Differences between the Leaf Mycobiome of Coffea arabica and Wild Coffee Species and Their Modulation by Caffeine/Chlorogenic Acid Content
Abstract
:1. Introduction
2. Methods
2.1. Biological Material
2.2. Microbes Collection, DNA Extraction, and Sequencing
2.3. Pre-sequence Analysis
2.4. Identification of Differentially Abundant Taxonomic Groups
2.5. Interaction Networks between Mycobiomes
2.6. Estimation of Fungal Abundance
2.7. Caffeine and Chlorogenic Acid Analysis
2.8. Statistical Analysis
3. Results
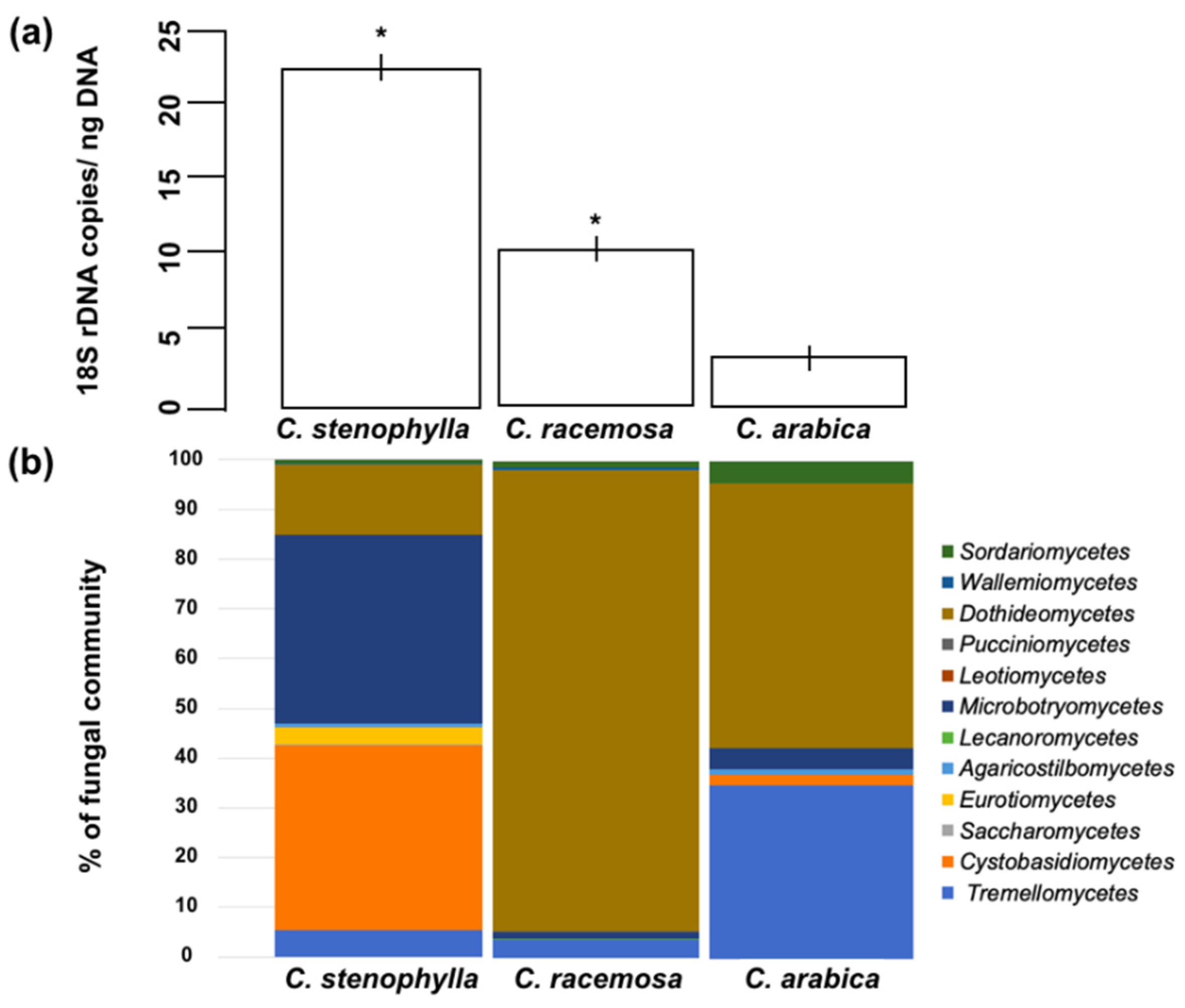
3.1. Fungal Abundance
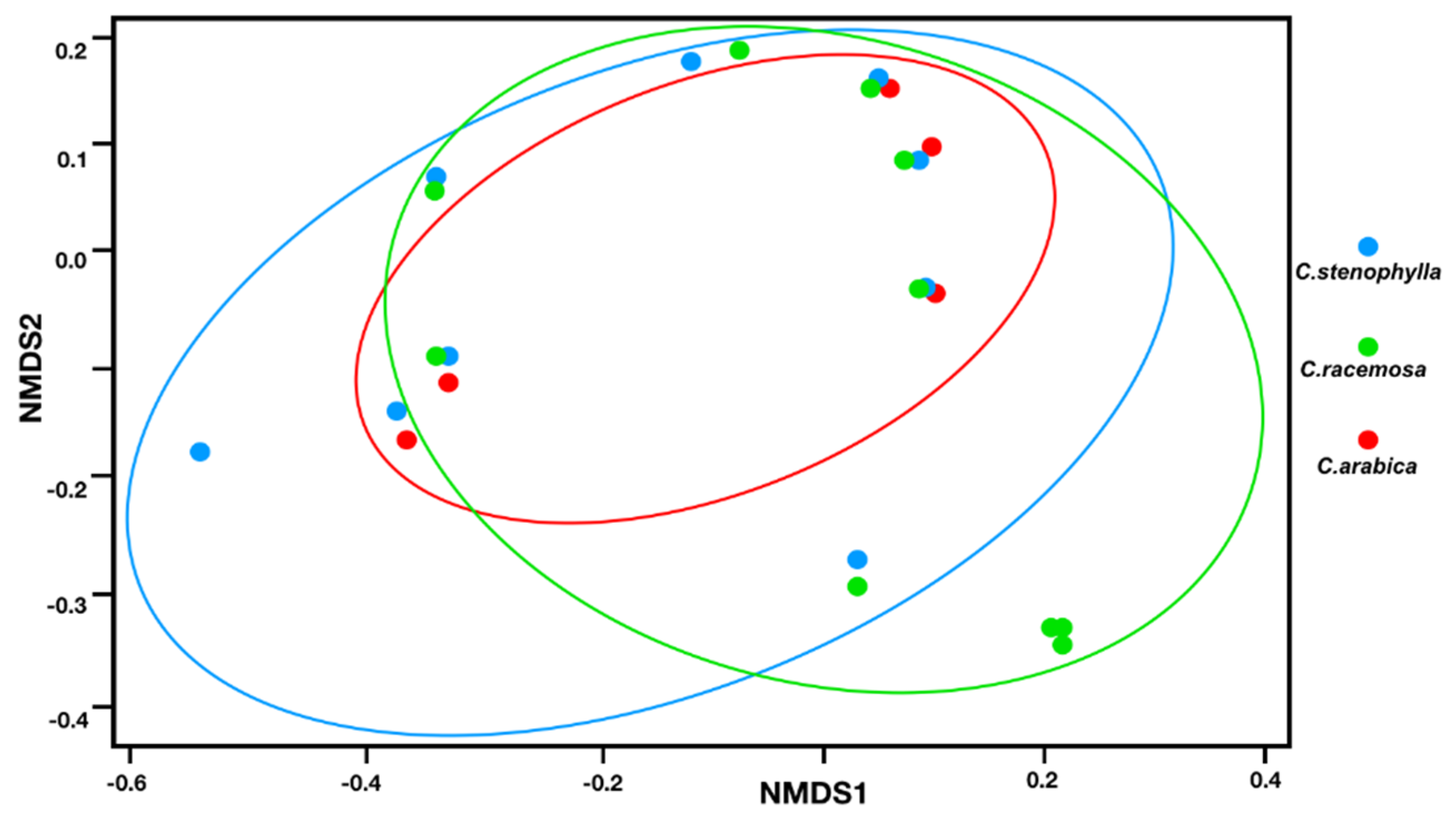
3.2. Fungal Species Composition
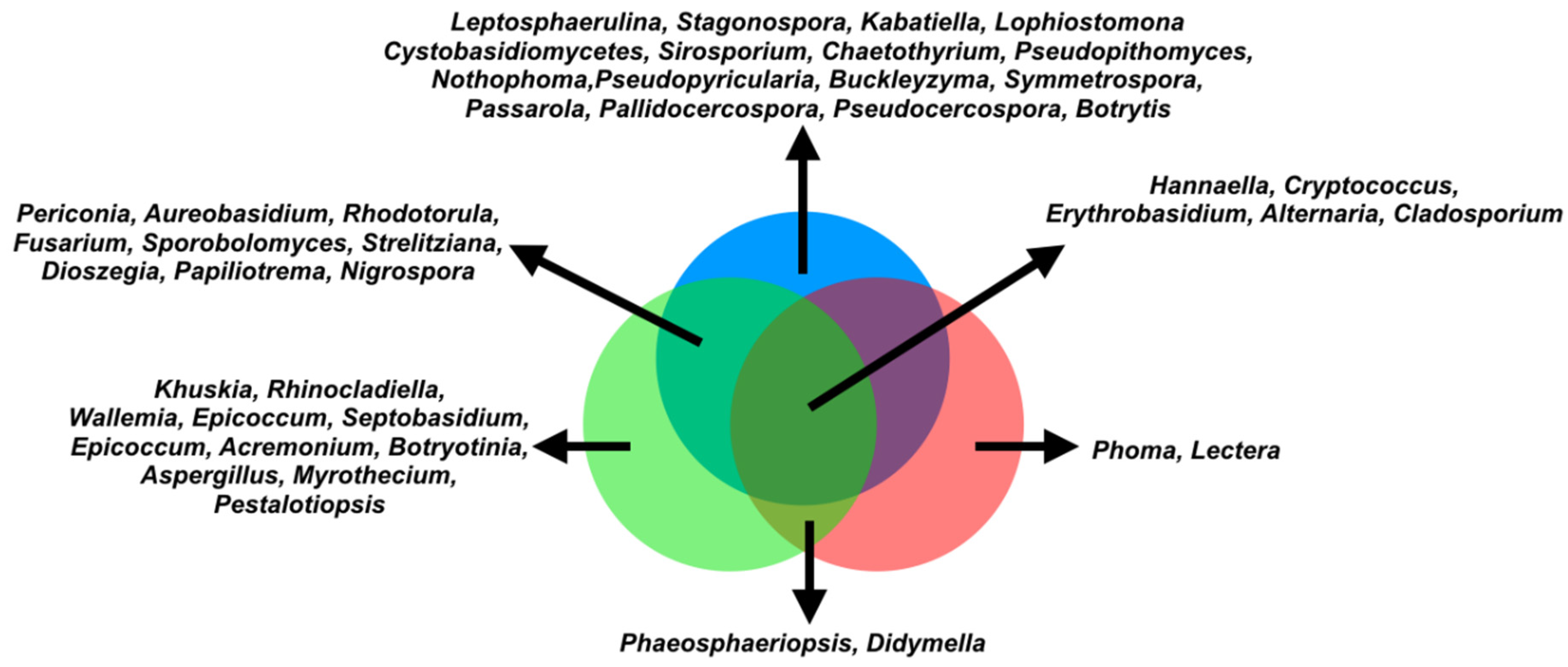
3.3. Differentially Abundant Fungal Genera in the Phyllosphere
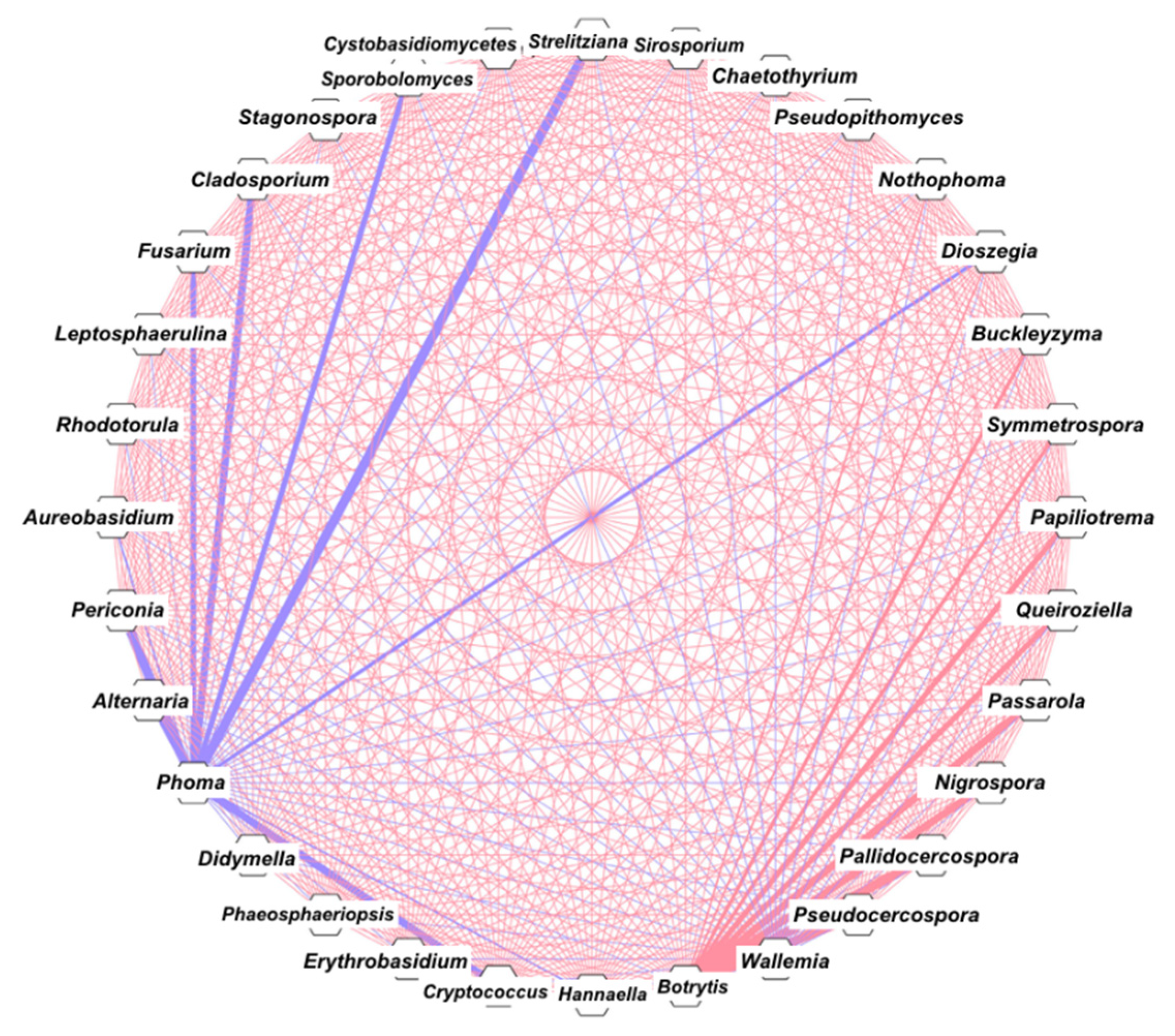
3.4. Correlation and Network Analysis
3.5. Caffeine and Chlorogenic Acids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Andrews, J.H. Biological control in the phyllosphere. Annu. Rev. Phytopathol. 1992, 30, 603–635. [Google Scholar] [CrossRef]
- Kong, X.; Han, Z.; Tai, X.; Jin, D.; Ai, S.; Zheng, X.; Bai, Z. Maize (Zea mays L. Sp.) varieties significantly influence bacterial and fungal community in bulk soil, rhizosphere soil and phyllosphere. FEMS Microbiol. Ecol. 2020, 96, fiaa020. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Liu, W.; Ke, M.; Qu, Q.; Zhou, Z.; Lu, T.; Qian, H. Phyllosphere bacterial assemblage is affected by plant genotypes and growth stages. Microbiol. Res. 2021, 248, 126743. [Google Scholar] [CrossRef]
- Davis, A.P.; Govaerts, R.; Bridson, D.M.; Stoffelen, P. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot. J. Linn. Soc. 2006, 152, 465–512. [Google Scholar] [CrossRef] [Green Version]
- Lewin, B.; Giovannucci, D.; Varangis, P. Coffee Markets: New Paradigms in Global Supply and Demand. In Agricultural and Rural Development Discussion Paper No. 3; International Bank for Reconstruction and Development: Washington, DC, USA, 2004. [Google Scholar]
- Osorio, N. The Global Coffee Crisis: A Threat to Sustainable Development; International Coffee Organization: London, UK, 2002. [Google Scholar]
- Zonneveld, M.V.; Solano, W. Coffea eugenioides S. Moore and Coffea stenophylla G. Don: Distribution, Botany, Agronomic Traits and Growing Areas; CATIE: Turrialba, Costa Rica, 2016. [Google Scholar]
- Carducci, F.C.; De Batista Fonseca, I.C.; Dos Santos, W.G.; Pereira, C.T.M.; Junior, V.M.; Sera, T.; Sera, G.H. Resistance to red mite in ‘Coffea arabica’ genotype introgressed with ‘Coffea racemosa’ genes. Aust. J. Crop. Sci. 2019, 13, 683. [Google Scholar] [CrossRef]
- Santamaría, J.; Bayman, P. Fungal epiphytes and endophytes of coffee leaves (Coffea arabica). Microb. Ecol. 2005, 50, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, H.F.; Silva, H.S.A.; Melo, I.S.D.; Nunes, F.V.; Bettiol, W. Bioprospecting endophytic bacteria for biological control of coffee leaf rust. Sci. Agric. 2006, 63, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Vega, F.E.; Ripoll, M.P.; Posada, F.; Buyer, J.S. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 2005, 45, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-García, A.; Anaya, A.L.; Espinosa-García, F.J.; González, M.C. Diversity and communities of foliar endophytic fungi from different agroecosystems of Coffea arabica L. in two regions of Veracruz, Mexico. PLoS ONE 2014, 9, e98454. [Google Scholar] [CrossRef]
- Bongiorno, V.A.; Rhoden, S.A.; Garcia, A.; Polonio, J.C.; Azevedo, J.L.; Pereira, J.O.; Pamphile, J.A. Genetic diversity of endophytic fungi from Coffea arabica cv. IAPAR-59 in organic crops. Ann. Microbiol. 2016, 66, 855–865. [Google Scholar] [CrossRef]
- Fulthorpe, R.; Martin, A.R.; Isaac, M.E. Root endophytes of coffee (Coffea arabica): Variation across climatic gradients and relationships with functional traits. Phytobiomes J. 2020, 4, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Veloso, T.G.R.; Da Silva, M.D.C.S.; Cardoso, W.S.; Guarçoni, R.C.; Kasuya, M.C.M.; Pereira, L.L. Effects of environmental factors on microbiota of fruits and soil of Coffea arabica in Brazil. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.A.; Bálint, M.; Greshake, B.; Bandow, C.; Römbke, J.; Schmitt, I. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013, 65, 128–132. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vernier, C.L.; Chin, I.M.; Adu-Oppong, B.; Krupp, J.J.; Levine, J.; Dantas, G.; Ben-Shahar, Y. The gut microbiome defines social group membership in honey bee colonies. Sci. Adv. 2020, 6, eabd3431. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.E.; Bates, S.T.; Knights, D.; Lauber, C.L.; Stombaugh, J.; Knight, R.; Fierer, N. Microbial biogeography of public restroom surfaces. PLoS ONE 2011, 6, e28132. [Google Scholar] [CrossRef] [PubMed]
- Prévost-Bouré, N.C.; Christen, R.; Dequiedt, S.; Mougel, C.; Lelievre, M.; Jolivet, C.; Shahbazkia, H.R.; Guillou, L.; Arrouays, D.; Ranjard, L. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS ONE 2011, 6, e24166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzafera, P.; Silvarolla, M.B.; Lima, M.M.A.; Medina Filho, H.P. Caffeine content in diploid coffee species. Cienc. Cult. 1997, 49, 216–218. [Google Scholar]
- Ramiro, D.A.; Guerreiro-Filho, O.; Mazzafera, P. Phenol contents, oxidase activities, and the resistance of coffee to the leaf miner Leucoptera coffeella. J. Chem. Ecol. 2006, 32, 1977–1988. [Google Scholar] [CrossRef]
- Kim, Y.S.; Choi, Y.E.; Sano, H. Plant vaccination: Stimulation of defense system by caffeine production in planta. Plant. Signal. Behav. 2010, 5, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Sung, W.S.; Lee, D.G. Antifungal action of chlorogenic acid against pathogenic fungi, mediated by membrane disruption. Pure Appl. Chem. 2010, 82, 219–226. [Google Scholar] [CrossRef]
- Sugiyama, A.; Sano, C.M.; Yazaki, K.; Sano, H. Caffeine fostering of mycoparasitic fungi against phytopathogens. Plant. Signal. Behav. 2016, 11, e1113362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, G.; Regente, M.; Jacobi, S.; Del Rio, M.; Pinedo, M.; De La Canal, L. Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Phys. 2017, 140, 30–35. [Google Scholar] [CrossRef]
- Hakil, M.; Denis, S.; Viniegra-Gonzalez, G.; Augur, C. Degradation and product analysis of caffeine and related dimethylxanthines by filamentous fungi. Enzyme Microb. Technol. 1998, 22, 355–359. [Google Scholar] [CrossRef]
- Mancera, M.T.T.; Peña, I.B.; Montero, A.F.; Serrano, G.R.; Zamora, E.G.; Torres, E.F.; Castañeda, G.S. Biotransformation and improved enzymatic extraction of chlorogenic acid from coffee pulp by filamentous fungi. Biotechnol. Prog. 2013, 29, 337–345. [Google Scholar] [CrossRef]
- Etienne, H.; Anthony, F.; Dussert, S.; Fernandez, D.; Lashermes, P.; Bertrand, B. Biotechnological applications for the improvement of coffee (Coffea arabica L.). In Vitro Cell. Dev. Biol. Plant 2002, 38, 129–138. [Google Scholar] [CrossRef]
- Yadav, R.K.P.; Karamanoli, K.; Vokou, D. Bacterial colonization of the phyllosphere of Mediterranean perennial species as influenced by leaf structural and chemical features. Microb. Ecol. 2005, 50, 185–196. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant. Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Catarino, A.D.M.; Pozza, E.A.; Pozza, A.A.A.; Vasco, G.B.; Souza, P.E.D. Calcium and potassium contents in nutrient solution on Phoma leaf spot intensity in coffee seedlings. Rev. Ceres 2016, 63, 486–491. [Google Scholar] [CrossRef] [Green Version]
- Geiser, D.M.; Lewis Ivey, M.L.; Hakiza, G.; Juba, J.H.; Miller, S.A. Gibberella xylarioides (anamorph: Fusarium xylarioides), a causative agent of coffee wilt disease in Africa, is a previously unrecognized member of the G. fujikuroi species complex. Mycologia 2005, 97, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Mondego, J.M.; Vidal, R.O.; Carazzolle, M.F.; Tokuda, E.K.; Parizzi, L.P.; Costa, G.G.; Pereira, L.F.; Andrade, A.C.; Colombo, C.A.; Vieira, L.G. An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Biol. 2011, 11, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, L.M.R.; Destéfano, S.A.L.; Almeida, I.M.G.D.; Beriam, L.O.S.; Braghini, M.T.; Guerreiro, O. Multiple resistances to bacterial halo blight and bacterial leaf spot in Coffea spp. Arq. Inst. Biol. 2019, 86. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.L.; Nascimento, G.O.; Lopes, G.S.; Matos, W.O.; Cunha, R.L.; Malta, M.R.; Liska, G.R.; Owen, R.W.; Trevisan, M.T.S. The concentration of polyphenolic compounds and trace elements in the Coffea arabica leaves: Potential chemometric pattern recognition of coffee leaf rust resistance. Food Res. Int. 2020, 134, 109221. [Google Scholar] [CrossRef]
- Osono, T.; Mori, A. Distribution of phyllosphere fungi within the canopy of giant dogwood. Mycoscience 2004, 45, 161–168. [Google Scholar] [CrossRef]
- Cordier, T.; Robin, C.; Capdevielle, X.; Desprez-Loustau, M.L.; Vacher, C. Spatial variability of phyllosphere fungal assemblages: Genetic distance predominates over geographic distance in a European beech stand (Fagus sylvatica). Fungal Ecol. 2012, 5, 509–520. [Google Scholar] [CrossRef]
| Caffeine (mg/g) | Chlorogenic Acid (mg/g) | |
|---|---|---|
| C. arabica | 7.4 ± 0.5 | 55 ± 3 |
| C. stenophylla | 0.26 ± 0.3 | 20 ± 2.5 |
| C. racemosa | 0.21 ± 0.1 | 23 ± 3.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sousa, L.P.; Guerreiro Filho, O.; Costa Mondego, J.M. Differences between the Leaf Mycobiome of Coffea arabica and Wild Coffee Species and Their Modulation by Caffeine/Chlorogenic Acid Content. Microorganisms 2021, 9, 2296. https://doi.org/10.3390/microorganisms9112296
de Sousa LP, Guerreiro Filho O, Costa Mondego JM. Differences between the Leaf Mycobiome of Coffea arabica and Wild Coffee Species and Their Modulation by Caffeine/Chlorogenic Acid Content. Microorganisms. 2021; 9(11):2296. https://doi.org/10.3390/microorganisms9112296
Chicago/Turabian Stylede Sousa, Leandro Pio, Oliveiro Guerreiro Filho, and Jorge Maurício Costa Mondego. 2021. "Differences between the Leaf Mycobiome of Coffea arabica and Wild Coffee Species and Their Modulation by Caffeine/Chlorogenic Acid Content" Microorganisms 9, no. 11: 2296. https://doi.org/10.3390/microorganisms9112296
APA Stylede Sousa, L. P., Guerreiro Filho, O., & Costa Mondego, J. M. (2021). Differences between the Leaf Mycobiome of Coffea arabica and Wild Coffee Species and Their Modulation by Caffeine/Chlorogenic Acid Content. Microorganisms, 9(11), 2296. https://doi.org/10.3390/microorganisms9112296





