Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Neonicotinoid Preparation and Quantification
2.3. Chromatography and Quantitative Analyses
2.4. Feeding Rate
2.5. Survival Analysis
2.6. RNA Extraction
2.7. 16S rRNA Gene Sequencing
2.7.1. cDNA Synthesis
2.7.2. Two-Step 16S rDNA Amplicon Library Preparation
2.7.3. Paired End Illumina Sequencing
2.7.4. Disclaimer
2.8. Bioinformatics Analysis
2.8.1. Sequence Clustering
2.8.2. Taxonomic Assignment
2.8.3. Alpha Diversity
2.8.4. Network Analysis
3. Results
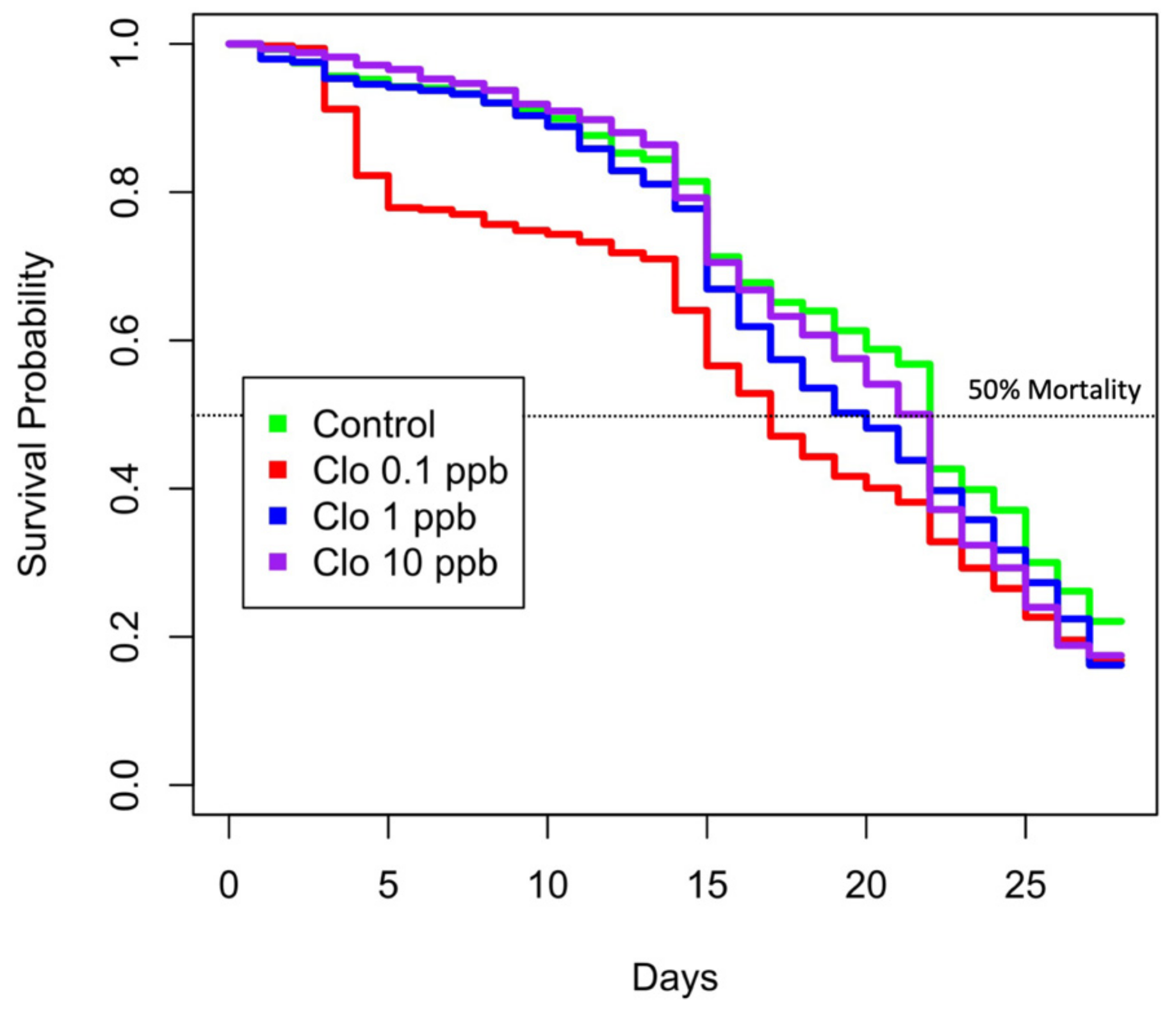
3.1. Honeybee Survival Is Unexpectedly Lower When Exposed to Lesser Concentrations of Clothianidin
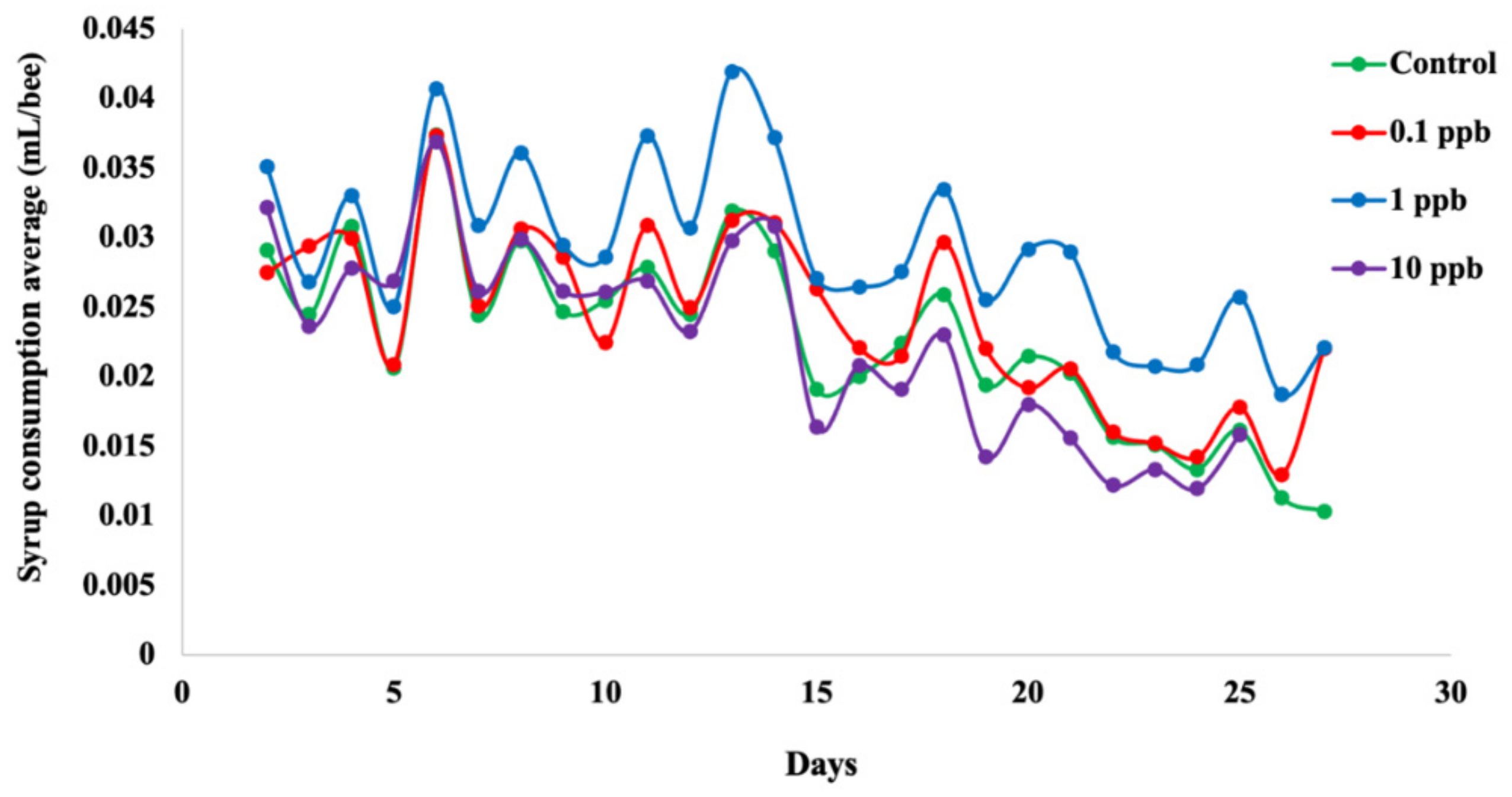
3.2. Feeding Rate of Honeybees Depends on Sublethal Clothianidin Concentration
3.3. Mean Clothianidin Quantification in Experimental Honeybees
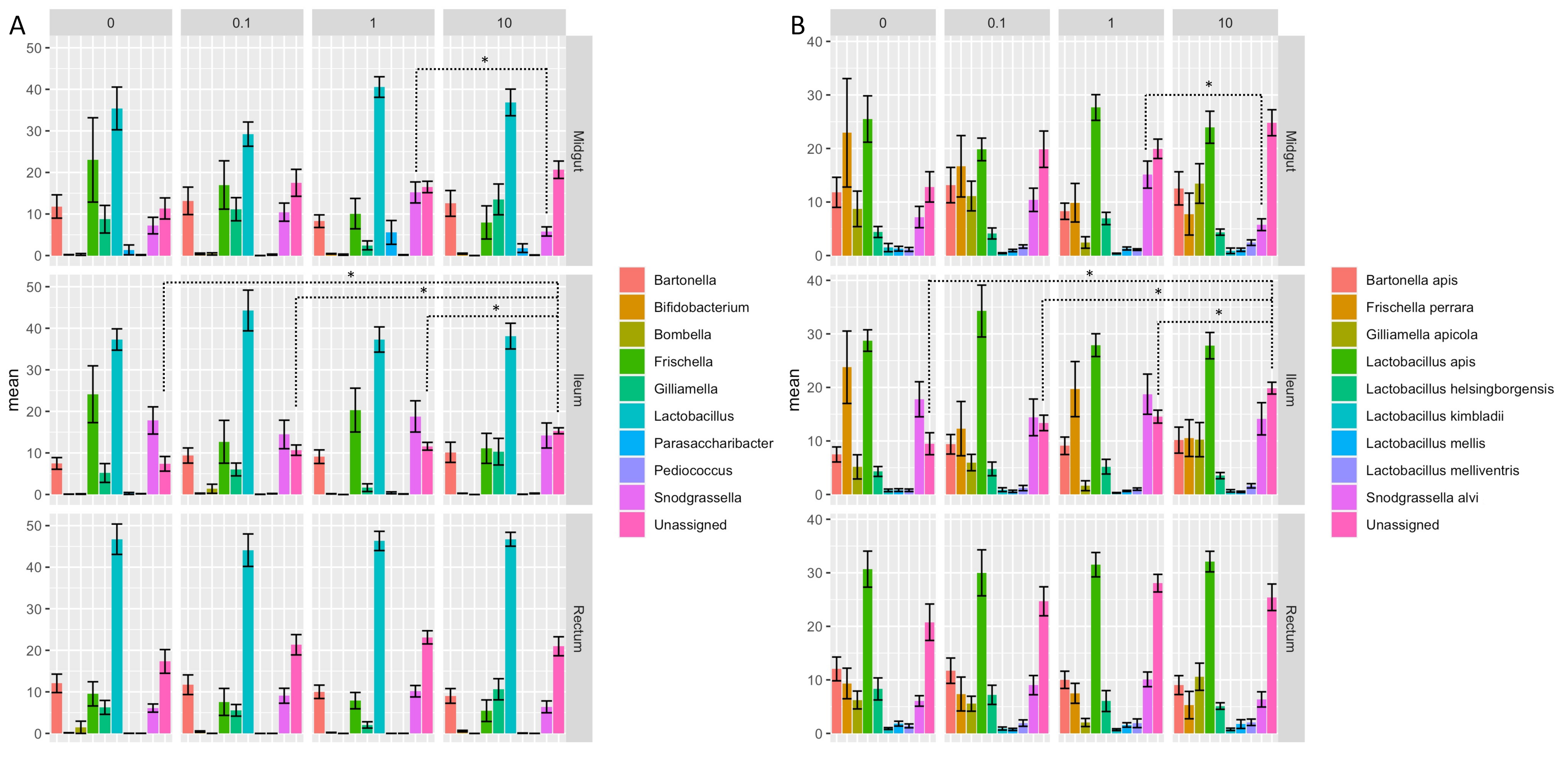
3.4. Clothianidin-Induced Changes to the Taxonomic Distribution of Intestinal Microbiota Activity
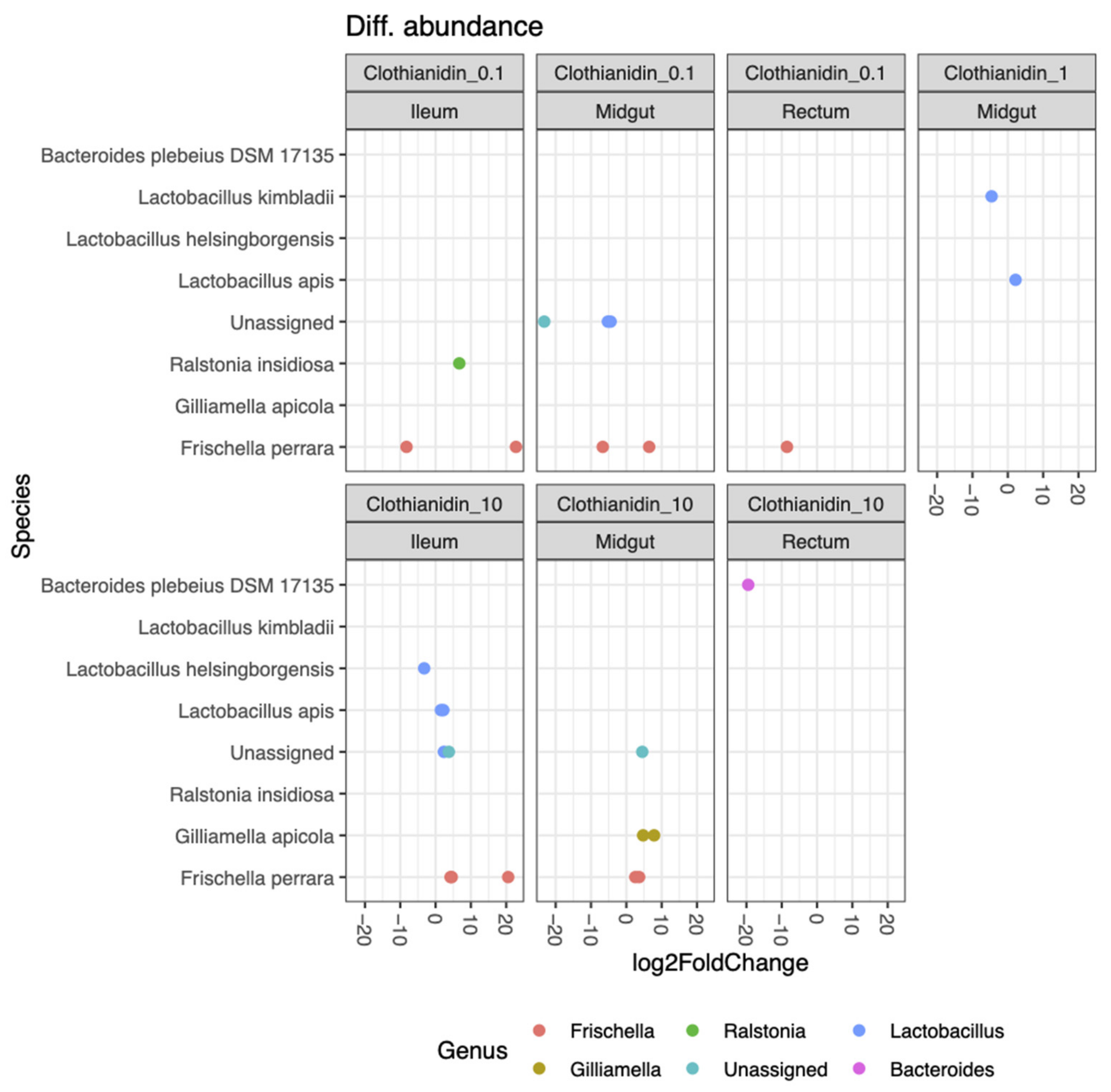
3.5. Clothianidin Differentially Impacts the Activity of Certain Taxa at Specific Concentrations
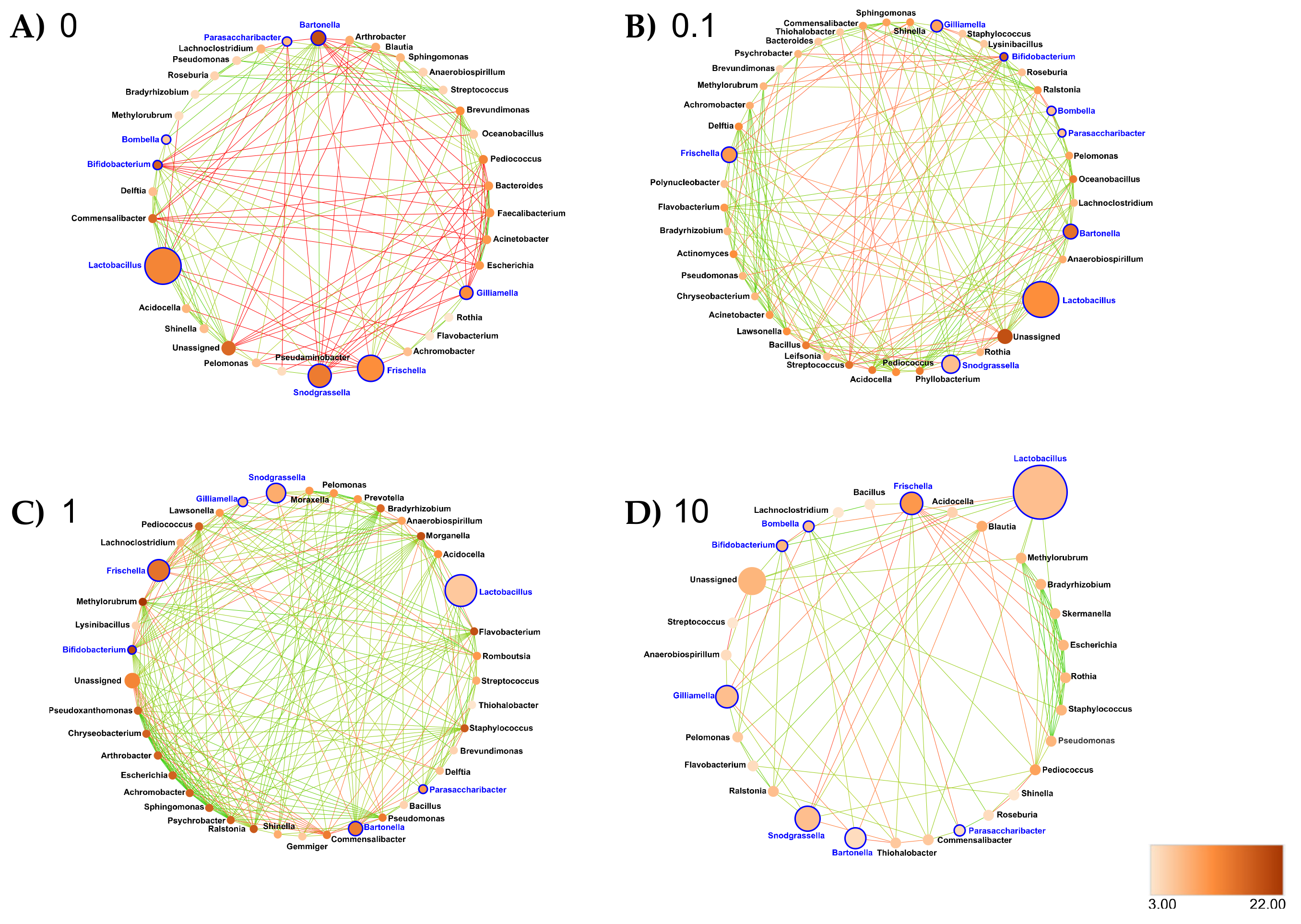
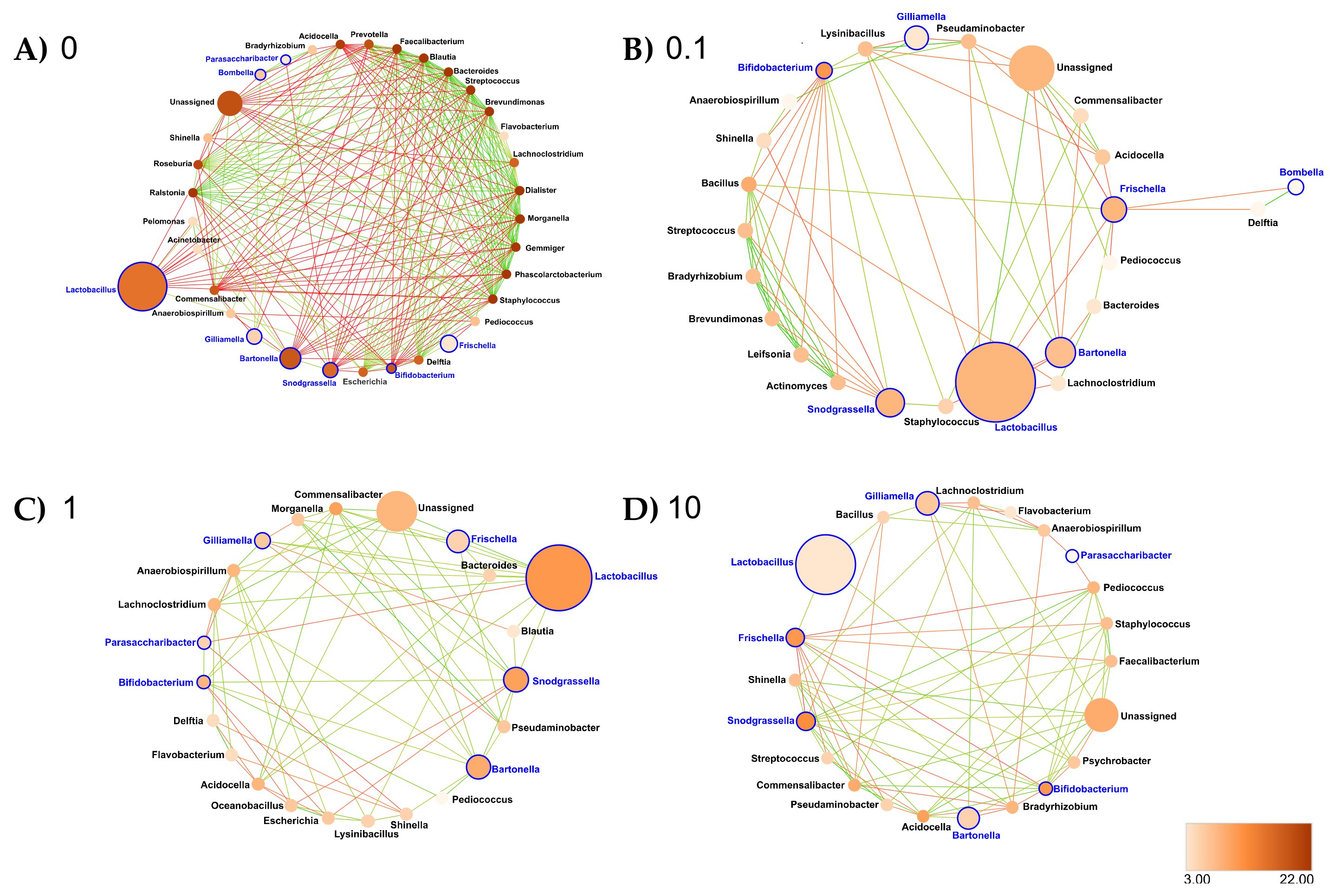
3.6. Clothianidin Disturbs Taxon–Taxon Interactions in the Honeybee Gut Microbiota
- Midgut: At the genus taxonomic rank, the number of interacting ASVs varied from 45 (control midgut) to 35, 36 and 33, respectively, after 0.1, 1 and 10 ppb of exposure to clothianidin (Figure 6). Significant positive correlations decreased from 96 (control midgut) to 28, 30 and 40, respectively, at 0.1, 1 and 10 ppb of exposure to clothianidin, and significant negative correlations increased from 1 (control midgut) to 5, 5 and 4, respectively, at 0.1, 1 and 10 ppb of exposure to clothianidin (Supplementary Materials Table S30). Concerning honeybee gut core members: Lactobacillus, Snodgrassella and Gilliamella activity was variable across concentrations. For all clothianidin treatment concentrations, Bifidobacterium (core members) activity increased, Frischella (core member) activity decreased and Flavobacterium (low activity taxa) decreased. Then, there was a gain in significant correlations with the following low activity ASV Ralstonia (0.1 and 10 ppb). Finally, there was a loss of correlations with Devosia (0.1 and 1 ppb) and Leifsonia (for all three concentrations).
- Ileum: At the genus taxonomic rank, the number of interacting ASVs varied from 35 (control ileum) to 42, 40 and 30, respectively, after 0.1, 1 and 10 ppb of exposure to clothianidin (Figure 7). Significant positive correlations increased from 27 (control ileum) to 52, 73 and 29 at 0.1, 1 and 10 ppb (Figure 7, Supplementary Materials Table S30). Regarding negative correlations, significant negative correlations decreased from 6 (control ileum) to 5, 4 and 2, respectively, at 0.1, 1 and 10 ppb of exposure to clothianidin (Figure 7, Supplementary Materials Table S30). Concerning honeybee gut core members, Lactobacillus and Gilliamella activity was variable across all treatment concentrations. For all concentrations, Bifidobacterium (core members) activity increased and Snodgrassella and Frischella (core members) activity decreased. We observed a gain in significant correlations with low activity ASVs: Moraxella and Prevotella (1 ppb); Lawsonella (0.1 and 1 ppb) and Ralstonia (for all three concentrations). At 0.1 ppb, we observed a slight increase in Pseudomonas and Flavobacterium taxa activity.
- Rectum: At the genus taxonomic rank, the number of interacting ASVs varied from 34 (control rectum) to 25, 23 and 22, respectively, after 0.1, 1 and 10 ppb of exposure to clothianidin (Figure 8). We observed a strong decrease in significant positive correlations, from 108 (control rectum) to 21, 13 and 17, respectively, at 0.1, 1 and 10 ppb of exposure to clothianidin, and a decrease in significant negative correlations from 5 (control rectum) to 1, 0 and 3, respectively, at 0.1, 1 and 10 ppb of exposure to clothianidin (Figure 8, Supplementary Materials Table S30). Concerning honeybee gut members: Lactobacillus, Snodgrassella and Gilliamella (core members) activity was variable across treatment concentrations. For all concentrations, Bifidobacterium (core member) activity increased, while Frischella (core member) activity decreased.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willmer, P.G.; Cunnold, H.; Ballantyne, G. Insights from Measuring Pollen Deposition: Quantifying the Pre-Eminence of Bees as Flower Visitors and Effective Pollinators. Arthropod Plant Interact. 2017, 11, 411–425. [Google Scholar] [CrossRef] [Green Version]
- Mancini, F.; Woodcock, B.A.; Isaac, N.J.B. Agrochemicals in the Wild: Identifying Links between Pesticide Use and Declines of Non-Target Organisms. Curr. Opin. Environ. Sci. Health 2019, 11, 53–58. [Google Scholar] [CrossRef]
- Casida, J.E. Neonicotinoids and Other Insect Nicotinic Receptor Competitive Modulators: Progress and Prospects. Annu. Rev. Entomol. 2018, 63, 125–144. [Google Scholar] [CrossRef]
- Alkassab, A.T.; Kirchner, W.H. Sublethal Exposure to Neonicotinoids and Related Side Effects on Insect Pollinators: Honeybees, Bumblebees, and Solitary Bees. J. Plant. Dis. Prot. 2017, 124, 1–30. [Google Scholar] [CrossRef]
- Iancu, V.-I.; Petre, J.; Galaon, T.; Radu, G.L. Occurrence of Neonicotinoid Residues in Danube River and Tributaries. Rev. De Chim. 2019, 70, 313–318. [Google Scholar] [CrossRef]
- Chrétien, F.; Giroux, I.; Thériault, G.; Gagnon, P.; Corriveau, J. Surface Runoff and Subsurface Tile Drain Losses of Neonicotinoids and Companion Herbicides at Edge-of-Field. Environ. Pollut. 2017, 224, 255–264. [Google Scholar] [CrossRef]
- Porrini, C.; Ghini, S.; Girotti, S.; Sabatini, A.G. 11 Use of Honey Bees as Bioindicators of Environmental Pollution in Italy. In Honey Bees: Estimating The Environmental Impact of Chemicals; Taylor & Francis: London, UK, 2002. [Google Scholar]
- Rouzé, R.; Moné, A.; Delbac, F.; Belzunces, L.; Blot, N. The Honeybee Gut Microbiota Is Altered after Chronic Exposure to Different Families of Insecticides and Infection by Nosema Ceranae. Microbes Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef] [Green Version]
- Cook, S.C. Compound and Dose-Dependent Effects of Two Neonicotinoid Pesticides on Honey Bee (Apis mellifera) Metabolic Physiology. Insects 2019, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Forkpah, C.; Dixon, L.R.; Fahrbach, S.E.; Rueppell, O. Xenobiotic Effects on Intestinal Stem Cell Proliferation in Adult Honey Bee (Apis mellifera L) Workers. PLoS ONE 2014, 9, e91180. [Google Scholar] [CrossRef]
- Aliouane, Y.; El Hassani, A.K.; Gary, V.; Armengaud, C.; Lambin, M.; Gauthier, M. Subchronic Exposure of Honeybees to Sublethal Doses of Pesticides: Effects on Behavior. Environ. Toxicol. Chem. 2009, 28, 113–122. [Google Scholar] [CrossRef]
- Desneux, N.; Decourtye, A.; Delpuech, J.-M. The Sublethal Effects of Pesticides on Beneficial Arthropods. Annu. Rev. Entomol. 2007, 52, 81–106. [Google Scholar] [CrossRef]
- Palmer, M.J.; Moffat, C.; Saranzewa, N.; Harvey, J.; Wright, G.A.; Connolly, C.N. Cholinergic Pesticides Cause Mushroom Body Neuronal Inactivation in Honeybees. Nat. Commun. 2013, 4, 1634. [Google Scholar] [CrossRef] [Green Version]
- Staveley, J.P.; Law, S.A.; Fairbrother, A.; Menzie, C.A. A Causal Analysis of Observed Declines in Managed Honey Bees (Apis mellifera). Hum. Ecol. Risk Assess. 2014, 20, 566–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberoni, D.; Gaggìa, F.; Baffoni, L.; Di Gioia, D. Beneficial Microorganisms for Honey Bees: Problems and Progresses. Appl. Microbiol. Biotechnol. 2016, 100, 9469–9482. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional Diversity within the Simple Gut Microbiota of the Honey Bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberti, J.; Engel, P. The Gut Microbiota—Brain Axis of Insects. Curr. Opin. Insect Sci. 2020, 39, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The Role of the Gut Microbiome in Health and Disease of Adult Honey Bee Workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Qiao, N.-H.; Diao, Q.-Y.; Jing, Z.; Vukanti, R.; Dai, P.-L.; Ge, Y. Thiacloprid Exposure Perturbs the Gut Microbiota and Reduces the Survival Status in Honeybees. J. Hazard. Mater. 2019, 389, 121818. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram Disturbs Gut Microbiota and Influences Metabolic Homeostasis and Immunity in Honey Bee (Apis mellifera, L.). Environ. Pollut. 2019, 258, 113671. [Google Scholar] [CrossRef]
- Daisley, B.A.; Trinder, M.; McDowell, T.W.; Welle, H.; Dube, J.S.; Ali, S.N.; Leong, H.S.; Sumarah, M.W.; Reid, G. Neonicotinoid-Induced Pathogen Susceptibility Is Mitigated by Lactobacillus plantarum Immune Stimulation in a Drosophila melanogaster Model. Sci. Rep. 2017, 7, 2703. [Google Scholar] [CrossRef]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid Clothianidin Adversely Affects Insect Immunity and Promotes Replication of a Viral Pathogen in Honey Bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef] [Green Version]
- Nazzi, F.; Pennacchio, F. Honey Bee Antiviral Immune Barriers as Affected by Multiple Stress Factors: A Novel Paradigm to Interpret Colony Health Decline and Collapse. Viruses 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yachi, S.; Loreau, M. Biodiversity and Ecosystem Productivity in a Fluctuating Environment: The Insurance Hypothesis. Proc. Natl. Acad. Sci. USA 1999, 96, 1463–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.-H.; Ha, E.-M.; Lee, W.-J. Innate Immunity and Gut–microbe Mutualism in Drosophila. Dev. Comp. Immunol. 2010, 34, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune Pathways and Defence Mechanisms in Honey Bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [Green Version]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive Gut Microbiota of Honey Bees Assessed Using Deep Sampling from Individual Worker Bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef] [Green Version]
- Kwong, W.K.; Moran, N.A. Gut Microbial Communities of Social Bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Pester, M.; Bittner, N.; Deevong, P.; Wagner, M.; Loy, A. A “rare Biosphere”microorganism Contributes to Sulfate Reduction in a Peatland. ISME J. 2010, 4, 1591–1602. [Google Scholar] [CrossRef]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey Bees as Models for Gut Microbiota Research. Lab. Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and Evolutionary Patterns of Bacterial Gut Associates of Corbiculate Bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, H.; Han, L.; Cui, Y.; Xue, Y.; Cai, L.; Yu, Y. Changes in Soil Microbial Community Structure and Function Associated with Degradation and Resistance of Carbendazim and Chlortetracycline during Repeated Treatments. Sci. Total Environ. 2016, 572, 1203–1212. [Google Scholar] [CrossRef]
- Sułowicz, S.; Cycoń, M.; Piotrowska-Seget, Z. Non-Target Impact of Fungicide Tetraconazole on Microbial Communities in Soils with Different Agricultural Management. Ecotoxicology 2016, 25, 1047–1060. [Google Scholar] [CrossRef] [Green Version]
- DeLorenzo, M.E.; Scott, G.I.; Ross, P.E. Toxicity of Pesticides to Aquatic Microorganisms: A Review. Environ. Toxicol. Chem. 2001, 20, 84–98. [Google Scholar] [CrossRef]
- Russell, R.J.; Scott, C.; Jackson, C.J.; Pandey, R.; Pandey, G.; Taylor, M.C.; Coppin, C.W.; Liu, J.-W.; Oakeshott, J.G. The Evolution of New Enzyme Function: Lessons from Xenobiotic Metabolizing Bacteria versus Insecticide-Resistant Insects. Evol. Appl. 2011, 4, 225–248. [Google Scholar] [CrossRef]
- Widenfalk, A.; Bertilsson, S.; Sundh, I.; Goedkoop, W. Effects of Pesticides on Community Composition and Activity of Sediment Microbes—Responses at Various Levels of Microbial Community Organization. Environ. Pollut. 2008, 152, 576–584. [Google Scholar] [CrossRef]
- Wintermantel, D.; Locke, B.; Andersson, G.K.S.; Semberg, E.; Forsgren, E.; Osterman, J.; Rahbek Pedersen, T.; Bommarco, R.; Smith, H.G.; Rundlöf, M.; et al. Field-Level Clothianidin Exposure Affects Bumblebees but Generally Not Their Pathogens. Nat. Commun. 2018, 9, 5446. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; An, J.; Dang, Z.; Lv, H.; Pan, W.; Gao, Z. Treating Wheat Seeds with Neonicotinoid Insecticides Does Not Harm the Rhizosphere Microbial Community. PLoS ONE 2018, 13, e0205200. [Google Scholar] [CrossRef]
- Liu, J.; Song, Y.; Tang, M.; Lu, Q.; Zhong, G. Enhanced Dissipation of Xenobiotic Agrochemicals Harnessing Soil Microbiome in the Tillage-Reduced Rice-Dominated Agroecosystem. J. Hazard. Mater. 2020, 398, 122954. [Google Scholar] [CrossRef] [PubMed]
- Parte, S.G.; Kharat, A.S. Aerobic Degradation of Clothianidin to 2-Chloro-Methyl Thiazole and Methyl 3-(Thiazole-Yl) Methyl Guanidine Produced by Pseudomonas Stutzeri Smk. J. Environ. Public Health 2019, 2019, 4807913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulligan, R.A.; Tomco, P.L.; Howard, M.W.; Schempp, T.T.; Stewart, D.J.; Stacey, P.M.; Ball, D.B.; Tjeerdema, R.S. Aerobic versus Anaerobic Microbial Degradation of Clothianidin under Simulated California Rice Field Conditions. J. Agric. Food Chem. 2016, 64, 7059–7067. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Wang, J.; Tanaka, Y.; Nagai, K.; Kawagishi, H.; Hirai, H. Bioremediation of the Neonicotinoid Insecticide Clothianidin by the White-Rot Fungus Phanerochaete Sordida. J. Hazard. Mater. 2017, 321, 586–590. [Google Scholar] [CrossRef]
- Mohanta, D.; Ahmaruzzaman, M. A Novel Au-SnO2-rGO Ternary Nanoheterojunction Catalyst for UV-LED Induced Photocatalytic Degradation of Clothianidin: Identification of Reactive Intermediates, Degradation Pathway and in-Depth Mechanistic Insight. J. Hazard. Mater. 2020, 397, 122685. [Google Scholar] [CrossRef]
- Pang, S.; Lin, Z.; Zhang, W.; Mishra, S.; Bhatt, P.; Chen, S. Insights Into the Microbial Degradation and Biochemical Mechanisms of Neonicotinoids. Front. Microbiol. 2020, 11, 868. [Google Scholar] [CrossRef]
- Daisley, B.A.; Trinder, M.; McDowell, T.W.; Collins, S.L.; Sumarah, M.W.; Reid, G. Microbiota-Mediated Modulation of Organophosphate Insecticide Toxicity by Species-Dependent Interactions with Lactobacilli in a Drosophila Melanogaster Insect Model. Appl. Environ. Microbiol. 2018, 84, e02820-17. [Google Scholar] [CrossRef] [Green Version]
- Raymann, K.; Motta, E.V.S.; Girard, C.; Riddington, I.M.; Dinser, J.A.; Moran, N.A. Imidacloprid Decreases Honey Bee Survival Rates but Does Not Affect the Gut Microbiome. Appl. Environ. Microbiol. 2018, 84, e00545-18. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, Y.; Fu, J.; Hu, S.; Qu, J. Degradation of Acetochlor and Beneficial Effect of Phosphate-Solubilizing Bacillus Sp. ACD-9 on Maize Seedlings. 3 Biotech 2020, 10, 67. [Google Scholar] [CrossRef]
- Hussain, S.; Hartley, C.J.; Shettigar, M.; Pandey, G. Bacterial Biodegradation of Neonicotinoid Pesticides in Soil and Water Systems. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [Green Version]
- Bonilla-Rosso, G.; Engel, P. Functional Roles and Metabolic Niches in the Honey Bee Gut Microbiota. Curr. Opin. Microbiol. 2018, 43, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Boutin, S.; Bernatchez, L.; Audet, C.; Derôme, N. Network Analysis Highlights Complex Interactions between Pathogen, Host and Commensal Microbiota. PLoS ONE 2013, 8, e84772. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Cheaib, B.; Seghouani, H.; Ijaz, U.Z.; Derome, N. Community Recovery Dynamics in Yellow Perch Microbiome after Gradual and Constant Metallic Perturbations. Microbiome 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, S.; van Treuren, W.; Lozupone, C.; Faust, K.; Friedman, J.; Deng, Y.; Xia, L.C.; Xu, Z.Z.; Ursell, L.; Alm, E.J.; et al. Correlation Detection Strategies in Microbial Data Sets Vary Widely in Sensitivity and Precision. ISME J. 2016, 10, 1669–1681. [Google Scholar] [CrossRef]
- Hallmaier-Wacker, L.K.; Lüert, S.; Gronow, S.; Spröer, C.; Overmann, J.; Buller, N.; Vaughan-Higgins, R.J.; Knauf, S. A Metataxonomic Tool to Investigate the Diversity of Treponema. Front. Microbiol. 2019, 10, 2094. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal Microbiota Dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. Microbiome Survey of the Inflamed and Noninflamed Gut at Different Compartments Within the Gastrointestinal Tract of Inflammatory Bowel Disease Patients. Inflamm. Bowel Dis. 2016, 22, 817–825. [Google Scholar] [CrossRef] [Green Version]
- Williams, G.R.; Alaux, C.; Costa, C.; Csáki, T.; Doublet, V.; Eisenhardt, D.; Fries, I.; Kuhn, R.; McMahon, D.P.; Medrzycki, P.; et al. Standard Methods for Maintaining Adult Apis mellifera in Cages under in Vitro Laboratory Conditions. J. Apic. Res. 2013, 52, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Chen, Y.P.; di Prisco, G.; Pettis, J.; Williams, V. Bee Cups: Single-Use Cages for Honey Bee Experiments. J. Apic. Res. 2009, 48, 300–302. [Google Scholar] [CrossRef]
- Paradis, D.; Bérail, G.; Bonmatin, J.-M.; Belzunces, L.P. Sensitive Analytical Methods for 22 Relevant Insecticides of 3 Chemical Families in Honey by GC-MS/MS and LC-MS/MS. Anal. Bioanal. Chem. 2014, 406, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Marsaglia, G.; Tsang, W.W.; Wang, J. Others Evaluating Kolmogorov’s Distribution. J. Stat. Softw. 2003, 8, 1–4. [Google Scholar] [CrossRef]
- Royston, J.P. Shapiro-Wilk Normality Test and P-Value. Appl. Stat. 1995, 44, 547–551. [Google Scholar] [CrossRef]
- Bauer, D.F. Constructing Confidence Sets Using Rank Statistics. J. Am. Stat. Assoc. 1972, 67, 687–690. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Therneau, T.; A Package for Survival Analysis in S. Version 2.38; 2015. Available online: http://CRAN.R-project.org/package=survival (accessed on 20 September 2021).
- El Khoury, S.; Rousseau, A.; Lecoeur, A.; Cheaib, B.; Bouslama, S.; Mercier, P.-L.; Demey, V.; Castex, M.; Giovenazzo, P.; Derome, N. Deleterious Interaction between Honeybees (Apis mellifera) and Its Microsporidian Intracellular Parasite Nosema Ceranae Was Mitigated by Administrating Either Endogenous or Allochthonous Gut Microbiota Strains. Front. Ecol. Evol. 2018, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- qScriptTM cDNA SuperMix. Available online: https://www.dnature.co.nz/wp-content/uploads/95048-qScript-cDNA-SuperMix-PPS.pdf (accessed on 20 September 2017).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R. Taxonomy Annotation and Guide Tree Errors in 16S rRNA Databases. PeerJ 2018, 6, e5030. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Hmisc Package. Available online: http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc (accessed on 27 April 2018).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, W. Protein–protein Interaction Network Analysis of Insecticide Resistance Molecular Mechanism in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 2019, 100, e21523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, D.; Widder, S. Deciphering Microbial Interactions and Detecting Keystone Species with Co-Occurrence Networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehmuth, K.; Ziviani, A. Daccer: Distributed Assessment of the Closeness Centrality Ranking in Complex Networks. Comput. Netw. 2013, 57, 2536–2548. [Google Scholar] [CrossRef]
- Maslov, S.; Sneppen, K. Specificity and Stability in Topology of Protein Networks. Science 2002, 296, 910–913. [Google Scholar] [CrossRef] [Green Version]
- Ai, D.; Pan, H.; Li, X.; Wu, M.; Xia, L.C. Association Network Analysis Identifies Enzymatic Components of Gut Microbiota That Significantly Differ between Colorectal Cancer Patients and Healthy Controls. PeerJ 2019, 7, e7315. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Hyde, E.R.; Suchodolski, J.S.; Knight, R. Dog and Human Inflammatory Bowel Disease Rely on Overlapping yet Distinct Dysbiosis Networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef] [PubMed]
- Rohr, J.R.; Kerby, J.L.; Sih, A. Community Ecology as a Framework for Predicting Contaminant Effects. Trends Ecol. Evol. 2006, 21, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Babendreier, D.; Joller, D.; Romeis, J.; Bigler, F.; Widmer, F. Bacterial Community Structures in Honeybee Intestines and Their Response to Two Insecticidal Proteins. FEMS Microbiol. Ecol. 2007, 59, 600–610. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Cultivation and Characterization of the Gut Symbionts of Honey Bees and Bumble Bees: Description of Snodgrassella Alvi Gen. Nov., Sp. Nov., a Member of the Family Neisseriaceae of the Betaproteobacteria, and Gilliamella Apicola Gen. Nov., Sp. Nov., a Member of Orbaceae Fam. Nov., Orbales Ord. Nov., a Sister Taxon to the Order “Enterobacteriales” of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 2008–2018. [Google Scholar] [PubMed]
- Olofsson, T.C.; Alsterfjord, M.; Nilson, B.; Butler, È.; Vásquez, A. Lactobacillus Apinorum Sp. Nov., Lactobacillus Mellifer Sp. Nov., Lactobacillus Mellis Sp. Nov., Lactobacillus Melliventris Sp. Nov., Lactobacillus Kimbladii Sp. Nov., Lactobacillus Helsingborgensis Sp. Nov. and Lactobacillus Kullabergensis Sp. Nov., Isolated from the Honey Stomach of the Honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2014, 64, 3109–3119. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic Exposure Perturbs the Gut Microbiota and Elevates Mortality in Honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Paris, L.; Peghaire, E.; Moné, A.; Diogon, M.; Debroas, D.; Delbac, F.; El Alaoui, H. Honeybee Gut Microbiota Dysbiosis in Pesticide/parasite Co-Exposures Is Mainly Induced by Nosema Ceranae. J. Invertebr. Pathol. 2020, 172, 107348. [Google Scholar] [CrossRef]
- Wu, S.-C.; Cao, Z.-S.; Chang, K.-M.; Juang, J.-L. Intestinal Microbial Dysbiosis Aggravates the Progression of Alzheimer’s Disease in Drosophila. Nat. Commun. 2017, 8, 24. [Google Scholar] [CrossRef]
- Meng, X.-L.; Li, S.; Qin, C.-B.; Zhu, Z.-X.; Hu, W.-P.; Yang, L.-P.; Lu, R.-H.; Li, W.-J.; Nie, G.-X. Intestinal Microbiota and Lipid Metabolism Responses in the Common Carp (Cyprinus carpio, L.) Following Copper Exposure. Ecotoxicol. Environ. Saf. 2018, 160, 257–264. [Google Scholar] [CrossRef]
- Fleeger, J.W.; Carman, K.R.; Nisbet, R.M. Indirect Effects of Contaminants in Aquatic Ecosystems. Sci. Total Environ. 2003, 317, 207–233. [Google Scholar] [CrossRef]
- Clements, W.H.; Rohr, J.R. Community Responses to Contaminants: Using Basic Ecological Principles to Predict Ecotoxicological Effects. Environ. Toxicol. Chem. 2009, 28, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-Related Gut Bacterial Dysbiosis Correlates with Impaired Development, Increased Mortality and Nosema Disease in the Honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef]
- Emery, O.; Schmidt, K.; Engel, P. Immune System Stimulation by the Gut Symbiont Frischella Perrara in the Honey Bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone Taxa as Drivers of Microbiome Structure and Functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic Patterns of Co-Occurrence Network Topological Features for Soil Microbiota at Continental Scale in Eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Meriweather, M.; Matthews, S.; Rio, R.; Baucom, R.S. A 454 Survey Reveals the Community Composition and Core Microbiome of the Common Bed Bug (Cimex lectularius) across an Urban Landscape. PLoS ONE 2013, 8, e61465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapio, I.; Fischer, D.; Blasco, L.; Tapio, M.; Wallace, R.J.; Bayat, A.R.; Ventto, L.; Kahala, M.; Negussie, E.; Shingfield, K.J.; et al. Taxon Abundance, Diversity, Co-Occurrence and Network Analysis of the Ruminal Microbiota in Response to Dietary Changes in Dairy Cows. PLoS ONE 2017, 12, e0180260. [Google Scholar] [CrossRef] [Green Version]
- Piccini, C.; Antúnez, K.; Zunino, P. An Approach to the Characterization of the Honey Bee Hive Bacterial Flora. J. Apic. Res. 2004, 43, 101–104. [Google Scholar] [CrossRef]
- Anhalt, J.C.; Moorman, T.B.; Koskinen, W.C. Biodegradation of Imidacloprid by an Isolated Soil Microorganism. J. Environ. Sci. Health B 2007, 42, 509–514. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Lenaerts, M.; Brys, R.; Willems, K.; Honnay, O.; Lievens, B. Among-Population Variation in Microbial Community Structure in the Floral Nectar of the Bee-Pollinated Forest Herb Pulmonaria officinalis, L. PLoS ONE 2013, 8, e56917. [Google Scholar] [CrossRef]
- Dudek, K.; Humińska, K.; Wojciechowicz, J.; Tryjanowski, P. Metagenomic Survey of Bacteria Associated with the Invasive Ladybird Harmonia Axyridis (Coleoptera: Coccinellidae). Eur. J. Entomol. 2017, 117, 312–316. [Google Scholar] [CrossRef]
- Parente, C.E.T.; Brito, E.M.S.; Caretta, C.A.; Cervantes-Rodríguez, E.A.; Fábila-Canto, A.P.; Vollú, R.E.; Seldin, L.; Malm, O. Bacterial Diversity Changes in Agricultural Soils Influenced by Poultry Litter Fertilization. Braz. J. Microbiol. 2021, 52, 675–686. [Google Scholar] [CrossRef]
- Eilmus, S.; Heil, M. Bacterial Associates of Arboreal Ants and Their Putative Functions in an Obligate Ant-Plant Mutualism. Appl. Environ. Microbiol. 2009, 75, 4324–4332. [Google Scholar] [CrossRef] [Green Version]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin Study Indicates Loss of Interaction between Microbiota and Mucosa of Patients with Ulcerative Colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Johansson, A.; Sheehan, T.H.; Mott, B.M.; Corby-Harris, V.; Johnstone, L.; Sprissler, R.; Fitz, W. Draft Genome Sequences of Two Bifidobacterium Sp. from the Honey Bee (Apis mellifera). Gut Pathog. 2013, 5, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathialagan, M.; Johnson, Y.; Thangaraj, E. Isolation, Characterization and Identification of Probiotic Lactic Acid Bacteria (LAB) from Honey Bees. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 849–906. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Žiarovská, J.; Kowalczewski, P.Ł. In Vitro Antagonistic Effect of Gut Bacteriota Isolated from Indigenous Honey Bees and Essential Oils against Paenibacillus Larvae. Int. J. Mol. Sci. 2020, 21, 6736. [Google Scholar] [CrossRef]
- Ryu, J.-H.; Kim, S.-H.; Lee, H.-Y.; Bai, J.Y.; Nam, Y.-D.; Bae, J.-W.; Lee, D.G.; Shin, S.C.; Ha, E.-M.; Lee, W.-J. Innate Immune Homeostasis by the Homeobox Gene Caudal and Commensal-Gut Mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling Metabolic Functions of Bacteria in the Honey Bee Gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee Gut Microbiota Promotes Host Weight Gain via Bacterial Metabolism and Hormonal Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ptaszyńska, A.A.; Borsuk, G.; Mułenko, W.; Wilk, J. Others Impact of Vertebrate Probiotics on Honeybee Yeast Microbiota and on the Course of Nosemosis. Med. Weter. 2016, 72, 430–434. [Google Scholar]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baffoni, L.; Gaggìa, F.; Alberoni, D.; Cabbri, R. Effect of Dietary Supplementation of Bifidobacterium and Lactobacillus Strains in Apis mellifera, L. against Nosema ceranae. Benef. Microbes 2016, 7, 45–51. [Google Scholar] [CrossRef]
- Berríos, P.; Fuentes, J.A.; Salas, D.; Carreño, A. Inhibitory Effect of Biofilm-Forming Lactobacillus Kunkeei Strains against Virulent Pseudomonas aeruginosa in Vitro and in Honeycomb Moth (Galleria Mellonella). Benef. Microbes 2018, 9, 257–268. [Google Scholar] [CrossRef]
- Trinder, M.; McDowell, T.W.; Daisley, B.A.; Ali, S.N.; Leong, H.S.; Sumarah, M.W.; Reid, G. Probiotic Lactobacillus rhamnosus Reduces Organophosphate Pesticide Absorption and Toxicity to Drosophila melanogaster. Appl. Environ. Microbiol. 2016, 82, 6204–6213. [Google Scholar] [CrossRef] [Green Version]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Al, K.F.; Chernyshova, A.M.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Novel Probiotic Approach to Counter Paenibacillus Larvae Infection in Honey Bees. ISME J. 2020, 14, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Peghaire, E.; Moné, A.; Delbac, F.; Debroas, D.; Chaucheyras-Durand, F.; El Alaoui, H. A Pediococcus Strain to Rescue Honeybees by Decreasing Nosema ceranae- and Pesticide-Induced Adverse Effects. Pestic. Biochem. Physiol. 2020, 163, 138–146. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey Bee Gut Microbiome Is Altered by In-Hive Pesticide Exposures. Front. Microbiol. 2016, 7, 1255. [Google Scholar] [CrossRef] [Green Version]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune System Stimulation by the Native Gut Microbiota of Honey Bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [Green Version]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate Perturbs the Gut Microbiota of Honey Bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blot, N.; Veillat, L.; Rouzé, R.; Delatte, H. Glyphosate, but Not Its Metabolite AMPA, Alters the Honeybee Gut Microbiota. PLoS ONE 2019, 14, e0215466. [Google Scholar] [CrossRef]
- Alaux, C.; Brunet, J.-L.; Dussaubat, C.; Mondet, F.; Tchamitchan, S.; Cousin, M.; Brillard, J.; Baldy, A.; Belzunces, L.P.; Le Conte, Y. Interactions between Nosema microspores and a Neonicotinoid Weaken Honeybees (Apis mellifera). Environ. Microbiol. 2010, 12, 774–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettis, J.S.; Vanengelsdorp, D.; Johnson, J.; Dively, G. Pesticide Exposure in Honey Bees Results in Increased Levels of the Gut Pathogen Nosema. Naturwissenschaften 2012, 99, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Cutler, G.C.; Scott-Dupree, C.D.; Sultan, M.; McFarlane, A.D.; Brewer, L. A Large-Scale Field Study Examining Effects of Exposure to Clothianidin Seed-Treated Canola on Honey Bee Colony Health, Development, and Overwintering Success. PeerJ 2014, 2, e652. [Google Scholar] [CrossRef] [PubMed]
- Rolke, D.; Fuchs, S.; Grünewald, B.; Gao, Z.; Blenau, W. Large-Scale Monitoring of Effects of Clothianidin-Dressed Oilseed Rape Seeds on Pollinating Insects in Northern Germany: Effects on Honey Bees (Apis mellifera). Ecotoxicology 2016, 25, 1648–1665. [Google Scholar] [CrossRef] [Green Version]
- Lemanski, N.J.; Bansal, S.; Fefferman, N.H. The Sensitivity of a Honeybee Colony to Worker Mortality Depends on Season and Resource Availability. BMC Evol. Biol. 2020, 20, 139. [Google Scholar] [CrossRef]
- Osterman, J.; Wintermantel, D.; Locke, B.; Jonsson, O.; Semberg, E.; Onorati, P.; Forsgren, E.; Rosenkranz, P.; Rahbek-Pedersen, T.; Bommarco, R.; et al. Clothianidin Seed-Treatment Has No Detectable Negative Impact on Honeybee Colonies and Their Pathogens. Nat. Commun. 2019, 10, 692. [Google Scholar] [CrossRef] [Green Version]
- Mattson, M.P. Hormesis Defined. Ageing Res. Rev. 2008, 7, 1–7. [Google Scholar] [CrossRef]
- Brandt, A.; Gorenflo, A.; Siede, R.; Meixner, M.; Büchler, R. The Neonicotinoids Thiacloprid, Imidacloprid, and Clothianidin Affect the Immunocompetence of Honey Bees (Apis mellifera, L.). J. Insect Physiol. 2016, 86, 40–47. [Google Scholar] [CrossRef]
- Kessler, S.; Tiedeken, E.J.; Simcock, K.L.; Derveau, S.; Mitchell, J.; Softley, S.; Stout, J.C.; Wright, G.A. Bees Prefer Foods Containing Neonicotinoid Pesticides. Nature 2015, 521, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Erkosar, B.; Leulier, F. Transient Adult Microbiota, Gut Homeostasis and Longevity: Novel Insights from the Drosophila Model. FEBS Lett. 2014, 588, 4250–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branchiccela, B.; Castelli, L.; Corona, M.; Díaz-Cetti, S.; Invernizzi, C.; Martínez de la Escalera, G.; Mendoza, Y.; Santos, E.; Silva, C.; Zunino, P.; et al. Impact of Nutritional Stress on the Honeybee Colony Health. Sci. Rep. 2019, 9, 10156. [Google Scholar] [CrossRef] [PubMed]


| Coef | Exp(Coef) | Se(Coef) | Robust Se | z | p | |
|---|---|---|---|---|---|---|
| 0.1 ppb | 1.64479 | 5.17992 | 0.22153 | 0.57321 | 2.869 | 0.00411 ** |
| 1 ppb | 0.16670 | 1.18140 | 0.24819 | 0.78791 | 0.212 | 0.83244 |
| 10 ppb | −0.67473 | 0.50929 | 0.28351 | 0.41147 | −1.640 | 0.10105 |
| Logtime 0.1 ppb | −0.49352 | 0.61048 | 0.08382 | 0.21253 | −2.322 | 0.02023 * |
| Logtime 1 ppb | 0.01193 | 1.01200 | 0.09169 | 0.28505 | 0.042 | 0.96661 |
| Logtime 10 ppb | 0.30299 | 1.35390 | 0.10303 | 0.13766 | 2.201 | 0.02774 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Khoury, S.; Gauthier, J.; Bouslama, S.; Cheaib, B.; Giovenazzo, P.; Derome, N. Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract. Microorganisms 2021, 9, 2283. https://doi.org/10.3390/microorganisms9112283
El Khoury S, Gauthier J, Bouslama S, Cheaib B, Giovenazzo P, Derome N. Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract. Microorganisms. 2021; 9(11):2283. https://doi.org/10.3390/microorganisms9112283
Chicago/Turabian StyleEl Khoury, Sarah, Jeff Gauthier, Sidki Bouslama, Bachar Cheaib, Pierre Giovenazzo, and Nicolas Derome. 2021. "Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract" Microorganisms 9, no. 11: 2283. https://doi.org/10.3390/microorganisms9112283
APA StyleEl Khoury, S., Gauthier, J., Bouslama, S., Cheaib, B., Giovenazzo, P., & Derome, N. (2021). Dietary Contamination with a Neonicotinoid (Clothianidin) Gradient Triggers Specific Dysbiosis Signatures of Microbiota Activity along the Honeybee (Apis mellifera) Digestive Tract. Microorganisms, 9(11), 2283. https://doi.org/10.3390/microorganisms9112283







