluid microbiota and cytokine profile in periodontal host homeostasis. Front. Microbiol. 2017, 8, 2144. [Google Scholar] [CrossRef]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M. A new classification scheme for periodontal and peri-implant diseases and conditions–Introduction and key changes from the 1999 classification. J. Periodontol. 2018, 89, S1–S8. [Google Scholar] [CrossRef]
- Bostanci, N.; Emingil, G.; Afacan, B.; Han, B.; Ilgenli, T.; Atilla, G.; Hughes, F.J.; Belibasakis, G.N. Tumor necrosis factor-α-converting enzyme (TACE) levels in periodontal diseases. J. Dent. Res. 2008, 87, 273–277. [Google Scholar] [CrossRef]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. Gigascience 2012, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 15 November 2021).
- Wickham, H. ggplot2. Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 15 November 2021).
- Oksanen, J. Vegan: Community Ecology Package. R Package Version 1.17-9. 2011. Available online: https://cran.r-project.org/package=vegan (accessed on 15 November 2021).
- Lahti, L.; Shetty, S.; Blake, T.; Salojarvi, J. Tools for Microbiome Analysis in R. Version 1:504. 2017. Available online: https://microbiome.github.io/tutorials/ (accessed on 15 November 2021).
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [Green Version]
- McDonald, A.G.; Boyce, S.; Tipton, K.F. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009, 37, D593–D597. [Google Scholar] [CrossRef]
- Teles, R.; Moss, K.; Preisser, J.S.; Genco, R.; Giannobile, W.V.; Corby, P.; Garcia, N.; Jared, H.; Torresyap, G.; Salazar, E. Patterns of periodontal disease progression based on linear mixed models of clinical attachment loss. J. Clin. Periodontol. 2018, 45, 15–25. [Google Scholar] [CrossRef]
- Stadler, A.F.; Angst, D.M.; Arce, R.M.; Gomes, S.C.; Oppermann, R.V.; Susin, C. Gingival crevicular fluid levels of cytokines/chemokines in chronic periodontitis: A meta-analysis. J. Clin. Periodontol. 2016, 43, 727–745. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, X.; Li, R. Genetic Relationship Between IL-6 rs1800796 Polymorphism and Susceptibility to Periodontitis. Immunol. Investig. 2019, 48, 268–282. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Wu, X.; Sun, W. TNF-α polymorphisms might influence predisposition to periodontitis: A meta-analysis. Microb. Pathog. 2020, 143, 104113. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Ji, X.; Chen, X.; Xu, Y.; Zhong, L. TNF-α gene promoter polymorphisms contribute to periodontitis susceptibility: Evidence from 46 studies. J. Clin. Periodontol. 2014, 41, 748–759. [Google Scholar] [CrossRef]
- Van Dyke, T.E. Pro-resolving mediators in the regulation of periodontal disease. Mol. Asp. Med. 2017, 58, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Kistler, J.O.; Booth, V.; Bradshaw, D.J.; Wade, W.G. Bacterial community development in experimental gingivitis. PLoS ONE 2013, 8, e71227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camelo-Castillo, A.J.; Mira, A.; Pico, A.; Nibali, L.; Henderson, B.; Donos, N.; Tomás, I. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front. Microbiol. 2015, 6, 119. [Google Scholar] [CrossRef] [Green Version]
- Patini, R.; Staderini, E.; Lajolo, C.; Lopetuso, L.; Mohammed, H.; Rimondini, L.; Rocchetti, V.; Franceschi, F.; Cordaro, M.; Gallenzi, P. Relationship between oral microbiota and periodontal disease: A systematic review. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5775–5788. [Google Scholar]
- Cross, K.L.; Chirania, P.; Xiong, W.; Beall, C.J.; Elkins, J.G.; Giannone, R.J.; Griffen, A.L.; Guss, A.M.; Hettich, R.L.; Joshi, S.S. Insights into the evolution of host association through the isolation and characterization of a novel human periodontal pathobiont, Desulfobulbus oralis. MBio 2018, 9, e02061-17. [Google Scholar] [CrossRef] [Green Version]
- Aruni, A.W.; Roy, F.; Fletcher, H. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 2011, 79, 3872–3886. [Google Scholar] [CrossRef] [Green Version]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012, 6, 1176–1185. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Zang, S.; Wei, J.; Yu, H.; Yang, Z.; Wu, H.; Kang, Y.; Tao, H.; Yang, M.; Jin, L. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021, 113, 664–676. [Google Scholar] [CrossRef]
- Oliveira, R.; Fermiano, D.; Feres, M.; Figueiredo, L.; Teles, F.; Soares, G.; Faveri, M. Levels of candidate periodontal pathogens in subgingival biofilm. J. Dent. Res. 2016, 95, 711–718. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neut, C.; Guillemot, F.; Colombel, J.F. Nitrate-Reducing Bacteria in Diversion Colitis (A Clue to Inflammation?). Dig. Dis. Sci. 1997, 42, 2577–2580. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Qiao, S.; Shen, W.; Liu, Y.; Lu, Y.; Liangyu, H.; Guo, Z.; Gong, J.; Shui, G.; Li, Y. Disturbance of Fatty Acid Desaturation Mediated by FADS2 in Mesenteric Adipocytes Contributes to Chronic Inflammation of Crohn’s Disease. J. Crohn’s Colitis 2020, 14, 1581–1599. [Google Scholar] [CrossRef] [PubMed]
- BinShabaib, M.; ALHarthi, S.S.; Akram, Z.; Khan, J.; Rahman, I.; Romanos, G.E.; Javed, F. Clinical periodontal status and gingival crevicular fluid cytokine profile among cigarette-smokers, electronic-cigarette users and never-smokers. Arch. Oral Biol. 2019, 102, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.L.; Haffajee, A.D.; Socransky, S.S. Effects of periodontitis and smoking on the microbiota of oral mucous membranes and saliva in systemically healthy subjects. J. Clin. Periodontol. 2003, 30, 1031–1037. [Google Scholar] [CrossRef]
| Demographic and Clinical Parameters | Patients Characteristics (N = 24) |
|---|---|
| Age Mean ± SD (Min-Max) | 49.70 ± 13.26 (28–75) |
| BMI Mean ± SD (Min-Max) | 25.77 ± 4.17 (19.4–36.5) |
| Sex % Men (n/N) | 54.2% (13/24) |
| Teeth Number Mean ± SD (Min-Max) | 25.75 ± 3.29 (19–31) |
| Bacterial Plaque Index (FMPS) Mean ± SD (Min-Max) | 47.58 ± 18.41 (11–78) |
| Bleeding Index (FMBS) Mean ± SD (Min-Max) | 23.04 ± 10.57 (1–42) |
| Pocket Depth in mm Mean ± SD (Min-Max) | 3.67 ± 0.60 (2.6–5.3) |
| Clinical Attachment Level in mm Mean ± SD (Min-Max) | 4.57 ± 1.23 (3.2–8) |
| Tobacco Consumption Mean ± SD (Min-Max) | 8.7 ± 8.87 (0.29-30) |
| Periodontal Disease Severity (According PAPAPANOU Classification) % (n/N) | Stage III grade B—71% (17/24) |
| Stage III grade C—12.5% (3/24) | |
| Stage IV grade B—8.3% (2/24) | |
| Stage IV grade C—8.3% (2/24) |
| Cytokines Median (Min-Max) | Site/Ratio | Tobacco (N = 11) | No Tobacco (N = 13) | p-Value |
|---|---|---|---|---|
| IL-10 | Healthy | 4.76 (1.7–5.9) | 4.95 (3.2–6.3) | >0.05 |
| Diseased | 5.93 (4.8–15.1) | 6.71 (4.4–9.1) | >0.05 | |
| D/H ratio | 1.61 (1–4.7) | 1.29 (0.9–2.1) | >0.05 | |
| IL-17A | Healthy | 1.02 (0.2–2) | 1.12 (0.4–2.3) | >0.05 |
| Diseased | 1.31 (0.8–3.2) | 1.71 (1–4.8) | >0.05 | |
| D/H ratio | 1.41 (0.7–5.5) | 1.90 (1–11.4) | >0.05 | |
| IL-1β | Healthy | 193.4 (5.1–673.9) | 62.09 (3.3–449.5) | <0.05 |
| Diseased | 679.94 (322.7–911.8) | 382.75 (122.9–836.9) | <0.05 | |
| D/H ratio | 3.52 (0.7–179.1) | 6.28 (1.2–79.9) | >0.05 | |
| IL-4 | Healthy | 2.74 (2.4–3.8) | 2.40 (2.4–3.8) | >0.05 |
| Diseased | 3.09 (3–4.5) | 3.09 (2.4–9.5) | >0.05 | |
| D/H ratio | 1.28 (0.8–1.5) | 1.29 (1–3.9) | >0.05 | |
| IL-6 | Healthy | 1.76 (0.7–6.4) | 3.52 (0.6–40.1) | >0.05 |
| Diseased | 2.4 (0.6–15.6) | 2 (0.9–36.6) | >0.05 | |
| D/H ratio | 0.87 (0.2–21.4) | 0.65 (0.3–3.9) | >0.05 | |
| TNF-Alpha | Healthy | 2.91 (1.3–10.4) | 4.90 (2.2–25.7) | <0.05 |
| Diseased | 1.46 (2.1–15.7) | 6.31 (1.3–27.6) | >0.05 | |
| D/H ratio | 1.80 (0.3–6.7) | 0.64 (0.2–7.1) | >0.05 |
| Cytokines Median (Min-Max) | Healthy Sites (N = 24) | Diseased Sites (N = 24) | p-Value |
|---|---|---|---|
| IL-10 | 4.86 (1.7–6.3) | 6.71 (4.4–15.1) | <0.05 |
| IL-17a | 1.02 (0.2–2.3) | 1.53 (0.8–4.8) | <0.05 |
| IL-1β | 68.59 (3.3–673,9) | 500.26 (122.9–911.8) | <0.05 |
| IL-4 | 2.4 (2.4–3.8) | 3.09 (2.4–9.5) | <0.05 |
| IL-6 | 2.12 (0.6–40.1) | 2.20 (0.6–36.6) | >0.05 |
| TNFα | 4.54 (1.3–25.7) | 4.85 (1.3–27.5) | >0.05 |
| Concentration of Total Cytokines | 84.27 (14.7–685.4) | 529.24 (174.8–933.6) | <0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esparbès, P.; Legrand, A.; Bandiaky, O.N.; Chéraud-Carpentier, M.; Martin, H.; Montassier, E.; Soueidan, A. Subgingival Microbiota and Cytokines Profile Changes in Patients with Periodontitis: A Pilot Study Comparing Healthy and Diseased Sites in the Same Oral Cavities. Microorganisms 2021, 9, 2364. https://doi.org/10.3390/microorganisms9112364
Esparbès P, Legrand A, Bandiaky ON, Chéraud-Carpentier M, Martin H, Montassier E, Soueidan A. Subgingival Microbiota and Cytokines Profile Changes in Patients with Periodontitis: A Pilot Study Comparing Healthy and Diseased Sites in the Same Oral Cavities. Microorganisms. 2021; 9(11):2364. https://doi.org/10.3390/microorganisms9112364
Chicago/Turabian StyleEsparbès, Pauline, Arnaud Legrand, Octave Nadile Bandiaky, Marjorie Chéraud-Carpentier, Hamida Martin, Emmanuel Montassier, and Assem Soueidan. 2021. "Subgingival Microbiota and Cytokines Profile Changes in Patients with Periodontitis: A Pilot Study Comparing Healthy and Diseased Sites in the Same Oral Cavities" Microorganisms 9, no. 11: 2364. https://doi.org/10.3390/microorganisms9112364
APA StyleEsparbès, P., Legrand, A., Bandiaky, O. N., Chéraud-Carpentier, M., Martin, H., Montassier, E., & Soueidan, A. (2021). Subgingival Microbiota and Cytokines Profile Changes in Patients with Periodontitis: A Pilot Study Comparing Healthy and Diseased Sites in the Same Oral Cavities. Microorganisms, 9(11), 2364. https://doi.org/10.3390/microorganisms9112364






