First Report of CC5-MRSA-IV-SCCfus “Maltese Clone” in Bat Guano
Abstract
:1. Introduction
2. Materials and Methods
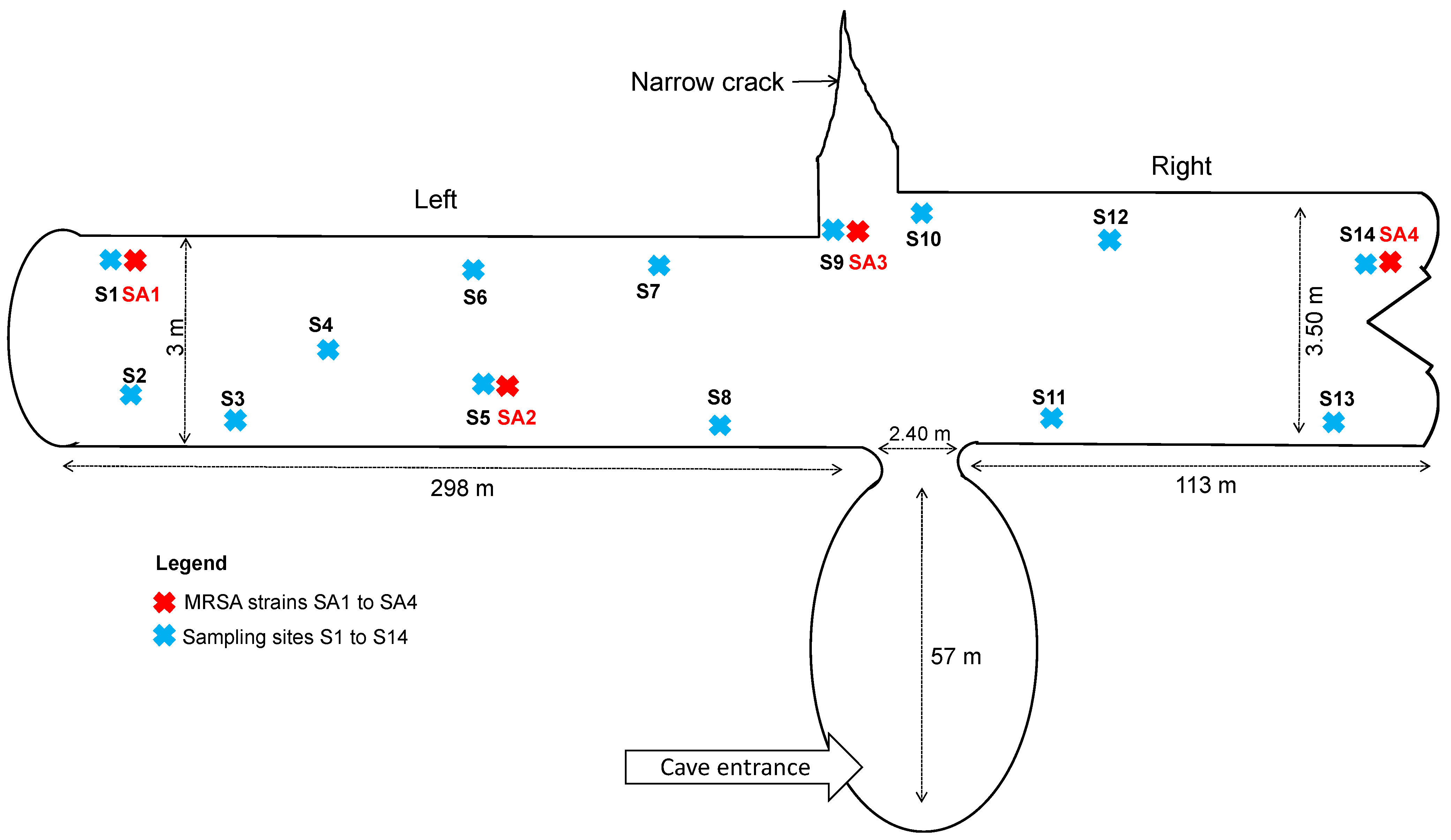
2.1. Fecal Samples
2.2. Bacterial Identification
2.3. Antimicrobial Susceptibility Testing
2.4. Whole-Genome Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Igrejas, G.; Correia, S.; Silva, V.; Hébraud, M.; Caniça, M.; Torres, C.; Gomes, C.; Nogueira, F.; Poeta, P. Planning a One Health Case Study to Evaluate Methicillin resistant Staphylococcus aureus and its economic burden in Portugal. Front. Microbiol. 2018, 9, 2964. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aires-de-Sousa, M. Methicillin-resistant Staphylococcus aureus among animals: Current overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Lancet Infect. Dis. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, M. Methicillin-resistant Staphylococcus aureus and animals: Zoonosis or humanosis? J. Antimicrob. Chemother. 2008, 62, 1181–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mairi, A.; Touati, A.; Pantel, A.; Zenati, K.; Yahiahoui-Martinez, A.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.P. Distribution of toxinogenic methicillin-resistant and methicillin-susceptible Staphylococcus aureus from different ecological niches in Algeria. Toxins 2019, 11, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moratelli, R.; Calisher, C.H. Bats and zoonotic viruses: Can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz 2015, 110, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Thiagavel, J.; Cechetto, C.; Santana, S.E.; Jakobsen, L.; Warrant, E.J.; Ratcliffe, J.M. Auditory opportunity and visual constraint enabled the evolution of echolocation in bats. Nat. Commun. 2018, 9, 98. [Google Scholar] [CrossRef]
- Boyles, J.G.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Conservation. Economic importance of bats in agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef]
- Kamani, J.; Baneth, G.; Mitchell, M.; Mumcuoglu, K.Y.; Gutiérrez, R.; Harrus, S. Bartonella species in bats (Chiroptera) and bat flies (Nycteribiidae) from Nigeria, West Africa. Vector Borne Zoonotic Dis. 2014, 14, 625–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allocati, N.; Petrucci, A.G.; Di Giovanni, P.; Masulli, M.; Di Ilio, C.; De Laurenzi, V. Bat-man disease transmission: Zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2016, 2, 16048. [Google Scholar] [CrossRef] [Green Version]
- Cicuttin, G.L.; De Salvo, M.N.; La Rosa, I.; Dohmen, F.E.G. Neoricketssia risticii, Rickettsia sp. and Bartonella sp. in Tadatida brasiliensis bats from Buenos Aires, Argentina. Comp. Immunol. Infect. Dis. 2017, 52, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Mühldorfer, K.; Tortosa, P.; Markotter, W. Leptospira and bats: Story of an emerging friendship. PLoS Pathog. 2015, 11, e1005176. [Google Scholar] [CrossRef] [PubMed]
- Akobi, B.; Aboderin, O.; Sasaki, T.; Shittu, A. Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 2012, 12, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galicia, M.M.; Buenrostro, A.; Garcia, J. Specific bacterial diversity in bats of different food guilds in Southern sierra Oaxaca, Mexico. Rev. Biol. Trop. 2014, 62, 1673–1681. [Google Scholar]
- Held, J.; Gmeiner, M.; Mordmüller, B.; Matsiégui, P.B.; Schaer, J.; Eckerle, I.; Weber, N.; Matuschewski, K.; Bletz, S.; Schaumburg, F. Bats are reservoirs of Staphylococcus aureus complex in Gabon. Infect. Genet. Evol. 2017, 47, 118–120. [Google Scholar] [CrossRef]
- Olatimehin, A.; Shittu, A.O.; Onwugamba, F.C.; Mellmann, A.; Becker, K.; Schaumburg, F. Staphylococcus aureus complex in the straw-colored fruit bat (Eidolon helvum) in Nigeria. Front. Microb. 2018, 9, 162. [Google Scholar] [CrossRef] [Green Version]
- Fountain, K.; Roberts, L.; Young, V.; Barbon, A.; Frosini, S.M.; Lloyd, D.H.; Loeffler, A. Diversity of staphylococcal species cultured from captive livingstone’s fruit bats (Pteropus livingstonii) and their environment. J. Zoo Wildl. Med. 2019, 50, 266–269. [Google Scholar] [PubMed]
- Shittu, A.O.; Mellmann, A.; Schaumburg, F. Molecular characterization of Staphylococcus aureus complex from fomites in Nigeria. Infect. Genet. Evol. 2020, 85, 104504. [Google Scholar] [CrossRef] [PubMed]
- Konieczna, I.; Durlik, M.; Kwinkowski, M.; Domański, J.; Markowski, J.; Kaca, W. Properties of bacterial microflora isolated from bat guano. Med. Weter. 2007, 63, 1626–1629. [Google Scholar]
- European Committee on Antimicrobial Susceptibility. Available online: www.eucast.org (accessed on 30 June 2019).
- Sahebnasagh, R.; Saderi, H.; Owlia, P. The prevalence of resistance to methicillin in Staphylococcus aureus strains isolated from patients by PCR method for detection of mecA and nuc genes. Iran. J. Public Health 2014, 43, 84–92. [Google Scholar]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ankrum, A.; Hall, B.G. Population dynamics of Staphylococcus aureus in cystic fibrosis patients to determine transmission events by use of whole-genome sequencing. J. Clin. Microbiol. 2017, 55, 2143–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppé, E.; Barbier, F.; Mesli, Y.; Maiga, A.; Cojocaru, R.; Benkhalfat, M.; Benchouk, S.; Hassaine, H.; Maiga, I.; Diallo, A.; et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 2009, 53, 442–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Titouche, Y.; Hakem, A.; Houali, K.; Meheut, T.; Vingadassalon, N.; Ruiz-Ripa, L.; Salmi, D.; Chergui, A.; Chenouf, N.; Hennekinne, J.A.; et al. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) ST8 in raw milk and traditional dairy products in the Tizi Ouzou area of Algeria. J. Dairy Sci. 2019, 102, 6876–6884. [Google Scholar] [CrossRef]
- Akkou, M.; Bouchiat, C.; Antri, K.; Bes, M.; Tristan, A.; Dauwalder, O.; Martins-Simoes, P.; Rasigade, J.P.; Etienne, J.; Vandenesch, F.; et al. New host shift from human to cows within Staphylococcus aureus involved in bovine mastitis and nasal carriage of animal’s caretakers. Vet. Microbiol. 2018, 223, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Agabou, A.; Ouchenane, Z.; Ngba Essebe, C.; Khemissi, S.; Chehboub, M.T.E.; Chehboub, I.B.; Sotto, A.; Dunyach-Remy, C.; Lavigne, J.P. Emergence of nasal carriage of ST80 and ST152 PVL+ Staphylococcus aureus isolates from Livestock in Algeria. Toxins 2017, 9, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achek, R.; Hotzel, H.; Nabi, I.; Kechida, S.; Mami, D.; Didouh, N.; Tomaso, H.; Neubauer, H.; Ehricht, R.; Monecke, S.; et al. Phenotypic and molecular detection of biofilm formation in Staphylococcus aureus isolated from different sources in Algeria. Pathogens 2020, 9, 153. [Google Scholar] [CrossRef] [Green Version]
- Scicluna, E.A.; Shore, A.C.; Thürmer, A.; Ehricht, R.; Slickers, P.; Borg, M.A.; Coleman, D.C.; Monecke, S. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Scerri, J.; Monecke, S.; Borg, M.A. Prevalence and characteristics of community carriage of methicillin-resistant Staphylococcus aureus in Malta. J. Epidemiol. Glob. Health 2013, 3, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senok, A.; Nassar, R.; Kaklamanos, E.G.; Belhoul, K.; Abu Fanas, S.; Nassar, M.; Azar, A.J.; Müller, E.; Reissig, A.; Gawlik, D.; et al. Molecular characterization of Staphylococcus aureus isolates associated with nasal colonization and environmental contamination in academic dental clinics. Microb. Drug Resist. 2020, 26, 661–669. [Google Scholar] [CrossRef]
- Roberts, M.C.; Joshi, P.R.; Monecke, S.; Ehricht, R.; Müller, E.; Gawlik, D.; Diezel, C.; Braun, S.D.; Paudel, S.; Acharya, M.; et al. Staphylococcus aureus and methicillin resistant S. aureus in Nepalese primates: Resistance to antimicrobials, virulence, and genetic lineages. Antibiotics 2020, 9, 689. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Alcalá, L.; Simón, C.; Gómez, P.; Mama, O.M.; Rezusta, A.; Zarazaga, M.; Torres, C. Diversity of Staphylococcus aureus clones in wild mammals in Aragon, Spain, with detection of MRSA ST130-mecC in wild rabbits. J. Appl. Microbiol. 2019, 127, 284–291. [Google Scholar] [CrossRef]
- Gómez, P.; Lozano, C.; González-Barrio, D.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet. Microbiol. 2015, 177, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.; Lozano, C.; Camacho, M.C.; Lima-Barbero, J.F.; Hernández, J.M.; Zarazaga, M.; Höfle, Ú.; Torre, C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016, 71, 53–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mairi, A.; Touati, A.; Lavigne, J.P. Methicillin-Resistant Staphylococcus aureus ST80 clone: A systematic review. Toxins 2020, 12, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antri, K.; Rouzic, N.; Boubekri, I.; Dauwalder, O.; Beloufa, A.; Ziane, H.; Djennane, F.; Neggazi, M.; Benhabyles, B.; Bes, M.; et al. High prevalence of community- and hospital-acquired infections of methicillin-resistant Staphylococcus aureus containing Panton-Valentine leukocidin gene in Algiers. Pathol. Biol. 2010, 58, e15–e20. [Google Scholar] [CrossRef] [PubMed]
- Chaalal, W.; Chaalal, N.; Bourafa, N.; Kihal, M.; Diene, S.M.; Rolain, J.M. Characterization of Staphylococcus aureus isolated from food products in Western Algeria. Foodborne Pathog. Dis. 2018, 15, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Gharout-Sait, A.; Touati, A.; Ahmim, M.; Brasme, L.; Guillard, T.; Agsous, A.; de Champs, C. Occurrence of carbapenemase-producing Klebsiella pneumoniae in Bat Guano. Microb. Drug Resist. 2019, 25, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Makowska, N.; Bresa, K.; Koczura, R.; Philips, A.; Nowis, K.; Mokracka, J. Urban wastewater as a conduit for pathogenic Gram-positive bacteria and genes encoding resistance to β-lactams and glycopeptides. Sci. Total Environ. 2021, 765, 144176. [Google Scholar] [CrossRef]
- Adesoji, T.O.; Egyir, B.; Shittu, A.O. Antibiotic-resistant staphylococci from the wastewater treatment plant and grey-water samples in Obafemi Awolowo University, Ile-Ife, Nigeria. J. Water Health 2020, 18, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Boopathy, R. Presence of Methicillin resistant Staphylococcus aureus (MRSA) in sewage treatment plant. Bioresour. Technol. 2017, 240, 144–148. [Google Scholar] [CrossRef]
- Daniel, D.S.; Ng, Y.K.; Chua, E.L.; Arumugam, Y.; Wong, W.L.; Kumaran, J.V. Isolation and identification of gastrointestinal microbiota from the short-nosed fruit bat Cynopterus brachyotis brachyotis. Microbiol. Res. 2013, 168, 485–496. [Google Scholar] [CrossRef]
- Ahmim, M. Current status, distribution and conservation status of Algerian bats (Mammalia: Chiroptera). J. Threat. Taxa 2017, 9, 9723–9733. [Google Scholar] [CrossRef]
- Onwugamba, F.C.; Fitzgerald, J.R.; Rochon, K.; Guardabassi, L.; Alabi, A.; Kühne, S.; Grobusch, M.P.; Schaumburg, F. The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018, 22, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Kalemaki, D. Antimicrobial overuse and misuse in the community in Greece and link to antimicrobial resistance using methicillin-resistant S. aureus as an example. J. Infect. Public Health 2019, 12, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, F.; Mugisha, L.; Peck, B.; Becker, K.; Gillespie, T.R.; Peters, G.; Leendertz, F.H. Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 2012, 74, 1071–1075. [Google Scholar] [CrossRef]
- Senghore, M.; Bayliss, S.C.; Kwambana-Adams, B.A.; Foster-Nyarko, E.; Manneh, J.; Dione, M.; Badji, H.; Ebruke, C.; Doughty, E.L.; Thorpe, H.A.; et al. Transmission of Staphylococcus aureus from humans to green monkeys in the Gambia as revealed by whole-genome sequencing. Appl. Environ. Microbiol. 2016, 82, 5910–5917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cláudio, V.C.; Gonzalez, I.; Barbosa, G.; Rocha, V.; Moratelli, R.; Rassy, F. Bacteria richness and antibiotic-resistance in bats from a protected area in the Atlantic Forest of Southeastern Brazil. PLoS ONE 2018, 13, e0203411. [Google Scholar] [CrossRef]
- Fu, Z.; Ma, Y.; Chen, C.; Guo, Y.; Hu, F.; Liu, Y.; Xu, X.; Wang, M. Prevalence of fosfomycin resistance and mutations in murA, glpT, and uhpT in methicillin-resistant Staphylococcus aureus strains isolated from blood and cerebrospinal fluid samples. Front. Microbiol. 2016, 6, 1544. [Google Scholar] [CrossRef] [PubMed]
- Fillgrove, K.; Pakhomova, S.; Schaab, M.R.; Newcomer, M.E.; Armstrong, R.N. Structure and mechanism of the genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes. Biochemistry 2007, 46, 8110–8120. [Google Scholar] [CrossRef]
- García, P.; Arca, P.; Evaristo, S.J. Product of fosC, a gene from Pseudomonas syringae, mediates fosfomycin resistance by using ATP as cosubstrate. Antimicrob. Agents Chemother. 1995, 39, 1569–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y.; Park, Y.J.; Yu, J.K.; Jung, S.; Kim, Y.; Jeong, S.H.; Arakawa, Y. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 2012, 67, 2843–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Features | B-AoC-SA1 | B-AoC-SA2 | B-AoC-SA3 | B-AoC-SA4 |
|---|---|---|---|---|
| Genome size (bp) | 2,777,157 | 2,734,323 | 2,795,689 | 2,776,793 |
| Genome Coverage | 64.36X | 85.14X | 72.08X | 60.20X |
| Contigs | 35 | 46 | 60 | 34 |
| G + C Content (%) | 32.67 | 32.70 | 32.67 | 32.67 |
| CDS | 2539 | 2498 | 2556 | 2541 |
| tRNA | 61 | 61 | 61 | 61 |
| MLST | 149 | 149 | 149 | 149 |
| Locality | Bejaia (Algeria) | Bejaia (Algeria) | Bejaia (Algeria) | Bejaia (Algeria) |
| Strain Type | Strain | Date Isolated and Isolation Site 1 | Antimicrobial Resistance Phenotype 2 | Antibiotic Resistance Genes | Specific Toxinogenic Profile | Leucocidins Genes | Enterotoxins Genes 3 | Hemolysins Genes | MSCRAMMs Genes | Immune Evasion Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| CC5-MRSA-IV (Maltese clone)/ ST149; t010, agr group II | B-AoC-SA1 | 17 March 2016 S1 | PEN, FOX, FUS | mecA, blaZ, blaI, blaR, fusC (Q6GD50) | tst1+, pvl-, etA-, etB-, etD- | lukF, lukS, lukD, lukE, lukY | sea, sec, sel, egc-cluster | hl, hla, hllll, hlb, hlgA | bbp, clfA, ebh, ebpS, eno, fib, fnbA, fnbB, map, sasG, sdrC, sdrD, vwb | sak, scn |
| CC5-MRSA-IV (Maltese clone)/ ST149; t010, agr group II | B-AoC-SA2 | 1 April 2016 S5 | PEN, FOX, FUS | mecA, blaZ, blaI, blaR, fusC (Q6GD50) | tst1+, pvl-, etA-, etB-, etD- | lukF, lukS, lukD, lukE, lukY | sea, sec, sel, egc-cluster | hl, hla, hllll, hlb, hlgA | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sasG, sdrC, sdrD, vwb | sak, scn |
| CC5-MRSA-IV (Maltese clone)/ ST149; t010, agr group II | B-AoC-SA3 | 16 April 2016 S9 | PEN, FOX, FUS | mecA, blaZ, blaI, blaR, fusC (Q6GD50) | tst1+, pvl-, etA-, etB-, etD- | lukF, lukS, lukD, lukE, lukY | sea, sec, sel, egc-cluster | hl, hla, hllll, hlb, hlgA | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sasG, sdrC, sdrD, vwb | sak, scn |
| CC5-MRSA-IV (Maltese clone)/ ST149; t010, agr group II | B-AoC-SA4 | 17 May 2016 S14 | PEN, FOX, FUS | mecA, blaZ, blaI, blaR, fusC (Q6GD50) | tst1+, pvl-, etA-, etB-, etD- | lukF, lukS, lukD, lukE, lukY | sea, sec, sel, egc-cluster | hl, hla, hllll, hlb, hlgA | bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sasG, sdrC, sdrD, vwb | sak, scn |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mairi, A.; Touati, A.; Pantel, A.; Yahiaoui Martinez, A.; Ahmim, M.; Sotto, A.; Dunyach-Remy, C.; Lavigne, J.-P. First Report of CC5-MRSA-IV-SCCfus “Maltese Clone” in Bat Guano. Microorganisms 2021, 9, 2264. https://doi.org/10.3390/microorganisms9112264
Mairi A, Touati A, Pantel A, Yahiaoui Martinez A, Ahmim M, Sotto A, Dunyach-Remy C, Lavigne J-P. First Report of CC5-MRSA-IV-SCCfus “Maltese Clone” in Bat Guano. Microorganisms. 2021; 9(11):2264. https://doi.org/10.3390/microorganisms9112264
Chicago/Turabian StyleMairi, Assia, Abdelaziz Touati, Alix Pantel, Alex Yahiaoui Martinez, Mourad Ahmim, Albert Sotto, Catherine Dunyach-Remy, and Jean-Philippe Lavigne. 2021. "First Report of CC5-MRSA-IV-SCCfus “Maltese Clone” in Bat Guano" Microorganisms 9, no. 11: 2264. https://doi.org/10.3390/microorganisms9112264
APA StyleMairi, A., Touati, A., Pantel, A., Yahiaoui Martinez, A., Ahmim, M., Sotto, A., Dunyach-Remy, C., & Lavigne, J.-P. (2021). First Report of CC5-MRSA-IV-SCCfus “Maltese Clone” in Bat Guano. Microorganisms, 9(11), 2264. https://doi.org/10.3390/microorganisms9112264







