Development and Evaluation of Alternative Methods to Identify the Three Most Common Serotypes of Salmonella enterica Causing Clinical Infections in Kazakhstan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms Strains
2.2. Salmonella Isolates Used for PCR Method Validation
2.3. Isolation and Identification of Salmonella
2.4. DNA Extraction
2.5. Specific Primers and Probes
2.6. Real-Time Polymerase Chain Reaction
2.7. PCR
2.8. Randomly Amplified Polymorphic DNA (RAPD) PCR
2.9. Electrophoretic Analysis of DNA Amplification Products
2.10. Statistical Analysis
3. Results
3.1. Specificity, Sensitivity, and Efficacy of Real-Time PCR and Conventional PCR
3.2. Typing of Salmonella enterica Strains by RAPD PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, S.S.; Pendrak, M.L.; Abela-Ridder, B.; Punderson, J.W.; Fedorko, D.P.; Foley, S.L. An overview of Salmonella typing: public health perspectives. Clin. Appl. Immunol. Rev. 2003, 4, 189–204. [Google Scholar] [CrossRef]
- Zhao, S.; McDermott, P.F.; Friedman, S.; Abbott, J.; Ayers, S.; Glenn, A.; Hall-Robinson, E.; Hubert, S.K.; Harbottle, H.; Walker, R.D.; et al. Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog Dis. 2006, 3, 106–117. [Google Scholar] [CrossRef]
- Messens, W.; Grijspeerdt, K.; De Reu, K.; De Ketelaere, B.; Mertens, K.; Bamelis, F.; Kemps, B.; De Baerdemaeker, J.; Decuypere, E.; Herman, L. Eggshell penetration of various types of hens’ eggs by Salmonella enterica serovar Enteritidis. J. Food Prot. 2007, 70, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars. 9th Edition, World Health Organization Collaborating Center for Reference and Research on Salmonella. Institut. Pasteur. Paris 2007, 9, 3. [Google Scholar]
- Guibourdenche, M.; Roggentin, P.; Mikoleit, M.; Fields, P.I.; Bockemuhl, J.; Grimont, P.A.; Weill, F.X. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2010, 161, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Vieira, A.R.; Karlsmose, S.; Lo Fo Wong, D.M.; Jensen, A.B.; Wegener, H.C.; Aarestrup, F.M. Global monitoring of Salmonella serovar distribution from the world health organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011, 8, 887–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herikstad, H.; Motarjemi, Y.; Tauxe, R.V. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 2002, 129, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Smagulova, M.K.; Sataeva, A.M. Analysis of the incidence of intestinal infections in the Republic of Kazakhstan for 2019, tasks for 2020. Environ. Public Health 2020, 1, 24–32. [Google Scholar]
- Yasin, R.M.; Tiew, C.C.; Jegathesan, M. Human salmonellosis in Malaysia for the period 1989–July 1994. Southeast Asian J. Trop. Med. Public Health 1995, 26, 457–460. [Google Scholar]
- Turki, Y.; Mehri, I.; Fhoula, I.; Hassen, A.; Ouzari, H. Comparison of five molecular subtyping methods for differentiation of Salmonella Kentucky isolates in Tunisia. World J. Microbiol. Biotechnol. 2014, 30, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Interstate standard GOST 31904-2012 Food products. Sampling methods for microbiological analyses. Available online: https://internet-law.ru/gosts/gost/53156/ (accessed on 1 July 2013).
- U.S. Food and Drug Administration. Food Sampling and Preparation of Sample Homogenate, Chap. 1. In Bacteriological Analytical Manual. Available online: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm063335.htm (accessed on 15 June 2016).
- Sanitary rules “Sanitary and epidemiological requirements for laboratories using potentially hazardous chemical and biologi-cal substances”. Order of the Minister of Health of the Republic of Kazakhstan. Available online: https://adilet.zan.kz/rus/docs/V1700015990 (accessed on 15 October 2021).
- Laboratory Diagnostics of Salmonella, Detection of Salmonella in Food and Environmental Objects: Guidelines—M.: Federal Center for Hygiene and Epidemiology of Rospotrebnadzor. 2011. 111p. Available online: https://files.stroyinf.ru/Data2/1/4293790/4293790005.pdf (accessed on 2 February 2010).
- Interstate Standard GOST 31659-2012 (ISO 6579: 2002) Food Products. Method for the Detection of Bacteria of the Genus Salmonella (ISO 6579: 2002, MOD). Available online: https://docs.cntd.ru/document/1200098239 (accessed on 1 July 2013).
- U.S. Food and Drug Administration. Bacteriological Analytical Manual (BAM) Chapter 5: Salmonella. Available online: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm (accessed on 15 December 2015).
- Clavijo, R.I.; Loui, C.; Andersen, G.L.; Riley, L.W.; Lu, S. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl. Environ. Microbiol. 2006, 72, 1055–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.S.; Tsen, H.Y. Development and Use of Polymerase Chain Reaction for the Specific Detection of Salmonella Typhimurium in Stool and Food Samples. J. Food Protection. 1999, 62, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, N.L.; Petty, N.K.; Zakour, N.L.B.; Szubert, J.M.; Savill, J.; Beatson, S.A. Genome analysis and CRISPR typing of Salmonella enterica serovar Virchow. BMC Genomics 2014, 15, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Engelsdorp, D.; Lengerich, E.; Spleen, A.; Dainat, B.; Cresswell, J.; Baylis, K.; Nguyen, B.K.; Soroker, V.; Underwood, R.; Human, H. Standard epidemiological methods to understand and improve Apis mellifera health. J. Apic. Res. 2013, 52, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wilson, E.B. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 1927, 22, 209–212. [Google Scholar] [CrossRef]
- Malorny, B.; Bunge, C.; Hoorfar, J.; Helmuth, R. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microb. 2003, 69, 290–296. [Google Scholar] [CrossRef] [Green Version]
- Kasturi, K.N.; Drgon, T. Real-Time PCR Method for Detection of Salmonella spp. in Environmental Samples. Appl. Environ. Microbiol. 2017, 83, e00644-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, I.; Flekna, G.; Krassnig, M.; Wagner, M. Real-time PCR for the detection of Salmonella spp. in food: An alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 2006, 66, 538–547. [Google Scholar] [CrossRef]
- Malorny, B.; Hoorfar, J.; Hugas, M.; Heuvelink, A.; Fach, P.; Ellerbroek, L.; Bunge, C.; Dorn, C.; Helmuth, R. Inter-laboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 2003, 89, 241–249. [Google Scholar] [CrossRef]
- Siala, M.; Barbana, A.; Smaoui, S.; Hachicha, S.; Marouane, C.; Kammoun, S.; Gdoura, R.; Messadi-Akrout, F. Screening and Detecting Salmonella in Different Food Matrices in Southern Tunisia Using a Combined Enrichment/Real-Time PCR Method: Correlation with Conventional Culture Method. Front. Microbiol. 2017, 8, 2416. [Google Scholar] [CrossRef] [PubMed]
- Lampel, K.A.; Keasler, S.P.; Hanes, D.E. Specific detection of Salmonella enterica serotype Enteritidis using the polymerase chain reaction. Epidemiol. Infect. 1996, 116, 137–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, K.; Itoh, K.; Nakajima, H.; Kurazono, T.; Yamaguchi, M.; Moriya, K.; Ezaki, T.; Kawamura, Y.; Tamura, K.; Watanabe, H. Selective Amplification of Tyv (Rfbe), Prt (Rfbs), Viab, and fliC Genes by Multiplex PCR for Identification of Salmonella enterica Serovars Typhi and Paratyphi A. J. Clin. Microbiol. 2002, 40, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Liu, T.; Lee, M.D.; Hofacre, C.L.; Maier, M.; White, D.G.; Ayers, S.; Wang, L.; Berghaus, R.; Maurer, J.J. Rapid screening of Salmonella enterica serovars Enteritidis, Hadar, Heidelberg and Typhimurium using a serologically-correlative allelotyping PCR targeting the O and H antigen alleles. BMC Microbiol. 2008, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirmomeni, M.H.; Sisakhtnezhad, S.; Sharifi, A. Rapid Detection of Salmonella Enteritidis by PCR Amplification of The SefA Gene and It’s Cloning. Pak. J. Biol. Sci. 2008, 11, 428–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss, R., III; Gyles, C.L. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 1992, 271–279. [Google Scholar] [CrossRef]
- Cohen, H.J.; Mechanda, S.M.; Lin, W. PCR amplification of the fimA gene sequence of Salmonella Typhimurium, a specific method for detection of Salmonella spp. Appl Environ. Microbiol. 1996, 62, 4303–4308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Liu, Z.; Li, Y.; Yin, C.; Hu, Y.; Xie, X.; Li, Q.; Jiao, X. A Rapid method to identify Salmonella enterica serovar Gallinarum biovar pullorum using a specific target gene ipaJ. Avian Pathol. 2018, 47, 238–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginocchio, C.C.; Rahn, K.; Clarke, R.C.; Galan, J.E. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect. Immun. 1997, 65, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [Green Version]


| n | Name | Number of Tested Samples | Number of Isolated Salmonella Isolates | ||
|---|---|---|---|---|---|
| 2018 | 2019 | 2018 | 2019 | ||
| 1 | Clinical samples | 137 | 0 | 65 | 0 |
| 2 | Meat and meat products | 63 | 55 | 7 | 1 |
| 3 | Fish and fish products | 38 | 32 | 1 | 4 |
| 4 | Vegetables | 67 | 35 | 1 | 0 |
| 5 | Berries | 23 | 17 | 0 | 0 |
| 6 | Bird | 35 | 34 | 8 | 2 |
| 7 | Milk and dairy products | 94 | 65 | 3 | 1 |
| 8 | Mushrooms | 27 | 16 | 0 | 0 |
| 9 | Salads | 20 | 13 | 0 | 0 |
| 10 | Dried fruits | 36 | 25 | 0 | 0 |
| 11 | Fruits | 24 | 27 | 0 | 0 |
| 12 | Confectionery | 23 | 21 | 0 | 0 |
| 13 | Eggs | 50 | 43 | 5 | 1 |
| Total | 637 | 383 | 90 | 9 | |
| Type | Primer or Probe | Sequence | PCR Product Size |
|---|---|---|---|
| Salmonella enterica | SE-F | AGGTGACGCTATTGCCGGCAT | 155 |
| SE-R | ATGCGGGGATCTGGGCGA | ||
| SE-Probe | FAM-ATTTCGGTGGGGATGACTCGCCAT-BHQ-1 | ||
| S. Enteritidis | SEE-F | CGTCGTTGCTGCTTCCGGGA | 176 |
| SEE-R | GCTACAGAGAGTCACACTAA | ||
| SEE-Probe | FAM- TGCTGTAGATGCAAGGGTGCCTAA-BHQ-1 | ||
| S. Typhimurium | SET-F | GAAGTTGAAGTGCCGGTGAT | 251 |
| SET-R | CATTCCACCACGCCCTTCT | ||
| SET-Probe | FAM- CAGATTCCAGGCGTAAGTTTTA-BHQ-1 | ||
| S. Virchow | SEV-F | ACACCAGTACGACGATCTGCG | 105 |
| SEV-R | ATAAACCGGGCAACTGGG | ||
| SEV-Probe | FAM-GGAACACATAAACAGCGCCCAGAT-BHQ-1 | ||
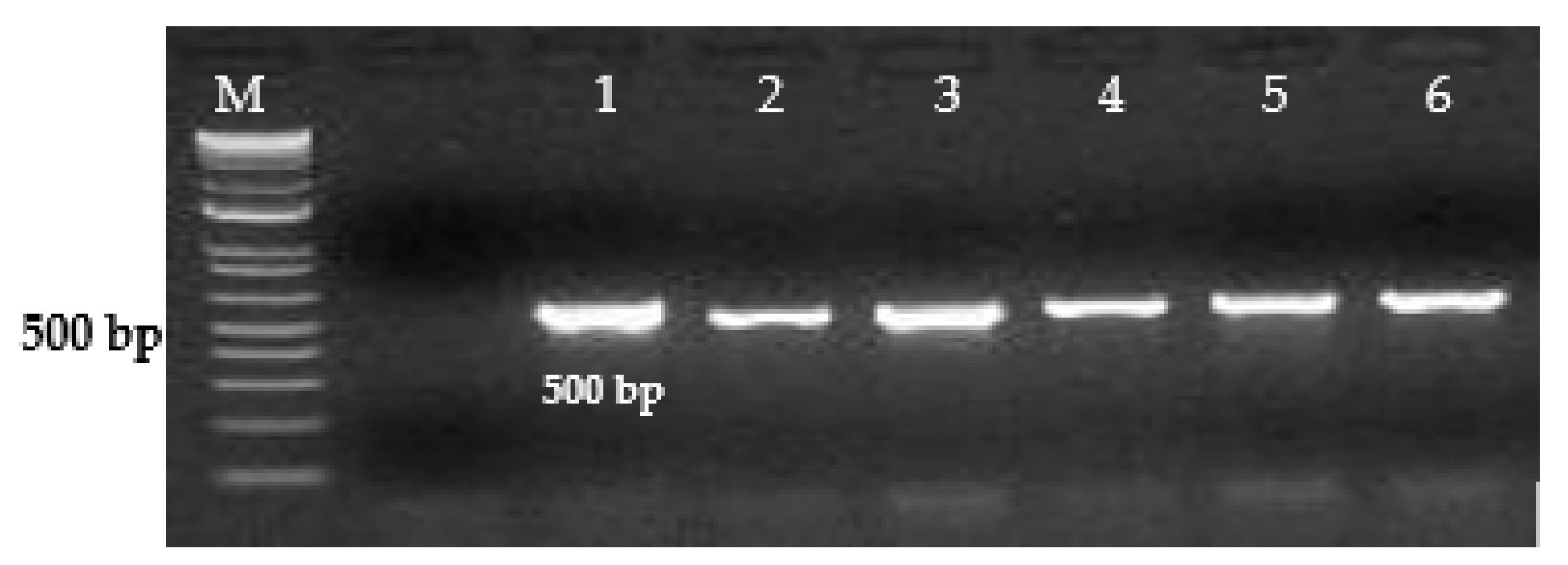
| Salmonella enterica | SE Inv-1F | GTGAAATTATCGCCACGTTCGG | 500 |
| SE Inv-1R | ATCGCCATTTACGCGGGTCA | ||
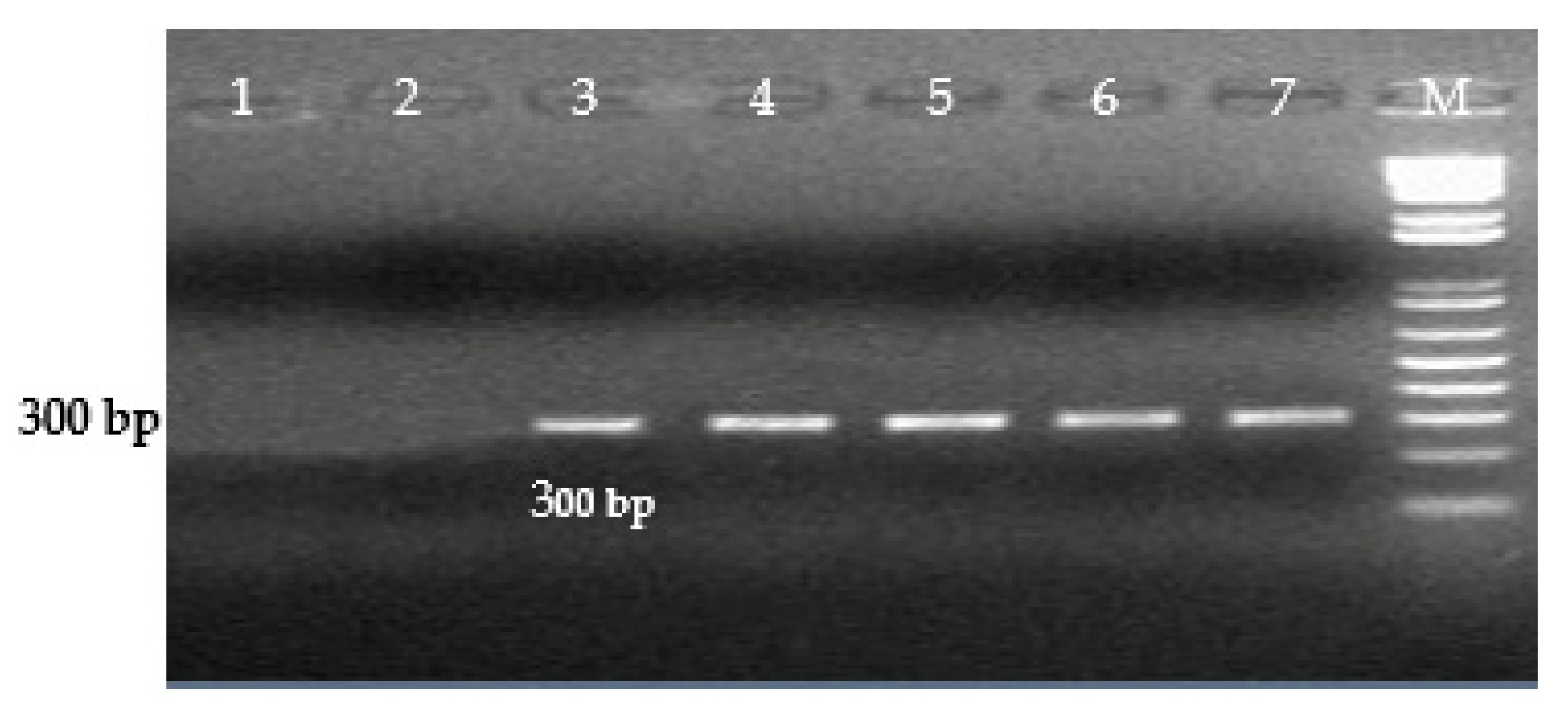
| S. Enteritidis | SE Prot6e-1F | TAACCGGAGAGGCGCTCATC | 300 |
| SE Prot6e-1R | AACCATGCTCAGCTGCTCCA | ||
| S. Typhimurium | ST mdh-1F | GTGCCGGTGATTGGCGGGCA | 243 |
| ST mdh-1R | CGCATTCCACCACGCCCTTC | ||
| S. Virchow | SV CRISPR–1F | GATCTGCGCGAACAATATCA | 269 |
| SV CRISPR–1R | CCGTTGTACTGATCATCTTC | ||
| S. Enteritidis S. Typhimurium S. Virchow | RAPD-A | GCGGGAATGCTGAAGATAAG | – |
| Control Organism | Real-Time PCR | Conventional PCR | ||||||
|---|---|---|---|---|---|---|---|---|
| S. enterica | S. Enteritidis | S. Typhimurium | S. Virchow | S. enterica | S. Enteritidis | S. Typhimurium | S. Virchow | |
| S. Enteritidis (S.e-0071) | Pos | Pos | Neg | Neg | Pos | Pos | Neg | Neg |
| S. Typhimurium TA 98 (reference strain) | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg |
| S. Typhimurium (S.t-0072) | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg |
| S. Virchow (reference strain) | Pos | Neg | Neg | Pos | Pos | Neg | Neg | Pos |
| S. Infantis (S.i-0073) | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| S. Abortusovis 37 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| S. Gallinarum 65 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| S. Abortus equi 17 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| S. Cholera suis 51 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| S. Dublin 31 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| Pasterella multocida subsp. multocida (ATCC-10544) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Clostridium perfringens Strain S 107 (ATCC-13124) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Clostridium sporogenes NCTC 532 (ATCC-19404) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Escherichia coli (ATCC-25922) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Bacillus cereus (ATCC-11778) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Bacillus subtilis subsp. spizizenii (ATCC-6633) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Staphylococcus aureus (ATCC-25923) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Staphylococcus aureus subsp. aureus (ATCC-6538P) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Pseudomonas aeruginosa Strain Boston 41501 (ATCC-27853) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Pseudomonas aeruginosa (ATCC-9027) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Candida albicans; 3147 (ATCC-10231) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Mycoplasma hyorhinis; BTS-7 (ATCC-17981) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Mycoplasma gallisepticum (ATCC-19610) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Mycoplasma synoviae; WVU 1853 [NCTC 10124] (ATCC-25204) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Klebsiella pneumoniae (ATCC-13883) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Aspergillus brasiliensis; formerly A. niger (ATCC-9642) | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Result | True Positive (TP) | False Positive (FP) | False Negative (FN) | True Negative (TN) | Total | |
|---|---|---|---|---|---|---|
| Test | ||||||
| Real-time PCR S. enterica | 99 (9.71%) | 0 | 0 | 921 (90.29%) | 1020 (100%) | |
| Real-time PCR S. Enteritidis | 20 (1.96%) | 0 | 1 (0.10%) | 999 (97.94%) | 1020 (100%) | |
| Real-time PCR S. Typhimurium | 42 (4.12%) | 0 | 1 (0.10%) | 977 (95.78%) | 1020 (100%) | |
| Real-time PCR S. Virchow | 24 (2.35%) | 1 (0.10%) | 1 (0.10%) | 994 (97.45%) | 1020 (100%) | |
| PCR S. enterica | 96 (9.41%) | 2 (0.20%) | 1 (0.10%) | 921 (90.29%) | 1020 (100%) | |
| PCR S. Enteritidis | 20 (1.96%) | 0 | 1 (0.10%) | 999 (97.94%) | 1020 (100%) | |
| PCR S. Typhimurium | 42 (4.12%) | 0 | 1 (0.10%) | 977 (95.78%) | 1020 (100%) | |
| PCR S. Virchow | 19 (1.86%) | 4 (0.40%) | 3 (0.29%) | 994 (97.45%) | 1020 (100%) | |
| Cultivating S. enterica | 99 (9.70%) | 0 | 0 | 921 (90.30%) | 1020 (100%) | |
| Result | SN, at 95% CI | SP, at 95% CI | PPV, at 95% CI | NPV, at 95% CI | Diagnostic Efficacy | |
|---|---|---|---|---|---|---|
| Test | ||||||
| Real-time PCR S. enterica | 100 | 100 | 100 | 100 | 100 | |
| Real-time PCR S. Enteritidis | 95.23 (93.93–96.53) | 100 | 100 | 99.90 (99.71–100) | 99.90 | |
| Real-time PCR S. Typhimurium | 97.67 (96.77–98.57) | 100 | 100 | 99.90 (99.71–100) | 99.90 | |
| Real-time PCR S. Virchow | 96.00 (94.8–97.2) | 99.90 (99.71–100) | 96.00 (94.8–97.2) | 99.90 (99.71–100) | 99.80 | |
| PCR S. enterica | 98.97 (98.35–99.59) | 99.78 (99.50–100) | 97.95 (97.15–98.75) | 99.89 (99.69–100) | 99.71 | |
| PCR S. Enteritidis | 95.24 (93.94–96.54) | 100 | 100 | 99.90 (99.71–100) | 99.90 | |
| PCR S. Typhimurium | 97.67 (96.77–98.57) | 100 | 100 | 99.89 (99.69–100) | 99.90 | |
| PCR S. Virchow | 86.36 (84.26–88.46) | 99.60 (99.30–99.90) | 82.61 (80.31–84.91) | 99.70 (99.40–100) | 99.31 | |
| Cultivating S. enterica | 100 | 100 | 100 | 100 | 100 | |
| Serovar | Food Product | Clinical Sample | ||||||
|---|---|---|---|---|---|---|---|---|
| Group | Number of Isolates | Number of Amplicons | Size of Amplicons, bp | Group | Number of Isolates | Number of Amplicons | Size of Amplicons, bp | |
| S. Enteritidis | A | 8 | 6 | 250, 350, 650, 1000, 1250, 3000 | A | 13 | 6 | 250, 350, 650, 1000, 1250, 3000 |
| S. Typhimurium | B | 12 | 4 | 250, 350, 1000, 1250 | B | 29 | 4 | 250, 350, 1000, 1250 |
| C | 2 | 3 | 250, 350, 1000 | |||||
| S. Virchow | D | 3 | 3 | 200, 650, 1200 | I | 19 | 3 | 300, 650, 1200 |
| F | 2 | 2 | 400, 650 | |||||
| G | 2 | 4 | 300, 500, 650, 1200 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barmak, S.M.; Sinyavskiy, Y.A.; Berdygaliev, A.B.; Sharmanov, T.S.; Savitskaya, I.S.; Sultankulova, G.T.; Zholdybayeva, E.V. Development and Evaluation of Alternative Methods to Identify the Three Most Common Serotypes of Salmonella enterica Causing Clinical Infections in Kazakhstan. Microorganisms 2021, 9, 2319. https://doi.org/10.3390/microorganisms9112319
Barmak SM, Sinyavskiy YA, Berdygaliev AB, Sharmanov TS, Savitskaya IS, Sultankulova GT, Zholdybayeva EV. Development and Evaluation of Alternative Methods to Identify the Three Most Common Serotypes of Salmonella enterica Causing Clinical Infections in Kazakhstan. Microorganisms. 2021; 9(11):2319. https://doi.org/10.3390/microorganisms9112319
Chicago/Turabian StyleBarmak, Sabyrkhan M., Yuriy A. Sinyavskiy, Aidar B. Berdygaliev, Turegeldy Sh. Sharmanov, Irina S. Savitskaya, Gulmira T. Sultankulova, and Elena V. Zholdybayeva. 2021. "Development and Evaluation of Alternative Methods to Identify the Three Most Common Serotypes of Salmonella enterica Causing Clinical Infections in Kazakhstan" Microorganisms 9, no. 11: 2319. https://doi.org/10.3390/microorganisms9112319
APA StyleBarmak, S. M., Sinyavskiy, Y. A., Berdygaliev, A. B., Sharmanov, T. S., Savitskaya, I. S., Sultankulova, G. T., & Zholdybayeva, E. V. (2021). Development and Evaluation of Alternative Methods to Identify the Three Most Common Serotypes of Salmonella enterica Causing Clinical Infections in Kazakhstan. Microorganisms, 9(11), 2319. https://doi.org/10.3390/microorganisms9112319






