Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides distasonis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Single-Stranded DNA (ssDNA) Library and Primers
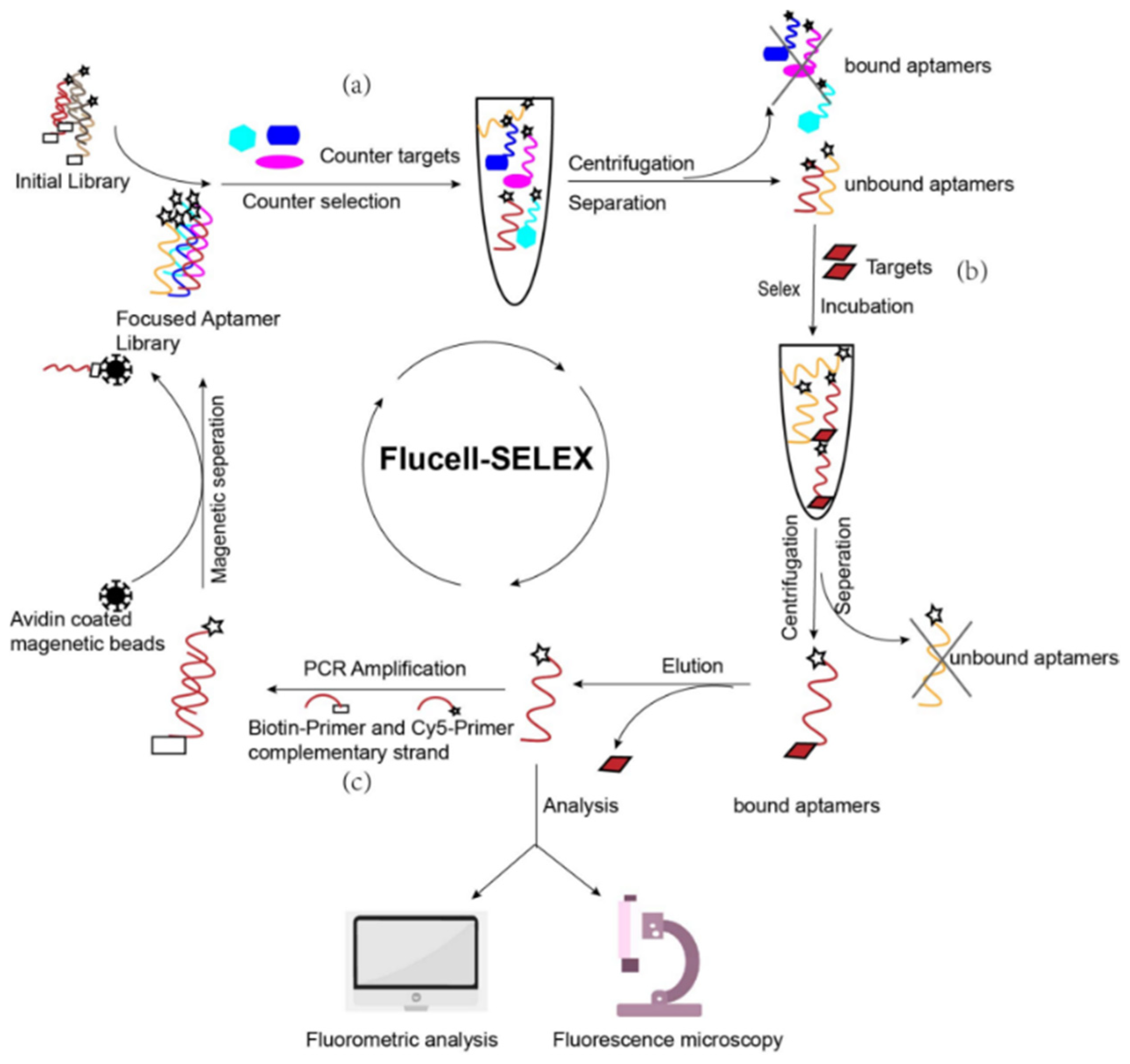
2.3. Cell-SELEX
2.3.1. Cell Pretreatment
2.3.2. Aptamer Library Activation
2.3.3. Screening
2.3.4. Elution
2.3.5. Acquisition of Secondary Libraries
2.3.6. Binding Assay
2.4. Determination of High Specificity Aptamer Libraries
2.4.1. Specificity Analysis
2.4.2. Affinity Analysis
2.4.3. Fluorescence Microscopy
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sakamoto, M.; Benno, Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 1599–1605. [Google Scholar] [CrossRef]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013, 21, E607–E615. [Google Scholar] [CrossRef]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Baranzini, S.E. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, E8943. [Google Scholar] [CrossRef] [Green Version]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandona, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Koh, G.Y.; Kane, A.V.; Wu, X.; Crott, J.W. Parabacteroides distasonis attenuates tumorigenesis, modulates inflammatory markers and promotes intestinal barrier integrity in azoxymethane-treated A/J mice. Carcinogenesis 2020, 41, 909–917. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef] [Green Version]
- De Boer, J.F.; Bloks, V.W.; Verkade, E.; Heiner-Fokkema, M.R.; Kuipers, F. New insights in the multiple roles of bile acids and their signaling pathways in metabolic control. Curr. Opin. Lipidol. 2018, 29, 194–202. [Google Scholar] [CrossRef]
- Pfalzer, A.C.; Nesbeth, P.D.; Parnell, L.D.; Iyer, L.K.; Liu, Z.; Kane, A.V.; Chen, C.Y.; Tai, A.K.; Bowman, T.A.; Obin, M.S.; et al. Diet- and Genetically-Induced Obesity Differentially Affect the Fecal Microbiome and Metabolome in Apc1638N Mice. PLoS ONE 2015, 10, e0135758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, G.Y.; Kane, A.; Lee, K.; Xu, Q.; Wu, X.; Roper, J.; Mason, J.B.; Crott, J.W. Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice. Int. J. Cancer 2018, 143, 1797–1805. [Google Scholar] [CrossRef] [Green Version]
- Amato, K.R. An introduction to microbiome analysis for human biology applications. Am. J. Hum. Biol. 2017, 29, e22931. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [PubMed] [Green Version]
- Kubiczek, D.; Raber, H.; Bodenberger, N.; Oswald, T.; Sahan, M.; Mayer, D.; Wiese, S.; Stenger, S.; Weil, T.; Rosenau, F. The Diversity of a Polyclonal FluCell-SELEX Library Outperforms Individual Aptamers as Emerging Diagnostic Tools for the Identification of Carbapenem Resistant Pseudomonas aeruginosa. Chemistry 2020, 26, 14536–14545. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Kubiczek, D.B.N.; Rosenau, F. Aptamers as promising agents in diagnostic and therapeutic applications. In Antimicrobial Research: Novel Bioknowledge and Educational Programs; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2017; pp. 368–378. [Google Scholar]
- Marrazza, G. Aptamer Sensors. Biosensors 2017, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.P.; Scott, J.K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993, 217, 228–257. [Google Scholar]
- Eaton, B.E.; Gold, L.; Zichi, D.A. Let’s get specific: The relationship between specificity and affinity. Chem. Biol. 1995, 2, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.K. Monitoring Intact Viruses Using Aptamers. Biosensors 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Rong, Y.; Chen, H.; Zhou, X.F.; Yin, C.Q.; Wang, B.C.; Peng, C.W.; Liu, S.P.; Wang, F.B. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget 2016, 7, 8282–8294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Sefah, K.; Tang, L.; Zhao, Z.; Zhu, G.; Ye, M.; Sun, W.; Goodison, S.; Tan, W. A novel aptamer developed for breast cancer cell internalization. ChemMedChem 2012, 7, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Li, W.M.; Zhou, L.L.; Zheng, M.; Fang, J. Selection of Metastatic Breast Cancer Cell-Specific Aptamers for the Capture of CTCs with a Metastatic Phenotype by Cell-SELEX. Mol. Ther. Nucleic Acids 2018, 12, 707–717. [Google Scholar] [CrossRef]
- Shi, H.; Cui, W.; He, X.; Guo, Q.; Wang, K.; Ye, X.; Tang, J. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE 2013, 8, e70476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Quang, N.; Perret, G.; Duconge, F. Applications of High-Throughput Sequencing for In Vitro Selection and Characterization of Aptamers. Pharmaceuticals 2016, 9, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [Green Version]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [Green Version]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef]
- Liu, C.; Finegold, S.M.; Song, Y.; Lawson, P.A. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 1896–1902. [Google Scholar]
- Gao, S.; Zheng, X.; Tang, Y.; Cheng, Y.; Hu, X.; Wu, J. Development of a Fluorescently Labeled Aptamer Structure-Switching Assay for Sensitive and Rapid Detection of Gliotoxin. Anal. Chem. 2019, 91, 1610–1618. [Google Scholar] [CrossRef]
- Ohuchi, S.P.; Ohtsu, T.; Nakamura, Y. Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface. Biochimie 2006, 88, 897–904. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, E.Y.; Kim, S.Y.; Byun, S.K.; Lee, D.; Oh, K.J.; Kim, W.K.; Han, B.S.; Chi, S.W.; Lee, S.C.; et al. Identification of DNA aptamers toward epithelial cell adhesion molecule via cell-SELEX. Mol. Cells 2014, 37, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.G.; Marangoni, K.; Fujimura, P.T.; Alves, P.T.; Silva, M.J.; Bastos, V.A.; Goulart, L.R.; Goulart, V.A. 3D Cell-SELEX: Development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp. Cell Res. 2016, 341, 147–156. [Google Scholar] [CrossRef]
- Thiel, W.H.; Thiel, K.W.; Flenker, K.S.; Bair, T.; Dupuy, A.J.; McNamara, J.O., 2nd; Miller, F.J.; Giangrande, P.H. Cell-internalization SELEX: Method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2015, 1218, 187–199. [Google Scholar] [PubMed] [Green Version]
- Ziegler, C.; Göpel, W. Biosensor development. Curr. Opin. Chem. Biol. 1998, 2, 585–591. [Google Scholar] [CrossRef]
- Sergelen, K.; Liedberg, B.; Knoll, W.; Dostalek, J. A surface plasmon field-enhanced fluorescence reversible split aptamer biosensor. Analyst 2017, 142, 2995–3001. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. Engl. 2021, 60, 16800–16823. [Google Scholar] [CrossRef]
- Fan, V.Y.; Jamison, D.T.; Summers, L.H. Pandemic risk: How large are the expected losses? Bull. World Health Organ. 2018, 96, 129–134. [Google Scholar] [CrossRef]
- Munzar, J.D.; Ng, A.; Juncker, D. Duplexed aptamers: History, design, theory, and application to biosensing. Chem. Soc. Rev. 2019, 48, 1390–1419. [Google Scholar] [CrossRef]
- Mujica, M.L.; Zhang, Y.; Bédioui, F.; Gutiérrez, F.; Rivas, G. Label-free graphene oxide-based SPR genosensor for the quantification of microRNA21. Anal. Bioanal. Chem. 2020, 412, 3539–3546. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef]
- Schachter, J.; Martel, J.; Lin, C.S.; Chang, C.J.; Wu, T.R.; Lu, C.C.; Ko, Y.F.; Lai, H.C.; Ojcius, D.M.; Young, J.D. Effects of obesity on depression: A role for inflammation and the gut microbiota. Brain Behav. Immun. 2018, 69, 1–8. [Google Scholar] [CrossRef]
- Forsythe, P.; Sudo, N.; Dinan, T.; Taylor, V.H.; Bienenstock, J. Mood and gut feelings. Brain Behav. Immun. 2010, 24, 9–16. [Google Scholar] [CrossRef]
- Dickerson, F.; Severance, E.; Yolken, R. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav. Immun. 2017, 62, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, H.; Kissmann, A.-K.; Raber, H.F.; Krämer, M.; Amann, V.; Kohn, K.; Weil, T.; Rosenau, F. Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides distasonis. Microorganisms 2021, 9, 2284. https://doi.org/10.3390/microorganisms9112284
Xing H, Kissmann A-K, Raber HF, Krämer M, Amann V, Kohn K, Weil T, Rosenau F. Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides distasonis. Microorganisms. 2021; 9(11):2284. https://doi.org/10.3390/microorganisms9112284
Chicago/Turabian StyleXing, Hu, Ann-Kathrin Kissmann, Heinz Fabian Raber, Markus Krämer, Valerie Amann, Kathrin Kohn, Tanja Weil, and Frank Rosenau. 2021. "Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides distasonis" Microorganisms 9, no. 11: 2284. https://doi.org/10.3390/microorganisms9112284
APA StyleXing, H., Kissmann, A.-K., Raber, H. F., Krämer, M., Amann, V., Kohn, K., Weil, T., & Rosenau, F. (2021). Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides distasonis. Microorganisms, 9(11), 2284. https://doi.org/10.3390/microorganisms9112284





