Flagella, Type I Fimbriae and Curli of Uropathogenic Escherichia coli Promote the Release of Proinflammatory Cytokines in a Coculture System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Design and Synthesis of Primers for Gene Disruption
2.3. Generation and Verification of Isogenic Mutants
2.4. Transmission Electron Microscopy and Protein Purification
2.5. Standardization of Cultured TCCSUP (HTB-5™) Human Bladder Cells and HMC-1 Human Mast Cells
2.6. Analysis of the Cytokines Production in a Coculture System
2.7. Determination of the Cytokines Levels Using Flow Cytometry
2.8. Adherence to HTB-5 Cells
2.9. Statistical Analysis
3. Results
3.1. Visualization of Type I Fimbriae, Curli, and Flagella in Different UPEC Strains under TEM
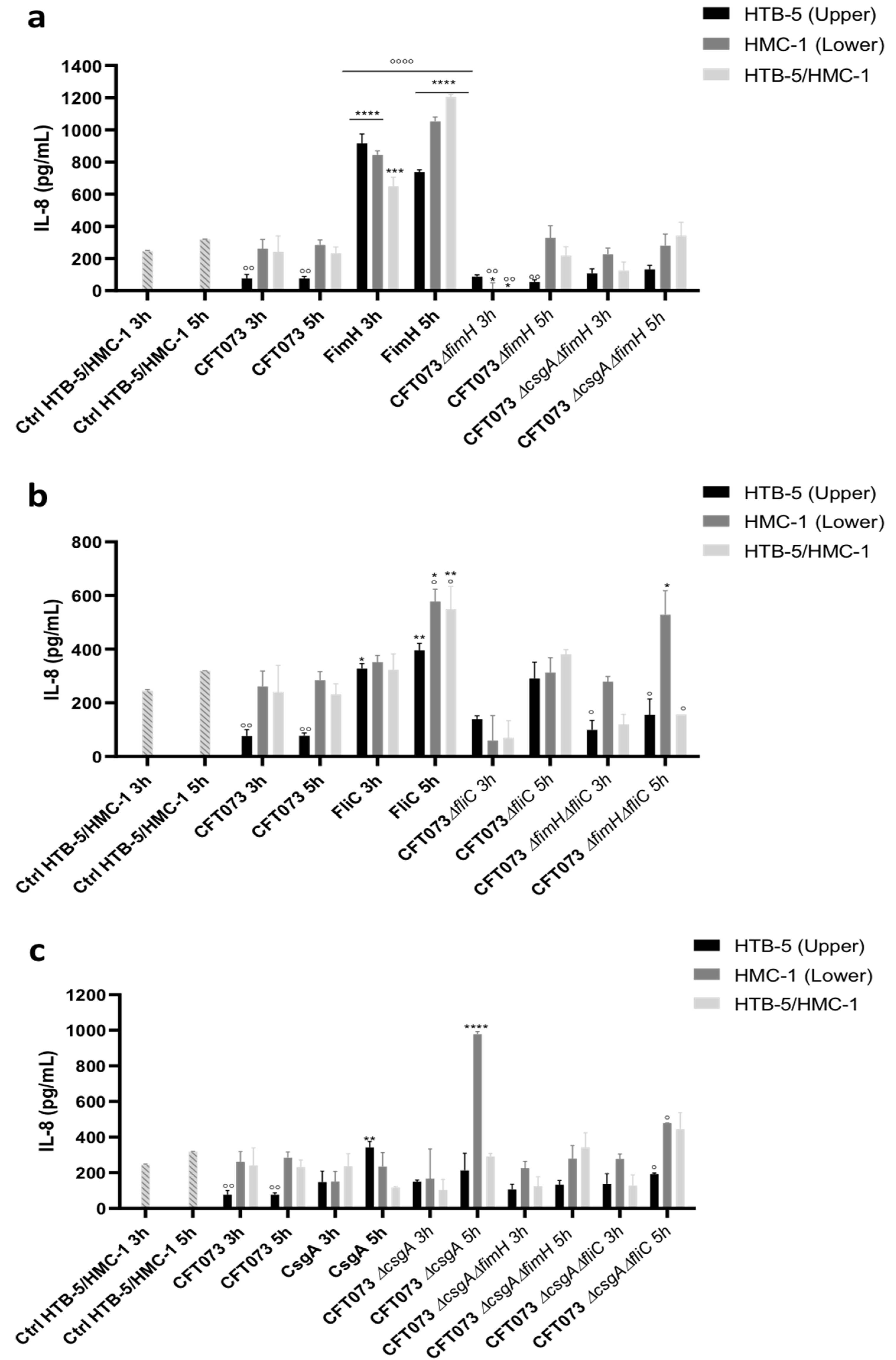
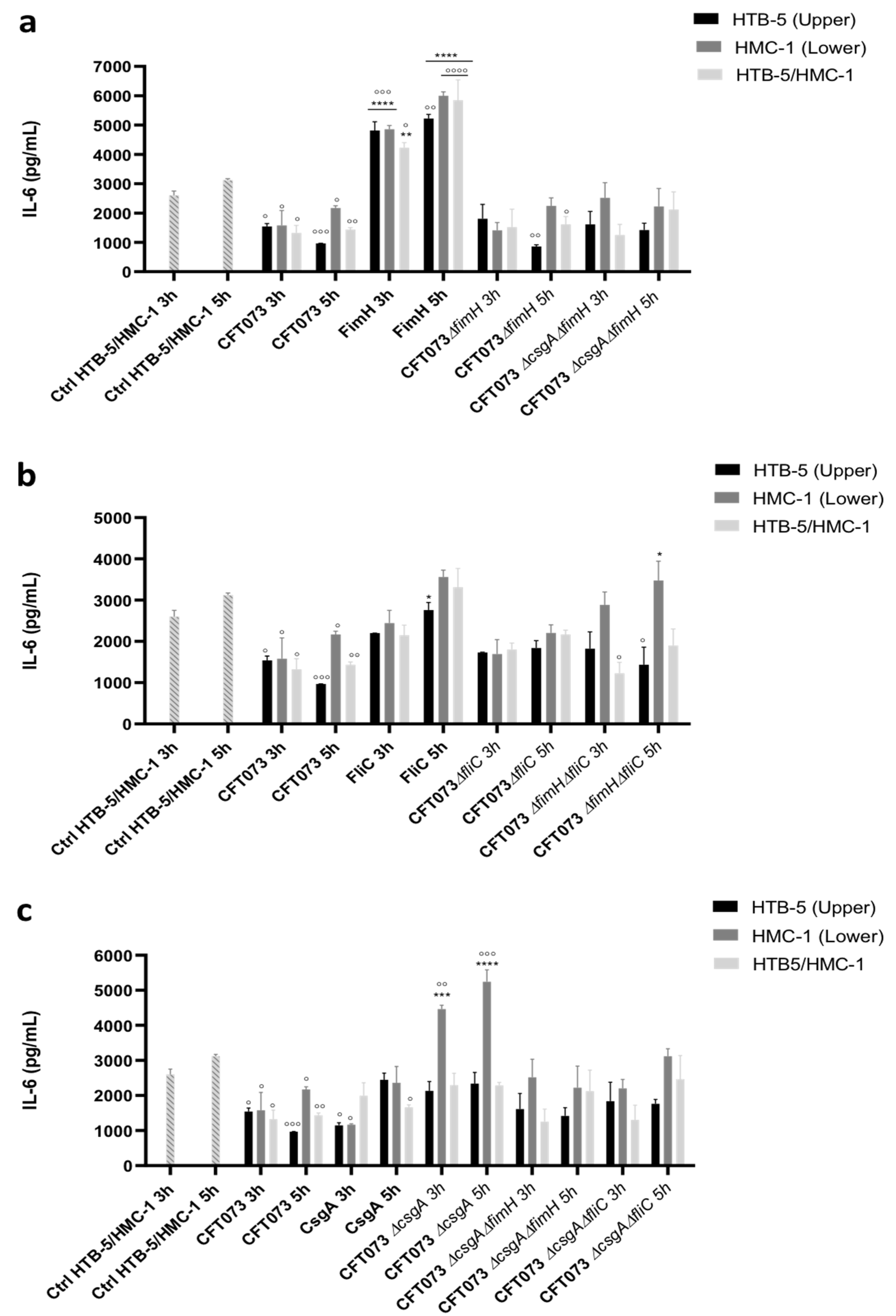
3.2. The Release of Proinflammatory Cytokines Is Induced in a Coculture System
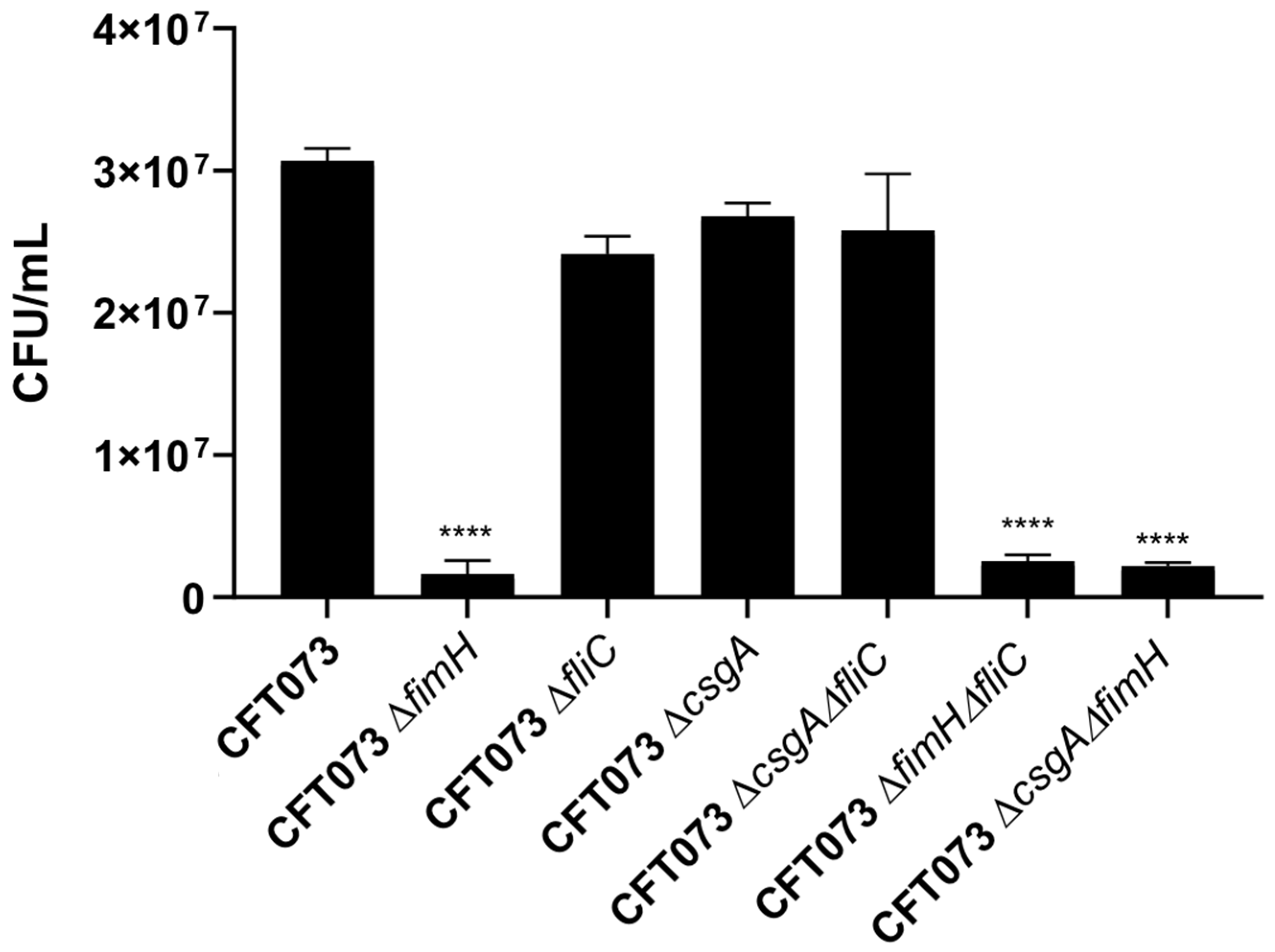
3.3. The Roles of the FimH, CsgA, and FliC Genes of UPEC in Adherence to HTB-5 Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Statement
References
- Brumbaugh, A.R.; Smith, S.N.; Mobley, H.L.T. Immunization with the yersiniabactin receptor, FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect. Immun. 2013, 81, 3309–3316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micali, S.; Isgro, G.; Bianchi, G.; Miceli, N.; Calapai, G.; Navarra, M. Cranberry and recurrent cystitis: More than marketing? Crit. Rev. Food Sci. Nutr. 2014, 54, 1063–1075. [Google Scholar] [CrossRef]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 175628721983217. [Google Scholar] [CrossRef] [Green Version]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Petca, R.C.; Mares, C.; Petca, A.; Negoita, A.; Popescu, R.I.; Bot, M.; Barabás, E.; Chibelean, C. Spectrum and Antibiotic Resistance of Uropathogens in Romanian Females. Antibiotics 2020, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Smelov, V.; Naber, K.; Bjerklund Johansen, T.E. Improved classification of urinary tract infection: Future considerations. Eur. Urol. Suppl. 2016, 15, 71–80. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Shaffer, K.; Bach, J.; Chun, R. Prospective study evaluating the incidence of bacteraemia and bacteruria in afebrile and febrile neutropaenic dogs undergoing chemotherapy. Vet. Med. Sci. 2016, 2, 281–294. [Google Scholar] [CrossRef] [Green Version]
- Aizen, E.; Shifrin, B.; Shugaev, I.; Potasman, I. Clinical and microbiological outcomes of asymptomatic bacteriuria in elderly stroke patients. Isr. Med. Assoc. J. 2017, 19, 147–151. [Google Scholar]
- Schaeffer, E.M. Re: Screening for asymptomatic bacteruria at one month after adult kidney transplantation: Clinical factors and implications. J. Urol. 2018, 200, 925. [Google Scholar] [CrossRef] [PubMed]
- Suthar, K.S.; Vanikar, A.V.; Nigam, L.A.; Patel, R.D.; Kanodia, K.V.; Thakkar, U.G.; Gandhi, P.A.; Chandak, S.A.; Prajapati, A.V.; Patel, M.H. Urinary screening for early detection of kidney diseases. Indian J. Pediatr. 2018, 85, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Kakkanat, A.; Totsika, M.; Schaale, K.; Duell, B.L.; Lo, A.W.; Phan, M.-D.; Moriel, D.G.; Beatson, S.A.; Sweet, M.J.; Ulett, G.C.; et al. The role of H4 flagella in Escherichia coli ST131 virulence. Sci. Rep. 2015, 5, 16149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, K.J.; Seed, P.C.; Hultgren, S.J. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 2005, 73, 7657–7668. [Google Scholar] [CrossRef] [Green Version]
- Hasanpour, M.; Najafi, A. Development of a multiplex real-time PCR assay for phylogenetic analysis of uropathogenic Escherichia coli. J. Microbiol. Methods 2017, 137, 25–29. [Google Scholar] [CrossRef]
- Ochoa, S.A.; Cruz-Córdova, A.; Luna-Pineda, V.M.; Reyes-Grajeda, J.P.; Cázares-Domínguez, V.; Escalona, G.; Sepúlveda-González, M.E.; López-Montiel, F.; Arellano-Galindo, J.; López-Martínez, B.; et al. Multidrug- and extensively drug-resistant uropathogenic Escherichia coli clinical strains: Phylogenetic groups widely associated with integrons maintain high genetic diversity. Front. Microbiol. 2016, 7, 2042. [Google Scholar] [CrossRef] [Green Version]
- Bartoletti, R.; Cai, T.; Wagenlehner, F.M.; Naber, K.; Bjerklund Johansen, T.E. Treatment of urinary tract infections and antibiotic stewardship. Eur. Urol. Suppl. 2016, 15, 81–87. [Google Scholar] [CrossRef]
- Sanchez, G.V.; Babiker, A.; Master, R.N.; Luu, T.; Mathur, A.; Bordon, J. Antibiotic resistance among urinary isolates from female outpatients in the United States in 2003 and 2012. Antimicrob. Agents Chemother. 2016, 60, 2680–2683. [Google Scholar] [CrossRef] [Green Version]
- Behzadi, P.; Urbán, E.; Gajdács, M. Association between Biofilm-Production and Antibiotic Resistance in Uropathogenic Escherichia coli (UPEC): An In Vitro Study. Diseases 2020, 8, 17. [Google Scholar] [CrossRef]
- Luna-Pineda, V.M.; Ochoa, S.A.; Cruz-Córdova, A.; Cázares-Domínguez, V.; Reyes-Grajeda, J.P.; Flores-Oropeza, M.A.; Arellano-Galindo, J.; Hernández-Castro, R.; Flores-Encarnación, M.; Ramírez-Vargas, A.; et al. Correction: Features of urinary Escherichia coli isolated from children with complicated and uncomplicated urinary tract infections in Mexico. PLoS ONE 2018, 13, e0204934. [Google Scholar] [CrossRef] [Green Version]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Subashchandrabose, S.; Smith, S.N.; Spurbeck, R.R.; Kole, M.M.; Mobley, H.L.T. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 2013, 9, e1003788. [Google Scholar] [CrossRef] [PubMed]
- Luna-Pineda, V.M.; Moreno-Fierros, L.; Cázares-Domínguez, V.; Ilhuicatzi-Alvarado, D.; Ochoa, S.A.; Cruz-Córdova, A.; Valencia-Mayoral, P.; Rodríguez-Leviz, A.; Xicohtencatl-Cortes, J. Curli of uropathogenic Escherichia coli enhance urinary tract colonization as a fitness factor. Front. Microbiol. 2019, 10, 2063. [Google Scholar] [CrossRef]
- Bessaiah, H.; Pokharel, P.; Loucif, H.; Kulbay, M.; Sasseville, C.; Habouria, H.; Houle, S.; Bernier, J.; Massé, É.; Van Grevenynghe, J.; et al. The RyfA small RNA regulates oxidative and osmotic stress responses and virulence in uropathogenic Escherichia coli. PLoS Pathog. 2021, 17, e1009617. [Google Scholar] [CrossRef] [PubMed]
- Luna-Pineda, V.M.; Reyes-Grajeda, J.P.; Cruz-Córdova, A.; Saldaña-Ahuactzi, Z.; Ochoa, S.A.; Maldonado-Bernal, C.; Cázares-Domínguez, V.; Moreno-Fierros, L.; Arellano-Galindo, J.; Hernández-Castro, R.; et al. Dimeric and trimeric fusion proteins generated with fimbrial adhesins of uropathogenic Escherichia coli. Front. Cell. Infect. Microbiol. 2016, 6, 135. [Google Scholar] [CrossRef] [Green Version]
- Shah, C.; Baral, R.; Bartaula, B.; Shrestha, L.B. Virulence factors of uropathogenic Escherichia coli (UPEC) and correlation with antimicrobial resistance. BMC Microbiol. 2019, 19, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, J.A.; Haugen, B.J.; Lockatell, C.V.; Maroncle, N.; Hagan, E.C.; Johnson, D.E.; Welch, R.A.; Mobley, H.L.T. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 2005, 73, 7588–7596. [Google Scholar] [CrossRef] [Green Version]
- Duell, B.L.; Carey, A.J.; Tan, C.K.; Cui, X.; Webb, R.I.; Totsika, M.; Schembri, M.A.; Derrington, P.; Irving-Rodgers, H.; Brooks, A.J.; et al. Innate transcriptional networks activated in bladder in response to uropathogenic Escherichia coli drive diverse biological pathways and rapid synthesis of IL-10 for defense against bacterial urinary tract infection. J. Immunol. 2011, 188, 781–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbone, M.; Hasty, D.L.; Yi, K.C.; Rue, J.; Fera, M.T.; Torre, F.; Giannone, M.; Losi, E. Cytokine induction in murine bladder tissue by type 1 fimbriated Escherichia coli. Ann. N. Y. Acad. Sci. 2006, 963, 332–335. [Google Scholar] [CrossRef]
- Acharya, D.; Sullivan, M.J.; Duell, B.L.; Goh, K.G.K.; Katupitiya, L.; Gosling, D.; Chamoun, M.N.; Kakkanat, A.; Chattopadhyay, D.; Crowley, M.; et al. Rapid bladder interleukin-10 synthesis in response to uropathogenic Escherichia coli is part of a defense strategy triggered by the major bacterial flagellar filament FliC and contingent on TLR5. mSphere 2019, 4, e00545-19. [Google Scholar] [CrossRef] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Hannan, T.J.; Hunstad, D.A. A Murine model for Escherichia coli urinary tract infection. Methods Mol. Biol. 2016, 1333, 159–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bien, J.; Sokolova, O.; Bozko, P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int. J. Nephrol. 2012, 2012, 681473. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Ábrok, M.; Lázar, A.; Burián, K. Urinary Tract Infections in Elderly Patients: A 10-Year Study on Their Epidemiology and Antibiotic Resistance Based on the WHO Access, Watch, Reserve (AWaRe) Classification. Antibiotics 2021, 10, 1098. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, P.; Brauner, A. Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Bact. Pathog. Biol. 2014, 65, 337–372. [Google Scholar] [CrossRef]
- Kot, B. Antibiotic resistance among uropathogenic Escherichia coli. Pol. J. Microbiol. 2019, 68, 403–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odoki, M.; Almustapha Aliero, A.; Tibyangye, J.; Nyabayo Maniga, J.; Wampande, E.; Drago Kato, C.; Agwu, E.; Bazira, J. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi district, Uganda. Int. J. Microbiol. 2019, 2019, 4246780. [Google Scholar] [CrossRef]
- Piantino, C.B.; Salvadori, F.A.; Ayres, P.P.; Kato, R.B.; Srougi, V.; Leite, K.R.; Srougi, M. An evaluation of the anti-neoplastic activity of curcumin in prostate cancer cell lines. Int. Braz. J. Urol. 2009, 35, 354–361. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, G.; Blom, T.; Kusche-Gullberg, M.; Kjellen, L.; Butterfield, J.H.; Sundstrom, C.; Nilsson, K.; Hellman, L. Phenotypic characterization of the human mast-cell line HMC-1. Scand. J. Immunol. 1994, 39, 489–498. [Google Scholar] [CrossRef]
- Wesolowski, J.; Paumet, F. The impact of bacterial infection on mast cell degranulation. Immunol. Res. 2011, 51, 215–226. [Google Scholar] [CrossRef]
- Lin, T.-J.; Gao, Z.; Arock, M.; Abraham, S.N. Internalization of FimH+ Escherichia coli by the human mast cell line (HMC-1 5C6) involves protein kinase C. J. Leukoc. Biol. 1999, 66, 1031–1038. [Google Scholar] [CrossRef]
- Cruse, G.; Fernandes, V.E.; de Salort, J.; Pankhania, D.; Marinas, M.S.; Brewin, H.; Andrew, P.W.; Bradding, P.; Kadioglu, A. Human lung mast cells mediate pneumococcal cell death in response to activation by pneumolysin. J. Immunol. 2010, 184, 7108–7115. [Google Scholar] [CrossRef]
- Garcia-Rodriguez, K.M.; Bahri, R.; Sattentau, C.; Roberts, I.S.; Goenka, A.; Bulfone-Paus, S. Human mast cells exhibit an individualized pattern of antimicrobial responses. Immun. Inflamm. Dis. 2020, 8, 198–210. [Google Scholar] [CrossRef]
- Duell, B.L.; Cripps, A.W.; Schembri, M.A.; Ulett, G.C. Epithelial cell coculture models for studying infectious diseases: Benefits and limitations. J. Biomed. Biotechnol. 2011, 2011, 852419. [Google Scholar] [CrossRef] [Green Version]
- Smith, Y.C.; Grande, K.K.; Rasmussen, S.B.; O’Brien, A.D. Novel three-dimensional organoid model for evaluation of the interaction of uropathogenic Escherichia coli with terminally differentiated human urothelial cells. Infect. Immun. 2006, 74, 750–757. [Google Scholar] [CrossRef] [Green Version]
- Barrila, J.; Radtke, A.L.; Crabbé, A.; Sarker, S.F.; Herbst-Kralovetz, M.M.; Ott, C.M.; Nickerson, C.A. Organotypic 3D cell culture models: Using the rotating wall vessel to study host–pathogen interactions. Nat. Rev. Microbiol. 2010, 8, 791–801. [Google Scholar] [CrossRef]
- Malaviya, R.; Abraham, S.N. Interaction of bacteria with mast cells. Methods Enzymol. 1995, 253, 27–43. [Google Scholar] [CrossRef]
- Chan, C.Y.; St John, A.L.; Abraham, S.N. Mast cell interleukin-10 drives localized tolerance in chronic bladder infection. Immunity 2013, 38, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Tebroke, J.; Lieverse, J.E.; Säfholm, J.; Schulte, G.; Nilsson, G.; Rönnberg, E. Wnt-3a induces cytokine release in human mast cells. Cells 2019, 8, 1372. [Google Scholar] [CrossRef] [Green Version]
- Javaid, N.; Patra, M.C.; Seo, H.; Yasmeen, F.; Choi, S. A rational insight into the effect of dimethyl sulfoxide on TNF-α activity. Int. J. Mol. Sci. 2020, 21, 9450. [Google Scholar] [CrossRef]
- Chromek, M.; Slamová, Z.; Bergman, P.; Kovács, L.; Podracká, L.u.; Ehrén, I.; Hökfelt, T.; Gudmundsson, G.H.; Gallo, R.L.; Agerberth, B.; et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006, 12, 636–641. [Google Scholar] [CrossRef]
- Kai-Larsen, Y.; Lüthje, P.; Chromek, M.; Peters, V.; Wang, X.; Holm, Å.; Kádas, L.; Hedlund, K.-O.; Johansson, J.; Chapman, M.R.; et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010, 6, e1001010. [Google Scholar] [CrossRef] [PubMed]
- Supajatura, V.; Ushio, H.; Nakao, A.; Okumura, K.; Ra, C.; Ogawa, H. Protective roles of mast cells against enterobacterial infection are mediated by toll-like receptor 4. J. Immunol. 2001, 167, 2250–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, D.S.; Fitzgerald, S.M.; Pitts, S.; Cantor, K.; King, E.; Lee, S.A.; Huang, S.K.; Krishnaswamy, G. MAPK-dependent regulation of IL-1- and β- adrenoreceptor-induced inflammatory cytokine production from mast cells: Implications for the stress response. BMC Immunol. 2004, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Lippert, U.; Welker, P.; Krüger-Krasagakes, S.; Möller, A.; Henz, B.M. Modulation of in vitro cytokine release from human leukemic mast cells (HMC-1) by glucocorticoids. Skin Pharmacol. Physiol. 1996, 9, 93–98. [Google Scholar] [CrossRef]
- Barbuti, G.; Moschioni, M.; Censini, S.; Covacci, A.; Montecucco, C.; Montemurro, P. Streptococcus pneumoniae induces mast cell degranulation. Int. J. Med. Microbiol. 2006, 296, 325–329. [Google Scholar] [CrossRef]
- Duell, B.L.; Carey, A.J.; Dando, S.J.; Schembri, M.A.; Ulett, G.C. Human bladder uroepithelial cells synergize with monocytes to promote IL-10 synthesis and other cytokine responses to uropathogenic Escherichia coli. PLoS ONE 2013, 8, e78013. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.W.; Brooking-Dixon, R.; Neupane, S.; Lee, C.J.; Miao, E.A.; Staats, H.F.; Abraham, S.N. Salmonella typhimurium impedes innate immunity with a mast-cell-suppressing protein tyrosine phosphatase, SptP. Immunity 2013, 39, 1108–1120. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, S.; Hernández-Pando, R.; Abraham, S.N.; Enciso, J.A. Mast cell activation by Mycobacterium tuberculosis: Mediator release and role of CD48. J. Immunol. 2003, 170, 5590–5596. [Google Scholar] [CrossRef] [Green Version]
- Fischer, H.; Yamamoto, M.; Akira, S.; Beutler, B.; Svanborg, C. Mechanism of pathogen-specific TLR4 activation in the mucosa: Fimbriae, recognition receptors and adaptor protein selection. Eur. J. Immunol. 2006, 36, 267–277. [Google Scholar] [CrossRef]
- Mukai, K.; Tsai, M.; Saito, H.; Galli, S.J. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 2018, 282, 121–150. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Jung, M.Y.; Olivera, A.; Gilfillan, A.M.; Prussin, C.; Kirshenbaum, A.S.; Beaven, M.A.; Metcalfe, D.D. IL-6 promotes an increase in human mast cell numbers and reactivity through suppression of suppressor of cytokine signaling 3. J. Allergy Clin. Immunol. 2016, 137, 1863–1871.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
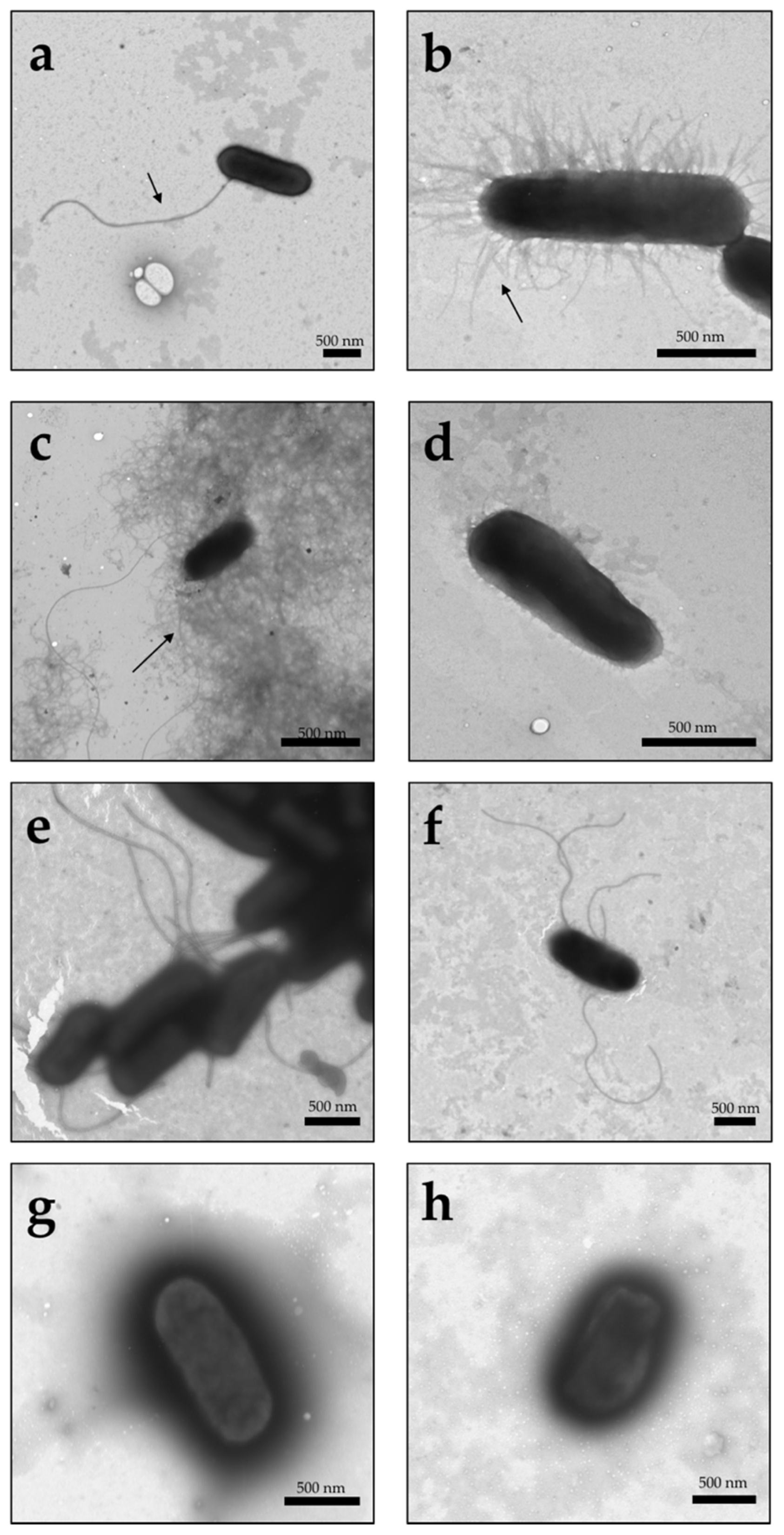
| Bacterial Strain and Plasmid. | Features | Reference |
|---|---|---|
| Bacterial Strains | ||
| CFT073 | UPEC Human clinical specimen: blood and urine from a woman with acute pyelonephritis. | ATCC |
| CFT073ΔfimH | fimH disruption, KmR | Luna-Pineda et al. [25] |
| CFT073ΔfliC | fliC disruption, KmR | This study |
| CFT073ΔcsgA | csgA disruption, KmR | Luna-Pineda et al. [25] |
| CFT073ΔfimHΔfliC | fimH and fliC disruption, KmR CmR | This study |
| CFT073ΔcsgAΔfimH | csgA and fimH disruption, KmR CmR | Luna-Pineda et al. [25] |
| CFT073ΔcsgAΔfliC | csgA and fliC disruption, KmR, CmR | This study |
| Plasmids | ||
| pKD46 | Plasmid expressing the λ phage recombination system (pBAD-λ-Red (γ β exo) ApR | Datsenko and Wanner [31] |
| pKD4 | Template vector for amplifying FRT-kan FRT; bla FRT km P1 P2 oriR6K KmR | Datsenko and Wanner [31] |
| pKD3 | Template vector for amplifying the cat, bla FRT cm P1 P2 oriR6K CmR gene | Datsenko and Wanner [31] |
| Primer | Sequence 5′–3′ | Resistance Cassette | Product Size (bp) |
|---|---|---|---|
| fliCm-F | ATGACGCCGCGGGTCAGGCGATTGCTAACCGTTTTACTTCTAACATTAAAGGCCTGACTCGTGTAGGCTGGAGCTGCTTC | pKD3 (CmR) | 1300 |
| fliCm-R | TCTGCGCTTTCGACATGTTGGACACTTCGGTCGCATAGTCGGCGTCCTGAATACGGGACTCATATGAATATCCTCCTTAG | pKD4 (KmR) | 1600 |
| fimHm-F | TATACCTACAGCTGAACCCAAAGAGATGATTGTAATGAAACGAGTTATTAGTGTAGGCTGGAGCTGCTTC | pKD3 (CmR) | 1300 |
| fimHm-R | CCTGCATTAGCAATGCCCTGTGATTTCTTTATTGATAAACAAAAGTCACGCCCATATGAATATCCTCCTTAG | pKD4 (KmR) | 1800 |
| csgAm-F | GTTTTACATGAAACTTTTAAAAGTAGCAGCAATTGCAGCAATCGTATTCGTGTAGGCTGGAGCTGCTTC | pKD3 (CmR) | 1300 |
| csgAm-R | GCGCCCTGTTTCTTTCATACTGATGATGTATTAGTACTGATGAGCGGTCGCATATGAATATCCTCCTTAG | pKD4 (KmR) | 1800 |
| Primer | Sequence 5′–3′ | Length | GC Content (%) | Tm (°C) | Product Size (bp) |
|---|---|---|---|---|---|
| fliCv-F | GGATCCCAGACGATAACAGGGTTGACGGC | 29 | 58.6 | 65.2 | 1923 |
| fliCv-R | GAGCTCTCAGGCAATTTGGCGTTGCCGTC | 29 | 58.6 | 65.2 | |
| fimHv-F | GAGCTACAGGATGACAGTGGC | 21 | 57.1 | 57.5 | 1237 |
| fimHv-R | GGAACAGACCAGCAAAGTGC | 20 | 55 | 56.8 | |
| csgAv-F | GCCAGTATTTCGCAAGGTGC | 20 | 55 | 57.1 | 789 |
| csgAv-R | GGTGTACATATCCCCTTGCTGG | 22 | 54.5 | 57.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vega-Hernández, R.; Ochoa, S.A.; Valle-Rios, R.; Jaimes-Ortega, G.A.; Arellano-Galindo, J.; Aparicio-Ozores, G.; Ibarra, J.A.; Hernández-Castro, R.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J. Flagella, Type I Fimbriae and Curli of Uropathogenic Escherichia coli Promote the Release of Proinflammatory Cytokines in a Coculture System. Microorganisms 2021, 9, 2233. https://doi.org/10.3390/microorganisms9112233
Vega-Hernández R, Ochoa SA, Valle-Rios R, Jaimes-Ortega GA, Arellano-Galindo J, Aparicio-Ozores G, Ibarra JA, Hernández-Castro R, Cruz-Córdova A, Xicohtencatl-Cortes J. Flagella, Type I Fimbriae and Curli of Uropathogenic Escherichia coli Promote the Release of Proinflammatory Cytokines in a Coculture System. Microorganisms. 2021; 9(11):2233. https://doi.org/10.3390/microorganisms9112233
Chicago/Turabian StyleVega-Hernández, Rubí, Sara A. Ochoa, Ricardo Valle-Rios, Gustavo A. Jaimes-Ortega, José Arellano-Galindo, Gerardo Aparicio-Ozores, José Antonio Ibarra, Rigoberto Hernández-Castro, Ariadnna Cruz-Córdova, and Juan Xicohtencatl-Cortes. 2021. "Flagella, Type I Fimbriae and Curli of Uropathogenic Escherichia coli Promote the Release of Proinflammatory Cytokines in a Coculture System" Microorganisms 9, no. 11: 2233. https://doi.org/10.3390/microorganisms9112233
APA StyleVega-Hernández, R., Ochoa, S. A., Valle-Rios, R., Jaimes-Ortega, G. A., Arellano-Galindo, J., Aparicio-Ozores, G., Ibarra, J. A., Hernández-Castro, R., Cruz-Córdova, A., & Xicohtencatl-Cortes, J. (2021). Flagella, Type I Fimbriae and Curli of Uropathogenic Escherichia coli Promote the Release of Proinflammatory Cytokines in a Coculture System. Microorganisms, 9(11), 2233. https://doi.org/10.3390/microorganisms9112233







