In-Vitro Characterization of Growth Inhibition against the Gut Pathogen of Potentially Probiotic Lactic Acid Bacteria Strains Isolated from Fermented Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples and LAB Strains
2.2. Selection and Characterization of Potentially Probiotic LAB Strains Using In-Vitro Assays
2.2.1. Acid Tolerance Test
2.2.2. Bile Salt Tolerance Test
2.2.3. Evaluation of Cell-Surface Hydrophobicity
2.2.4. Bacterial Adhesion to Caco-2 Cells
2.2.5. Evaluation of Antagonistic Activities
2.2.6. Molecular Identification and Phylogenetic Analysis Based on 16S rRNA Genes
2.2.7. Phenotypic Characterization
2.3. In-Vitro Characterization of Growth Inhibitory Activity against the Gut Pathogen of Selected LAB Strains using Microbiota Composition Analysis
2.3.1. In-Vitro Culture Condition of Fecal Microbial Populations and Sampling
2.3.2. Illumina MiSeq Paired-End Sequencing
2.3.3. Sequencing Processing and Bacterial Community Analysis
3. Results
3.1. Selection and Characterization of Potential Probiotic Strains
3.1.1. Selection of Five Potential Probiotic LAB Strains
3.1.2. Phylogenetic Affiliation of the Five Potential LAB Probiotic Strains
3.1.3. Phenotypic Characterization of the Five Potential Probiotic LAB Strains
3.2. In-Vitro Characterization of Growth Inhibitory Activity against the Gut Pathogen of Selected Five LAB Strains Using Microbiota Composition Analysis
3.2.1. Change in Growth Kinetics of the Fecal Microbiome by CFS Treatment
3.2.2. Analysis of Bacterial Diversity and Community Composition after CFS Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 57, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Djukovic, A.; Isaac, S. Roles of the intestinal microbiota in pathogen protection. Clin. Transl. Immunol. 2017, 6, e128. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Niederwerder, M.C. Fecal microbiota transplantation as a tool to treat and reduce susceptibility to disease in animals. Vet. Immunol. Immunopathol. 2018, 206, 65–72. [Google Scholar] [CrossRef]
- Kim, K.O.; Gluck, M. Fecal microbiota transplantation: An update on clinical practice. Clin. Endosc. 2019, 52, 137–143. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Divya Ganeshan, S.; Hosseinidoust, Z. Phage therapy with a focus on the human microbiota. Antibiotics 2019, 8, 131. [Google Scholar] [CrossRef]
- Rasmussen, T.S.; Koefoed, A.K.; Jakobsen, R.R.; Deng, L.; Castro-Mejía, J.L.; Brunse, A.; Neve, H.; Vogensen, F.K.; Nielsen, D.S. Bacteriophage-mediated manipulation of the gut microbiome—Promises and presents limitations. FEMS Microbiol. Rev. 2020, 44, 507–521. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Durand, H. Probiotics in animal nutrition and health. Benef. Microbes 2010, 1, 3–9. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, X.; Huang, L.; Shen, R.; Qin, H. Gut microbiota alteration after long-term consumption of probiotics in the elderly. Probiotics Antimicrob. Proteins 2019, 11, 655–666. [Google Scholar] [CrossRef]
- Liu, W.; Pang, H.; Zhang, H.; Cai, Y. Biodiversity of Lactic Acid Bacteria. In Lactic Acid Bacteria; Zhang, H., Cai, Y., Eds.; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. Biomed. Res. Int. 2018, 8, 9478630. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Hütt, P.; Shchepetova, J.; Lõivukene, K.; Kullisaar, T.; Mikelsaar, M. Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero- and uropathogens. J. Appl. Microbiol. 2006, 100, 1324–1332. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.J. Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 2015, 30, 126–132. [Google Scholar] [CrossRef]
- García-Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.L.; Nicoli, J.R.; Moreira-Silva, J.; Zoraya, R.; Héctor, F.; Odalys, N.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 2016, 108, 125–132. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, Y.; Zhou, H.; Tian, F.; Ni, Y. Antimicrobial activities and in vitro properties of cold-adapted Lactobacillus strains isolated from the intestinal tract of cold water fishes of high latitude water areas in Xinjiang, China. BMC Microbiol. 2019, 19, 247. [Google Scholar] [CrossRef]
- Walker, D.K.; Gilliland, S.E. Relationships among bile tolerance, bile salt deconjugation, and assimilation of cholesterol by Lactobacillus acidophilus. J. Dairy Sci. 1993, 76, 956–961. [Google Scholar] [CrossRef]
- Vinderola, C.G.; Reinheimer, J.A. Lactic acid starter and probiotic bacteria: A comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Cho, Y.J.; Kim, D.H.; Jeong, D.; Seo, K.H.; Jeong, H.S.; Lee, H.G.; Kim, H. Characterization of yeasts isolated from kefir as a probiotic and its synergic interaction with the wine byproduct grape seed flour/extract. Food Sci. Technol. 2018, 90, 535–539. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Burrows, J.R. Suggested procedure allowing use of plastic petri dishes in bacteriocin typing. Appl. Environ. Microbiol. 1978, 35, 970. [Google Scholar] [CrossRef]
- Jeon, H.H.; Jung, J.Y.; Chun, B.H.; Kim, M.D.; Baek, S.Y.; Moon, J.Y.; Yeo, S.H.; Jeon, C.O. Screening and characterization of potential Bacillus starter cultures for fermenting low-salt soybean paste (doenjang). J. Microbiol. Biotechnol. 2016, 26, 666–674. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fu, L.; Niu, B.; Wu, S.; Wooley, J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief. Bioinform. 2012, 13, 656–668. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalableand extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. R Package Version 3.1.1. 2020. Available online: https://cran.r-project.org/web/packages/gplots/ (accessed on 10 September 2021).
- Pandey, K.R.; Naik, S.R.; Vakil, B.V. Probiotics, prebiotics and synbiotics—A review. J. Food Sci. Technol. 2015, 52, 7577–7587. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73 (Suppl. S2), 386S–392S. [Google Scholar] [CrossRef]
- Solieri, L.; Bianchi, A.; Mottolese, G.; Lemmetti, F.; Giudici, P. Tailoring the probiotic potential of non-starter Lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol. 2014, 38, 240–249. [Google Scholar] [CrossRef]
- Chang, J.H.; Shim, Y.Y.; Cha, S.K.; Chee, K.M. Probiotic characteristics of lactic acid bacteria isolated from kimchi. J. Appl. Microbiol. 2010, 109, 220–230. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, H.; Ji, Y.; Kim, H.; Park, H.; Lee, J.; Shin, H.; Holzapfel, W. Functional properties of Lactobacillus strains isolated from kimchi. Int. J. Food Microbiol. 2011, 145, 155–161. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Y.; Tan, Z.; Li, Z.; Jiao, Z.; Huang, Q. Screening of probiotic activities of Lactobacilli strains isolated from traditional Tibetan Qula, a raw yak milk cheese. Asian-Australas. J. Anim. Sci. 2016, 29, 1490–1499. [Google Scholar] [CrossRef]
- Gilliland, S.E.; Staley, T.E.; Bush, L.J. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 1984, 67, 3045–3051. [Google Scholar] [CrossRef]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 1998, 84, 759–768. [Google Scholar] [CrossRef]
- Gotcheva, V.; Hristozova, E.; Hristozova, T.; Guo, M.; Roshkova, Z.; Angelov, A. Assessment of potential probiotic properties of lactic acid bacteria and yeast strains. Food Biotechnol. 2002, 16, 211–225. [Google Scholar] [CrossRef]
- Schillinger, U.; Guigas, C.; Holzapfel, W.H. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 2005, 15, 1289–1297. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Li, S.; Gan, R.Y.; Zhou, T.; Xu, D.P.; Li, H.B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Bengtsson, B.; Greko, C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Upsala J. Med. Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef]
- Cerniglia, C.E.; Pineiro, S.A.; Kotarski, S.F. An update discussion on the current assessment of the safety of veterinary antimicrobial drug residues in food with regard to their impact on the human intestinal microbiome. Drug Test. Anal. 2016, 8, 539–548. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S.T. Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef]
- Mandal, S.M.; Roy, A.; Ghosh, A.K.; Hazra, T.K.; Basak, A.; Franco, O.L. Challenges and future prospects of antibiotic therapy: From peptides to phages utilization. Front. Pharm. 2014, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Bebbington, C.; Yarranton, G. Antibodies for the treatment of bacterial infections: Current experience and future prospects. Curr. Opin. Biotechnol. 2008, 19, 613–619. [Google Scholar] [CrossRef]
- Scott, R.W.; Tew, G.N. Mimics of host defense proteins; strategies for translation to therapeutic applications. Curr. Top. Med. Chem. 2017, 17, 576–589. [Google Scholar] [CrossRef]
- Ayhan, D.H.; Tamer, Y.T.; Akbar, M.; Bailey, S.M.; Wong, M.; Daly, S.M.; Greenberg, D.E.; Toprak, E. Sequence-specific targeting of bacterial resistance genes increases antibiotic efficacy. PLoS Biol. 2016, 14, e1002552. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically engineered phages: A review of advances over the last decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar]
- Kokai-Kun, J.F.; Roberts, T.; Coughlin, O.; Sicard, E.; Rufiange, M.; Fedorak, R.; Carter, C.; Adams, M.H.; Longstreth, J.; Whalen, H.; et al. The oral β-lactamase SYN-004 (Ribaxamase) degrades ceftriaxone excreted into the intestine in phase 2a clinical studies. Antimicrob. Agents Chemother. 2017, 61, e02197-16. [Google Scholar] [CrossRef] [PubMed]
- Liévin-Le Moal, V.; Servin, A.L. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 2014, 27, 167–199. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.B.; Pamer, E.G. Microbiota-based therapies for Clostridium difficile and antibiotic-resistant enteric infections. Annu. Rev. Microbiol. 2017, 71, 157–178. [Google Scholar] [CrossRef] [PubMed]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Funke, G.; Hany, A.; Altwegg, M. Isolation of Escherichia fergusonii from four different sites in a patient with pancreatic carcinoma and cholangiosepsis. J. Clin. Microbiol. 1993, 31, 2201–2203. [Google Scholar] [CrossRef]
- Lai, C.C.; Cheng, A.; Huang, Y.T.; Chung, K.P.; Lee, M.R.; Liao, C.H.; Hsueh, P.R. Escherichia fergusonii bacteremia in a diabetic patient with pancreatic cancer. J. Clin. Microbiol. 2011, 49, 4001–4002. [Google Scholar] [CrossRef]
- Weiss, A.T.A.; Lübke-Becker, A.; Krenz, M.; van der Grinten, E. Enteritis and septicemia in a horse associated with infection by Escherichia fergusonii. J. Equine Vet. Sci. 2011, 31, 361–364. [Google Scholar] [CrossRef]
- Savini, V.; Catavitello, C.; Talia, M.; Manna, A.; Pompetti, F.; Favaro, M.; Fontana, C.; Febbo, F.; Balbinot, A.; Di Berardino, F.; et al. Multidrug-resistant Escherichia fergusonii: A case of acute cystitis. J. Clin. Microbiol. 2008, 46, 1551–1552. [Google Scholar] [CrossRef][Green Version]
- Forgetta, V.; Rempel, H.; Malouin, F.; Vaillancourt, R., Jr.; Topp, E.; Dewar, K.; Diarra, M.S. Pathogenic and multidrug-resistant Escherichia fergusonii from broiler chicken. Poult. Sci. 2012, 91, 512–525. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis 2016, 50, 10–17. [Google Scholar] [CrossRef]
- Jones, M.K.; Oliver, J.D. Vibrio vulnificus: Disease and pathogenesis. Infect. Immun. 2009, 77, 1723–1733. [Google Scholar] [CrossRef]
- Heng, S.P.; Letchumanan, V.; Deng, C.Y.; Ab Mutalib, N.S.; Khan, T.M.; Chuah, L.H.; Chan, K.G.; Goh, B.H.; Pusparajah, P.; Lee, L.H. Vibrio vulnificus: An environmental and clinical burden. Front. Microbiol. 2017, 8, 997. [Google Scholar] [CrossRef]
- Jadhav, K.P.; Pai, P.G. A rare infective endocarditis caused by Vagococcus fluvialis. J. Cardiol. Cases 2019, 20, 129–131. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, C.; Gu, H.J.; Zhang, J.; Sun, L. Characterization of the genome feature and toxic capacity of a Bacillus wiedmannii isolate from the hydrothermal field in Okinawa Trough. Front. Cell Infect. Microbiol. 2019, 9, 370. [Google Scholar] [CrossRef]
- Spiegelhauer, M.R.; Andersen, P.F.; Frandsen, T.H.; Nordestgaard, R.L.M.; Andersen, L.P. Leclercia adecarboxylata: A case report and literature review of 74 cases demonstrating its pathogenicity in immunocompromised patients. Infect. Dis. 2019, 51, 179–188. [Google Scholar] [CrossRef]
- Timothy, B.; Iliyasu, A.H.; Anvikar, A.R. Bacteriocins of lactic acid bacteria and their industrial application. Curr Top. Lact Acid Bact. Probiotics 2021, 7, 1–13. [Google Scholar] [CrossRef]
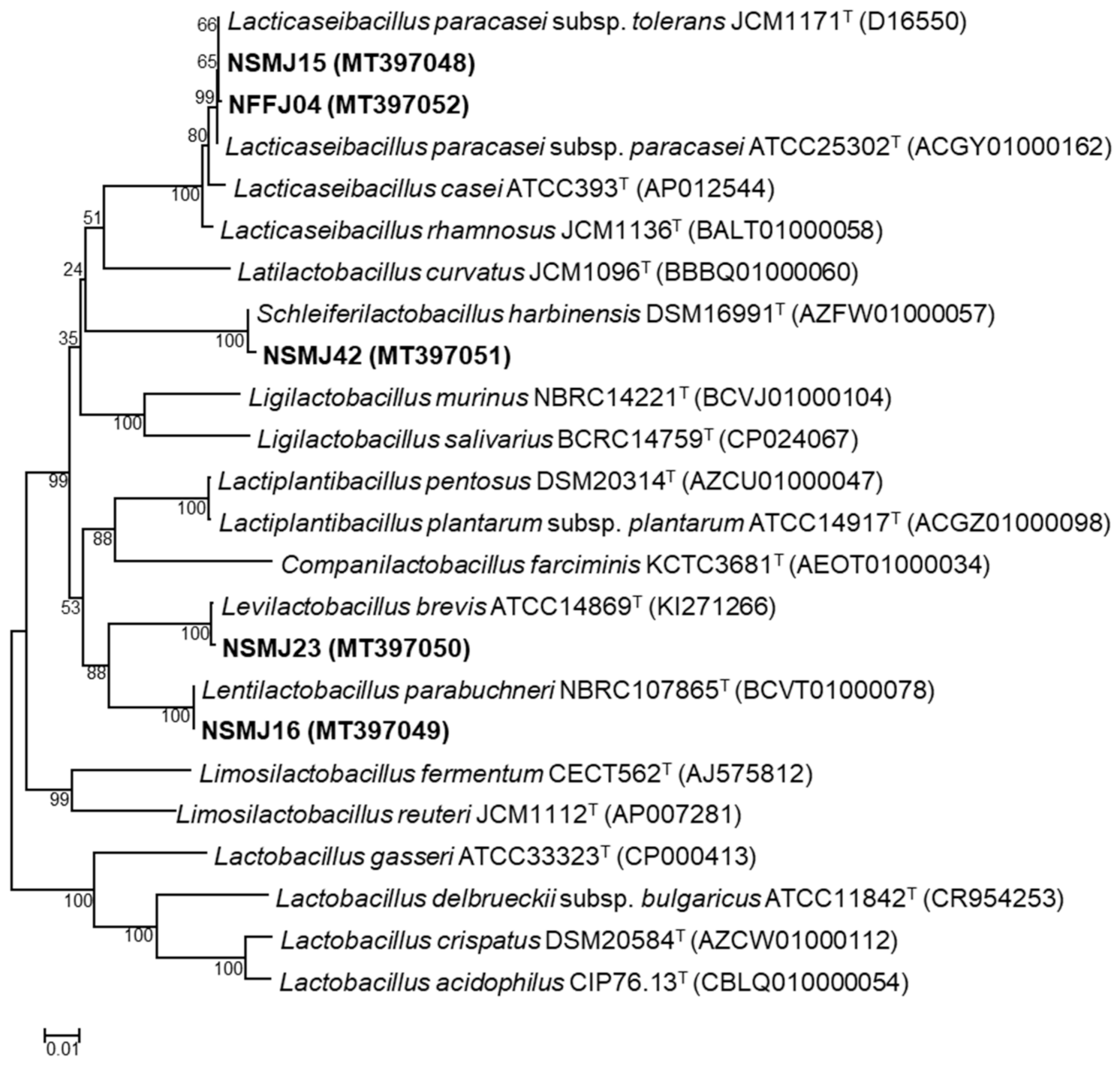
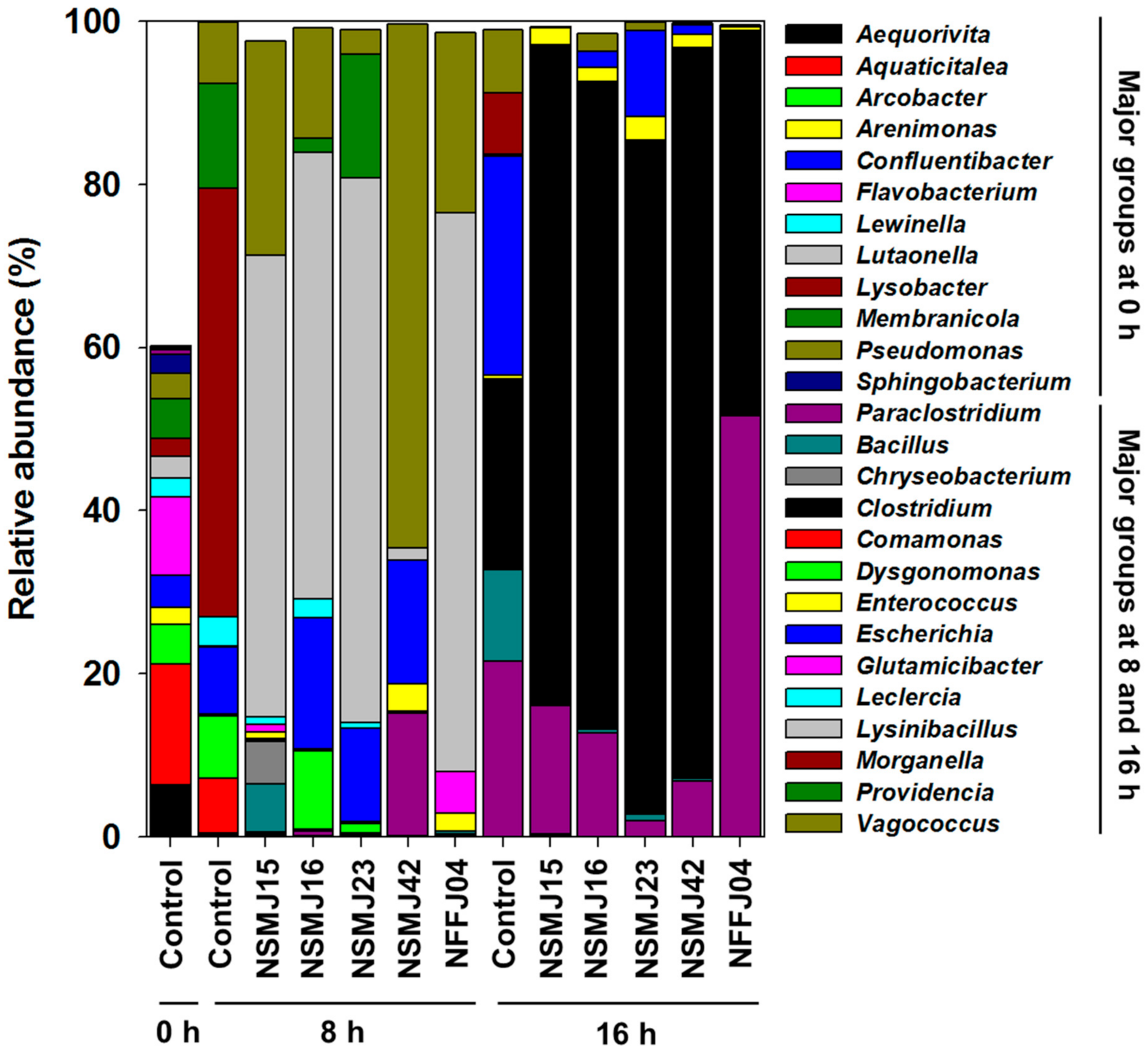
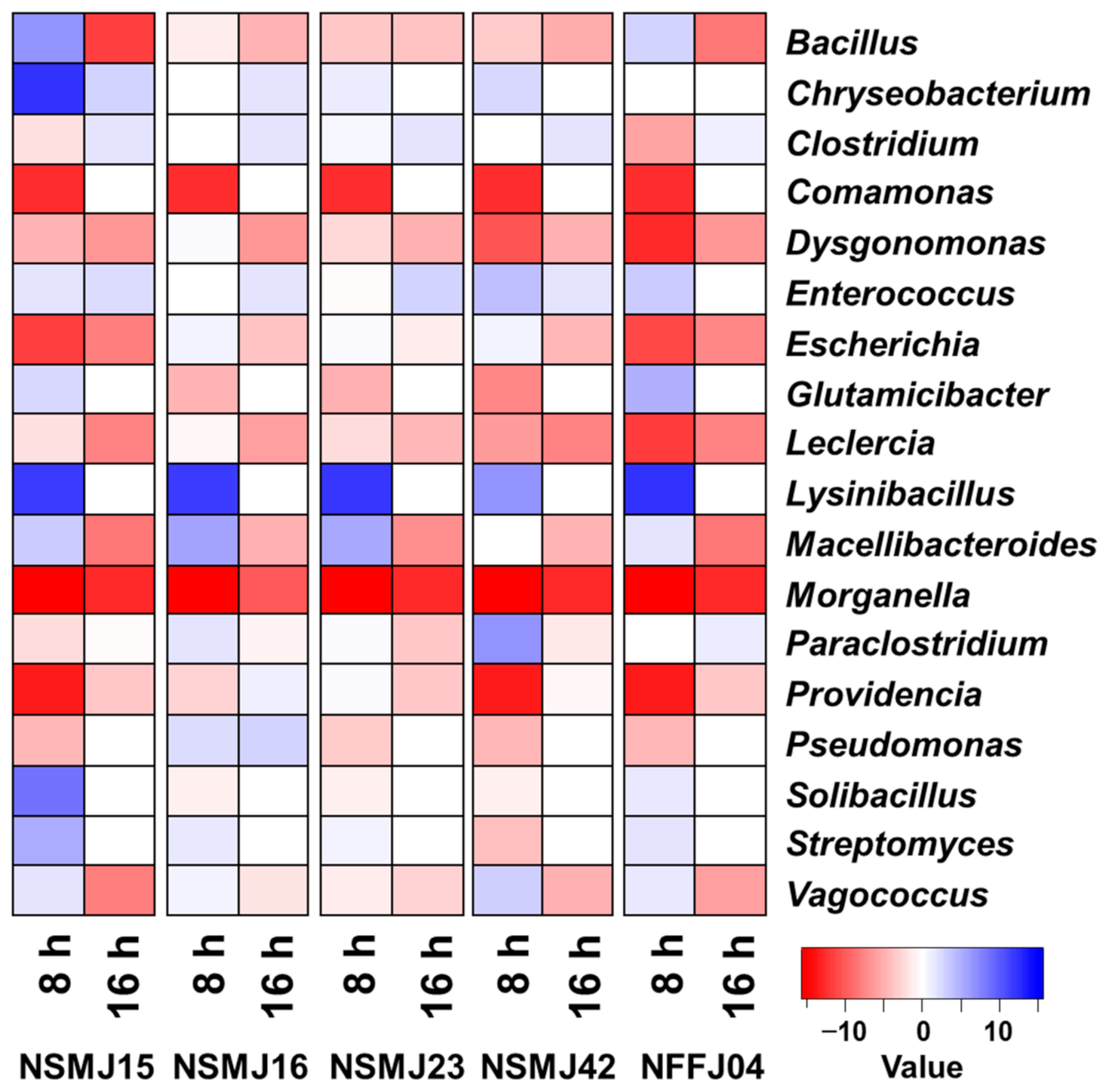
| Strain | Acid Tolerance | Bile Salt Tolerance | Adhesion to Caco-2 Cell (%) | Hydrophobicity (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Control (log CFU/mL) | pH 2.5 (log CFU/mL) | Survival Rate (%) | Control (log CFU/mL) | 0.3% (w/v) BS (log CFU/mL) | Survival Rate (%) | |||
| NSMJ15 | 8.65 ± 0.50 | 6.35 ± 1.17 | 73.11 | 9.96 ± 0.15 | 8.89 ± 0.10 | 89.23 | 14.93 ± 1.54 | 88.20 ± 7.18 |
| NSMJ16 | 8.58 ± 0.66 | 7.02 ± 0.68 | 81.77 | 9.58 ± 0.17 | 8.99 ± 0.14 | 93.82 | 8.08 ± 0.95 | 80.64 ± 6.16 |
| NSMJ23 | 8.16 ± 0.59 | 6.21 ± 0.65 | 76.08 | 9.61 ± 0.02 | 9.36 ± 0.08 | 97.52 | 26.19 ± 5.56 | 78.19 ± 13.81 |
| NSMJ42 | 9.97 ± 0.25 | 8.06 ± 0.25 | 80.86 | 10.26 ± 0.27 | 9.55 ± 0.08 | 93.29 | 26.73 ± 4.68 | 78.32 ± 5.20 |
| NFFJ04 | 9.79 ± 0.06 | 6.86 ± 0.52 | 70.02 | 10.24 ± 0.04 | 9.61 ± 0.01 | 93.97 | 7.62 ± 0.61 | 57.12 ± 3.23 |
| LGG | 9.79 ± 0.20 | 7.00 ± 0.67 | 71.60 | 10.11 ± 0.22 | 9.03 ± 0.24 | 89.32 | 19.50 ± 7.82 | 26.39 ± 5.01 |
| Strains | Antagonistic Activity a | |
|---|---|---|
| E. coli CCARM 1G440 | S. aureus CCARM 3A860 | |
| NSMJ15 | +++ | + |
| NSMJ16 | +++++ | ++++ |
| NSMJ23 | +++++ | +++++ |
| NSMJ42 | +++ | ++++ |
| NFFJ04 | +++ | +++ |
| Samples | No. of Reads | No. of High-Quality Reads | Average Read Length (bp) | OTUs a | Chao1 a | Shannon a | Inverse Simpson a | |
|---|---|---|---|---|---|---|---|---|
| 0 h | Non-treated | 100,495 | 11,623 | 457 | 332 | 359.51 | 6.13 | 0.96 |
| 8 h | Non-treated | 113,395 | 43,322 | 464 | 64 | 94.00 | 2.34 | 0.69 |
| NSMJ15 | 113,312 | 38,697 | 464 | 123 | 176.81 | 1.93 | 0.60 | |
| NSMJ16 | 97,465 | 36,225 | 464 | 60 | 79.46 | 2.03 | 0.65 | |
| NSMJ23 | 107,232 | 42,360 | 465 | 66 | 85.50 | 1.60 | 0.52 | |
| NSMJ42 | 86,614 | 33,807 | 460 | 83 | 131.75 | 1.58 | 0.54 | |
| NFFJ04 | 87,505 | 28,044 | 463 | 75 | 134.50 | 1.41 | 0.48 | |
| 16 h | Non-treated | 104,636 | 32,494 | 452 | 67 | 106.55 | 2.59 | 0.80 |
| NSMJ15 | 89,975 | 27,654 | 441 | 79 | 98.12 | 1.05 | 0.34 | |
| NSMJ16 | 102,863 | 29,971 | 441 | 55 | 132.50 | 1.21 | 0.36 | |
| NSMJ23 | 109,505 | 36,047 | 443 | 46 | 59.33 | 1.05 | 0.32 | |
| NSMJ42 | 113,993 | 37,162 | 441 | 69 | 92.63 | 0.69 | 0.20 | |
| NFFJ04 | 100,741 | 32,133 | 440 | 38 | 57.43 | 1.23 | 0.52 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, J.Y.; Han, S.-S.; Kim, Z.-H.; Kim, M.H.; Kang, H.K.; Jin, H.M.; Lee, M.H. In-Vitro Characterization of Growth Inhibition against the Gut Pathogen of Potentially Probiotic Lactic Acid Bacteria Strains Isolated from Fermented Products. Microorganisms 2021, 9, 2141. https://doi.org/10.3390/microorganisms9102141
Jung JY, Han S-S, Kim Z-H, Kim MH, Kang HK, Jin HM, Lee MH. In-Vitro Characterization of Growth Inhibition against the Gut Pathogen of Potentially Probiotic Lactic Acid Bacteria Strains Isolated from Fermented Products. Microorganisms. 2021; 9(10):2141. https://doi.org/10.3390/microorganisms9102141
Chicago/Turabian StyleJung, Ji Young, Sang-Soo Han, Z-Hun Kim, Myung Hoo Kim, Hye Kyeong Kang, Hyun Mi Jin, and Mi Hwa Lee. 2021. "In-Vitro Characterization of Growth Inhibition against the Gut Pathogen of Potentially Probiotic Lactic Acid Bacteria Strains Isolated from Fermented Products" Microorganisms 9, no. 10: 2141. https://doi.org/10.3390/microorganisms9102141
APA StyleJung, J. Y., Han, S.-S., Kim, Z.-H., Kim, M. H., Kang, H. K., Jin, H. M., & Lee, M. H. (2021). In-Vitro Characterization of Growth Inhibition against the Gut Pathogen of Potentially Probiotic Lactic Acid Bacteria Strains Isolated from Fermented Products. Microorganisms, 9(10), 2141. https://doi.org/10.3390/microorganisms9102141






