Ambient Air Pollution Shapes Bacterial and Fungal Ivy Leaf Communities
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site Characteristics
2.2. Collection and Preparation of Phylloplane Samples
2.3. Black Carbon Detection in Leaf Wash Suspensions
2.4. Metabarcoding of the Bacterial and Fungal Phylloplane
2.5. Analysis of Sequencing Data
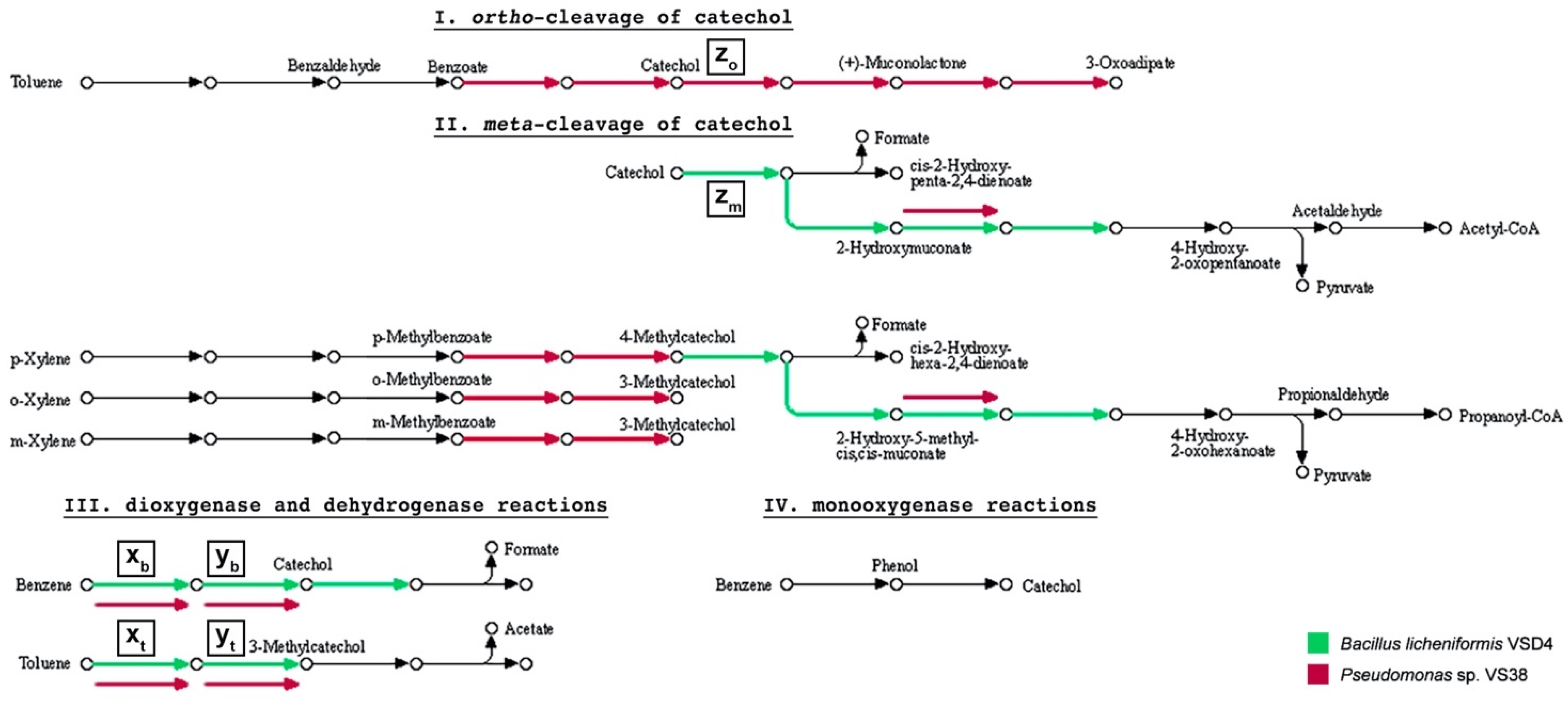
2.6. Benzene, Toluene, and Xylene Degradation by Phylloplane Isolates
2.7. Genome Sequencing and Analysis
3. Results
3.1. Ambient Air Pollution Increases Black Carbon Load in the Phylloplane
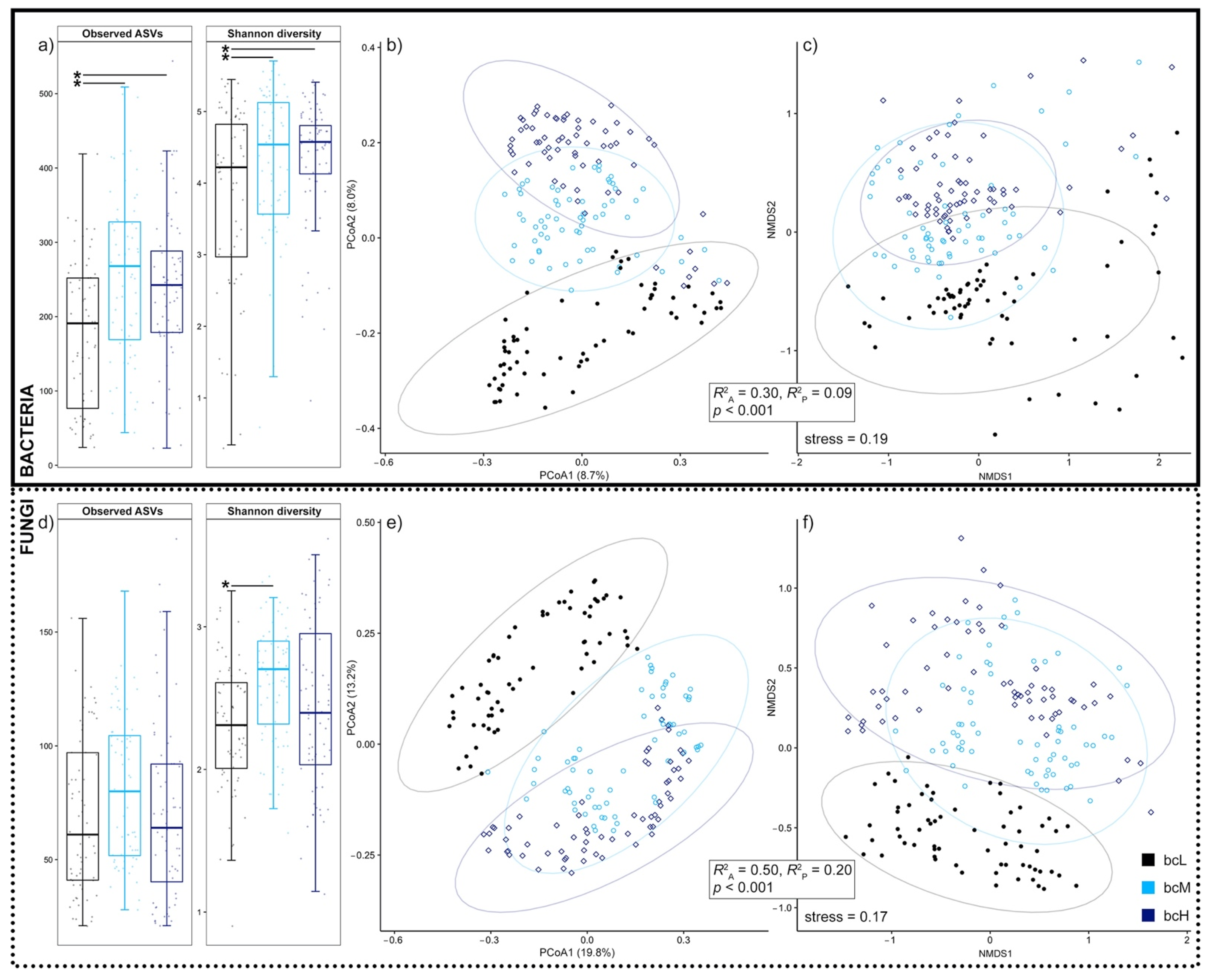
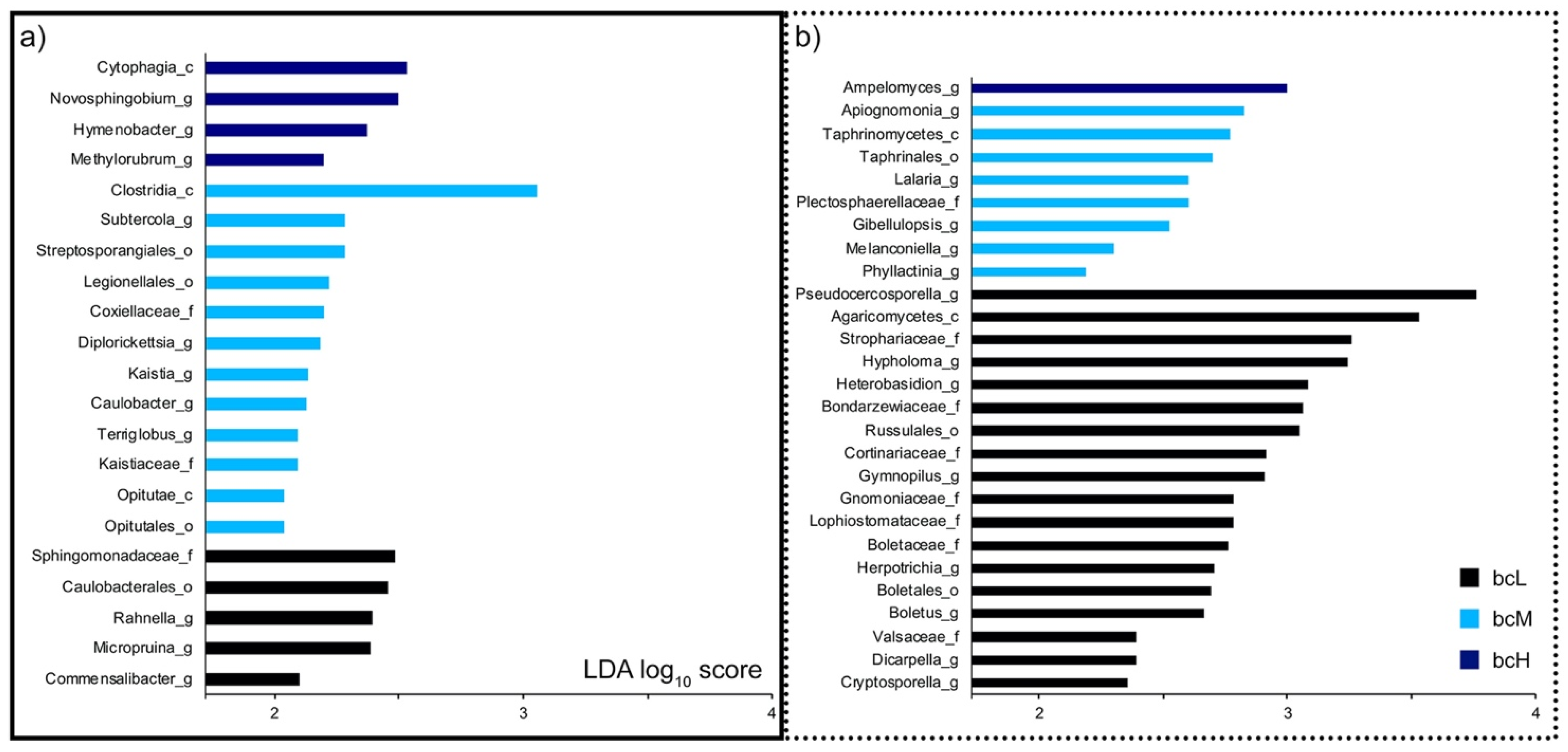
3.2. Phylloplane Black Carbon Load Correlates with Bacterial and Fungal Diversity
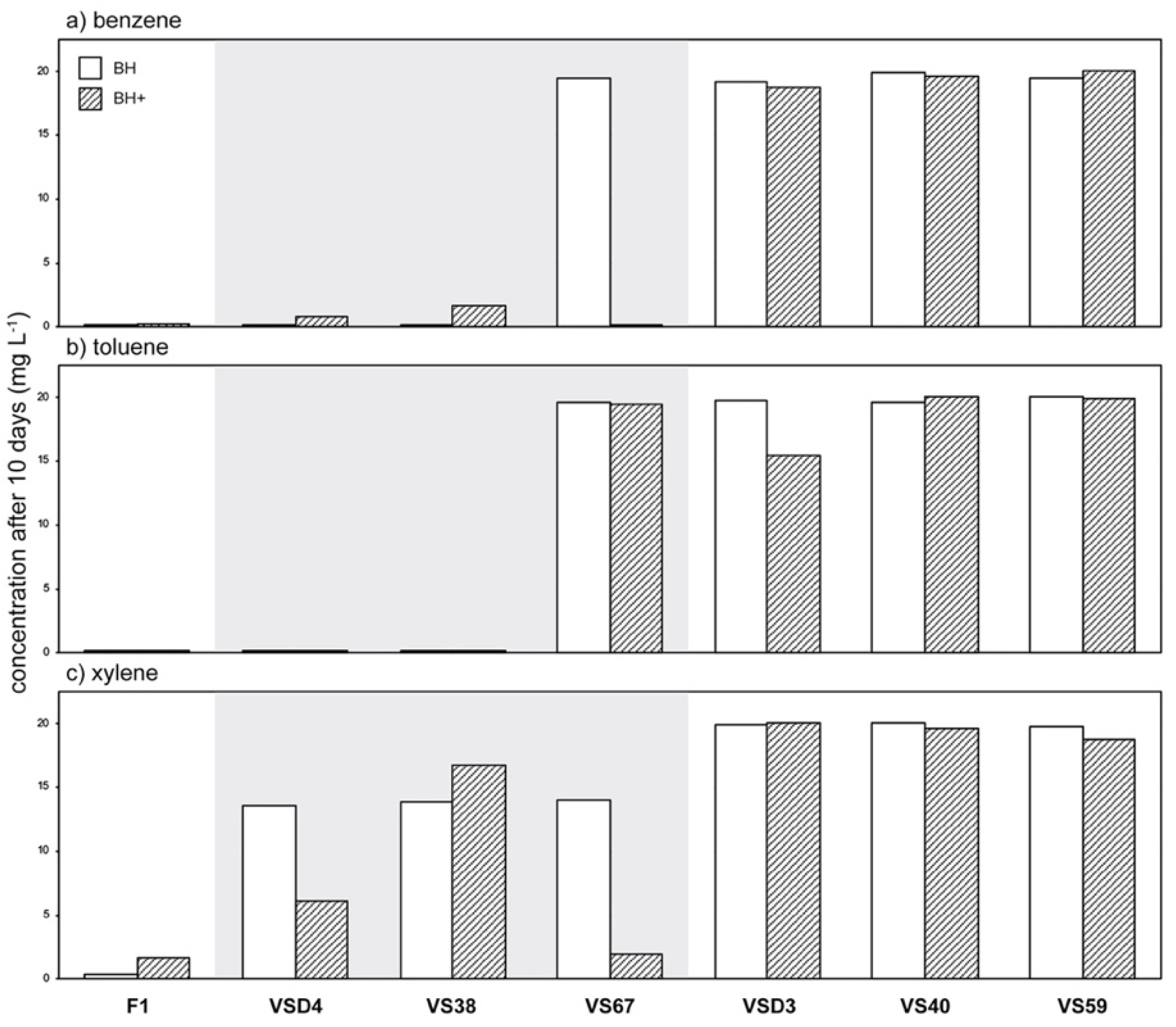
3.3. Phylloplane Isolates from an Air-Polluted Environment Degrade BTX
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Air Pollution. Available online: https://www.who.int/health-topics/air-pollution (accessed on 2 October 2021).
- Zhang, K.; Batterman, S.A. Air pollution and health risks due to vehicle traffic. Sci. Total Environ. 2013, 450–451, 307–316. [Google Scholar] [CrossRef]
- Anenberg, S.C.; Schwartz, J.; Shindell, D.; Amann, M.; Faluvegi, G.; Klimont, Z.; Janssens-Maenhout, G.; Pozzoli, L.; Van Dingenen, R.; Vignati, E.; et al. Global air quality and health co-benefits of mitigating near-term climate change through methane and black carbon emission controls. Environ. Health Perspect. 2012, 120, 831–839. [Google Scholar] [CrossRef]
- Grahame, T.J.; Klemm, R.; Schlesinger, R.B. Public health and components of particulate matter: The changing assessment of black carbon. J. Air Waste Manag. Assoc. 2014, 64, 620–660. [Google Scholar] [CrossRef]
- Rai, P.K. Impacts of particulate matter pollution on plants: Implications for environmental biomonitoring. Ecotoxicol. Environ. Saf. 2016, 129, 120–136. [Google Scholar] [CrossRef]
- Chin, J.Y.; Batterman, S.A. VOC composition of current motor vehicle fuels and vapors, and collinearity analyses for receptor modeling. Chemosphere 2012, 86, 951–958. [Google Scholar] [CrossRef]
- Zielinska, B.; Sagebiel, J.; McDonald, J.D.; Whitney, K.; Lawson, D.R. Emission rates and comparative chemical composition from selected in-use diesel and gasoline-fueled vehicles. J. Air Waste Manag. Assoc. 2004, 54, 1138–1150. [Google Scholar] [CrossRef]
- Nurmatov, U.B.; Tagiyeva, N.; Semple, S.; Devereux, G.; Sheikh, A. Volatile organic compounds and risk of asthma and allergy: A systematic review. Eur. Respir. Rev. 2015, 24, 92–101. [Google Scholar] [CrossRef]
- Kuykendall, J.R.; Shaw, S.L.; Paustenbach, D.; Fehling, K.; Kacew, S.; Kabay, V. Chemicals present in automobile traffic tunnels and the possible community health hazards: A review of the literature. Inhal. Toxicol. 2009, 21, 747–792. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Woodward, F.I.; Lomas, M.R. Vegetation dynamics—Simulating responses to climatic change. Biol. Rev. 2004, 79, 643–670. [Google Scholar] [CrossRef] [PubMed]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 2014, 87, 2–17. [Google Scholar] [CrossRef]
- Metcalfe, D.J. Hedera helix L. J. Ecol. 2005, 93, 632–648. [Google Scholar] [CrossRef]
- Dzierżanowski, K.; Popek, R.; Gawrońska, H.; Sæbø, A.; Gawroński, S.W. Deposition of particulate matter of different size fractions on leaf surfaces and in waxes of urban forest species. Int. J. Phytoremediat. 2011, 13, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, W.; Van Poppel, M.; Maiheu, B.; Janssen, S.; Dons, E. Evaluation of the RIO-IFDM-street canyon model chain. Atmos. Environ. 2013, 77, 325–337. [Google Scholar] [CrossRef]
- Janssen, S.; Dumont, G.; Fierens, F.; Mensink, C. Spatial interpolation of air pollution measurements using CORINE land cover data. Atmos. Environ. 2008, 42, 4884–4903. [Google Scholar] [CrossRef]
- Lefebvre, W.; Degrawe, B.; Beckx, C.; Vanhulsel, M.; Kochan, B.; Bellemans, T.; Janssens, D.; Wets, G.; Janssen, S.; De Vlieger, I.; et al. Presentation and evaluation of an integrated model chain to respond to traffic-and health-related policy questions. Environ. Model. Softw. 2013, 40, 160–170. [Google Scholar] [CrossRef]
- Lefebvre, W.; Vercauteren, J.; Schrooten, L.; Janssen, S.; Degraeuwe, B.; Maenhaut, W.; de Vlieger, I.; Vankerkom, J.; Cosemans, G.; Mensink, C.; et al. Validation of the MIMOSA-AURORA-IFDM model chain for policy support: Modeling concentrations of elemental carbon in Flanders. Atmos. Environ. 2011, 45, 6705–6713. [Google Scholar] [CrossRef]
- Bové, H.; Steuwe, C.; Fron, E.; Slenders, E.; D’Haen, J.; Fujita, Y.; Uji, I.H.; vandeVen, M.; Roeffaers, M.; Ameloot, M. Biocompatible label-free detection of carbon black particles by femtosecond pulsed laser microscopy. Nano Lett. 2016, 16, 3173–3178. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Vancov, T.; Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pages, H.; Gentleman, R. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 2009, 25, 2607–2608. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, V.; Wang, Q.; Greenfield, P.; Charleston, M.; Porras-Alfaro, A.; Kuske, C.R.; Cole, J.R.; Midgley, D.J.; Tran-Dinh, N. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 2016, 108, 1–5. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Kandlikar, G.S.; Gold, Z.J.; Cowen, M.C.; Meyer, R.S.; Freise, A.C.; Kraft, N.J.B.; Moberg-Parker, J.; Sprague, J.; Kushner, D.J.; Curd, E.E. ranacapa: An R package and Shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 October 2021).
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Bushnell, L.D.; Haas, H.F. The utilization of certain hydrocarbons by microorganisms. J. Bacteriol. 1941, 41, 653–673. [Google Scholar] [CrossRef]
- Bučková, M.; Puškarová, A.; Chovanová, K.; Kraková, L.; Ferianc, P.; Pangallo, D. A simple strategy for investigating the diversity and hydrocarbon degradation abilities of cultivable bacteria from contaminated soil. World J. Microbiol. Biotechnol. 2013, 29, 1085–1098. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; Vangronsveld, J. Diversity and plant growth-promoting potential of (un)culturable bacteria in the Hedera helix phylloplane. BMC Microbiol. 2021, 21, 66. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; McAmmond, B.; Langill, T.; Van Hamme, J.; Weyens, N.; Vangronsveld, J. Draft genome sequence of Bacillus licheniformis VSD4, a diesel fuel-degrading and plant growth-promoting phyllospheric bacterium. Genome Announc. 2017, 5, e00027-17. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; McAmmond, B.; Langill, T.; Van Hamme, J.; Weyens, N.; Vangronsveld, J. Draft genome sequence of Rhodococcus erythropolis VSD3, a diesel fuel-degrading and plant growth-promoting bacterium isolated from Hedera helix leaves. Genome Announc. 2017, 5, e01680-16. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.D.; White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Rivoire, C.; Auchincloss, A.H.; Coudert, E.; Keller, G.; de Castro, E.; Baratin, D.; Cuche, B.A.; Bougueleret, L.; Poux, S.; et al. HAMAP in 2015: Updates to the protein family classification and annotation system. Nucleic Acids Res. 2015, 43, D1064–D1070. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Hagenbjörk, A.; Malmqvist, E.; Mattisson, K.; Sommar, N.J.; Modig, L. The spatial variation of O3, NO, NO2 and NOx and the relation between them in two Swedish cities. Environ. Monit. Assess. 2017, 189, 161. [Google Scholar] [CrossRef] [PubMed]
- Vardoulakis, S.; Fisher, B.E.A.; Pericleous, K.; Gonzalez–Flesca, N. Modelling air quality in street canyons: A review. Atmos. Environ. 2003, 37, 155–182. [Google Scholar] [CrossRef]
- Janhäll, S. Review on urban vegetation and particle air pollution—Deposition and dispersion. Atmos. Environ. 2015, 105, 130–137. [Google Scholar] [CrossRef]
- Kumar, P.; Ketzel, M.; Vardoulakis, S.; Pirjola, L.; Britter, R. Dynamics and dispersion modelling of nanoparticles from road traffic in the urban atmospheric environment—A review. J. Aerosol Sci. 2011, 42, 580–603. [Google Scholar] [CrossRef]
- Karra, S.; Malki-Epshtein, L.; Neophytou, M. The dispersion of traffic related pollutants across a non-homogeneous street canyon. Procedia Environ. Sci. 2011, 4, 25–34. [Google Scholar] [CrossRef][Green Version]
- Wuyts, K.; Smets, W.; Lebeer, S.; Samson, R. Green infrastructure and atmospheric pollution shape diversity and composition of phyllosphere bacterial communities in an urban landscape. FEMS Microbiol. Ecol. 2020, 96, fiz173. [Google Scholar] [CrossRef] [PubMed]
- Laforest-Lapointe, I.; Messier, C.; Kembel, S.W. Tree leaf bacterial community structure and diversity differ along a gradient of urban intensity. mSystems 2017, 2, e00087-17. [Google Scholar] [CrossRef] [PubMed]
- Imperato, V.; Kowalkowski, Ł.; Portillo-Estrada, M.; Gawroński, S.W.; Vangronsveld, J.; Thijs, S. Characterisation of the Carpinus betulus L. phyllomicrobiome in urban and forest areas. Front. Microbiol. 2019, 10, 1110. [Google Scholar] [CrossRef]
- Redford, A.J.; Fierer, N. Bacterial succession on the leaf surface: A novel system for studying successional dynamics. Microb. Ecol. 2009, 58, 189–198. [Google Scholar] [CrossRef]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef]
- Copeland, J.K.; Yuan, L.; Layeghifard, M.; Wang, P.W.; Guttman, D.S. Seasonal community succession of the phyllosphere microbiome. Mol. Plant Microbe Interact. 2015, 28, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.W.G.; Jackson, C.R. Seasonal patterns contribute more towards phyllosphere bacterial community structure than short-term perturbations. Microb. Ecol. 2021, 81, 146–156. [Google Scholar] [CrossRef]
- Jackson, C.R.; Denney, W.C. Annual and seasonal variation in the phyllosphere bacterial community associated with leaves of the southern Magnolia (Magnolia grandiflora). Microb. Ecol. 2011, 61, 113–122. [Google Scholar] [CrossRef]
- Smets, W.; Wuyts, K.; Oerlemans, E.; Wuyts, S.; Denys, S.; Samson, R.; Lebeer, S. Impact of urban land use on the bacterial phyllosphere of ivy (Hedera sp.). Atmos. Environ. 2016, 147, 376–383. [Google Scholar] [CrossRef]
- Espenshade, J.; Thijs, S.; Gawroński, S.; Bové, H.; Weyens, N.; Vangronsveld, J. Influence of urbanization on epiphytic bacterial communities of the Platanus x hispanica tree leaves in a biennial study. Front. Microbiol. 2019, 10, 675. [Google Scholar] [CrossRef] [PubMed]
- Undugoda, L.J.S.; Kannangara, S.; Sirisena, D.M. Aromatic hydrocarbon degrading fungi inhabiting the phyllosphere of ornamental plants on roadsides of urban areas in Sri Lanka. J. Bioremediat. Biodegrad. 2016, 7, 328. [Google Scholar] [CrossRef]
- Sucharzewska, E.; Dynowska, M. Preliminary evaluation of the effect of Ampelomyces quisqualis on the degree of plant infestations with selected Erysiphales species proposed as potential bioindicators. Plant Protect. Sci. 2002, 38, 436–438. [Google Scholar] [CrossRef]
- Buswell, J.A. The meta-cleavage of catechol by a thermophilic Bacillus species. Biochem. Biophys. Res. Commun. 1974, 60, 934–941. [Google Scholar] [CrossRef]
- Banerjee, A.; Ghoshal, A.K. Phenol degradation by Bacillus cereus: Pathway and kinetic modeling. Bioresour. Technol. 2010, 101, 5501–5507. [Google Scholar] [CrossRef] [PubMed]
- Duffner, F.M.; Müller, R. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: Nucleotide sequence and analysis of the genes. FEMS Microbiol. Lett. 1998, 161, 37–45. [Google Scholar] [CrossRef]
- ChrisFelshia, S.; AshwinKarthick, N.; Thilagam, R.; Gnanamani, A. Elucidation of 2,4-dichlorophenol degradation by Bacillus licheniformis strain SL10. Environ. Technol. 2020, 41, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Bordoloi, N.K. Biodegradation of benzene, toluene, and xylene (BTX) in liquid culture and in soil by Bacillus subtilis and Pseudomonas aeruginosa strains and a formulated bacterial consortium. Environ. Sci. Pollut. Res. Int. 2012, 19, 3380–3388. [Google Scholar] [CrossRef]
- Wongbunmak, A.; Khiawjan, S.; Suphantharika, M.; Pongtharangkul, T. BTEX biodegradation by Bacillus amyloliquefaciens subsp. plantarum W1 and its proposed BTEX biodegradation pathways. Sci. Rep. 2020, 10, 17408. [Google Scholar] [CrossRef]
- Díaz, L.F.; Muñoz, R.; Bordel, S.; Villaverde, S. Toluene biodegradation by Pseudomonas putida F1: Targeting culture stability in long-term operation. Biodegradation 2008, 19, 197–208. [Google Scholar] [CrossRef]
- You, Y.; Shim, J.; Cho, C.H.; Ryu, M.H.; Shea, P.J.; Kamala-Kannan, S.; Chae, J.C.; Oh, B.T. Biodegradation of BTEX mixture by Pseudomonas putida YNS1 isolated from oil-contaminated soil. J. Basic Microbiol. 2013, 53, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, K.; Loh, K.C. Formulation of microbial cocktails for BTEX biodegradation. Biodegradation 2015, 26, 51–63. [Google Scholar] [CrossRef]
- Imperato, V.; Portillo-Estrada, M.; McAmmond, B.M.; Douwen, Y.; Van Hamme, J.D.; Gawroński, S.W.; Vangronsveld, J.; Thijs, S. Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of Bóbrka (Poland). Genes 2019, 10, 443. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Zhang, M.; Toyota, K. Biodegradation of volatile organic compounds and their effects on biodegradability under co-existing conditions. Microbes Environ. 2017, 32, 188–200. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Summerbell, R.; Sybren de Hoog, G. Fungi growing on aromatic hydrocarbons: Biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 2006, 30, 109–130. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Vervoort, J.; Grotenhuis, J.T.; Van Groenestijn, J.W. Substrate interactions during the biodegradation of benzene, toluene, ethylbenzene, and xylene (BTEX) hydrocarbons by the fungus Cladophialophora sp. strain T1. Appl. Environ. Microbiol. 2002, 68, 2660–2665. [Google Scholar] [CrossRef][Green Version]
- Blasi, B.; Tafer, H.; Kustor, C.; Poyntner, C.; Lopandic, K.; Sterflinger, K. Genomic and transcriptomic analysis of the toluene degrading black yeast Cladophialophora immunda. Sci. Rep. 2017, 7, 11436. [Google Scholar] [CrossRef] [PubMed]
- Luykx, D.M.; Prenafeta-Boldú, F.X.; de Bont, J.A. Toluene monooxygenase from the fungus Cladosporium sphaerospermum. Biochem. Biophys. Res. Commun. 2003, 312, 373–379. [Google Scholar] [CrossRef] [PubMed]
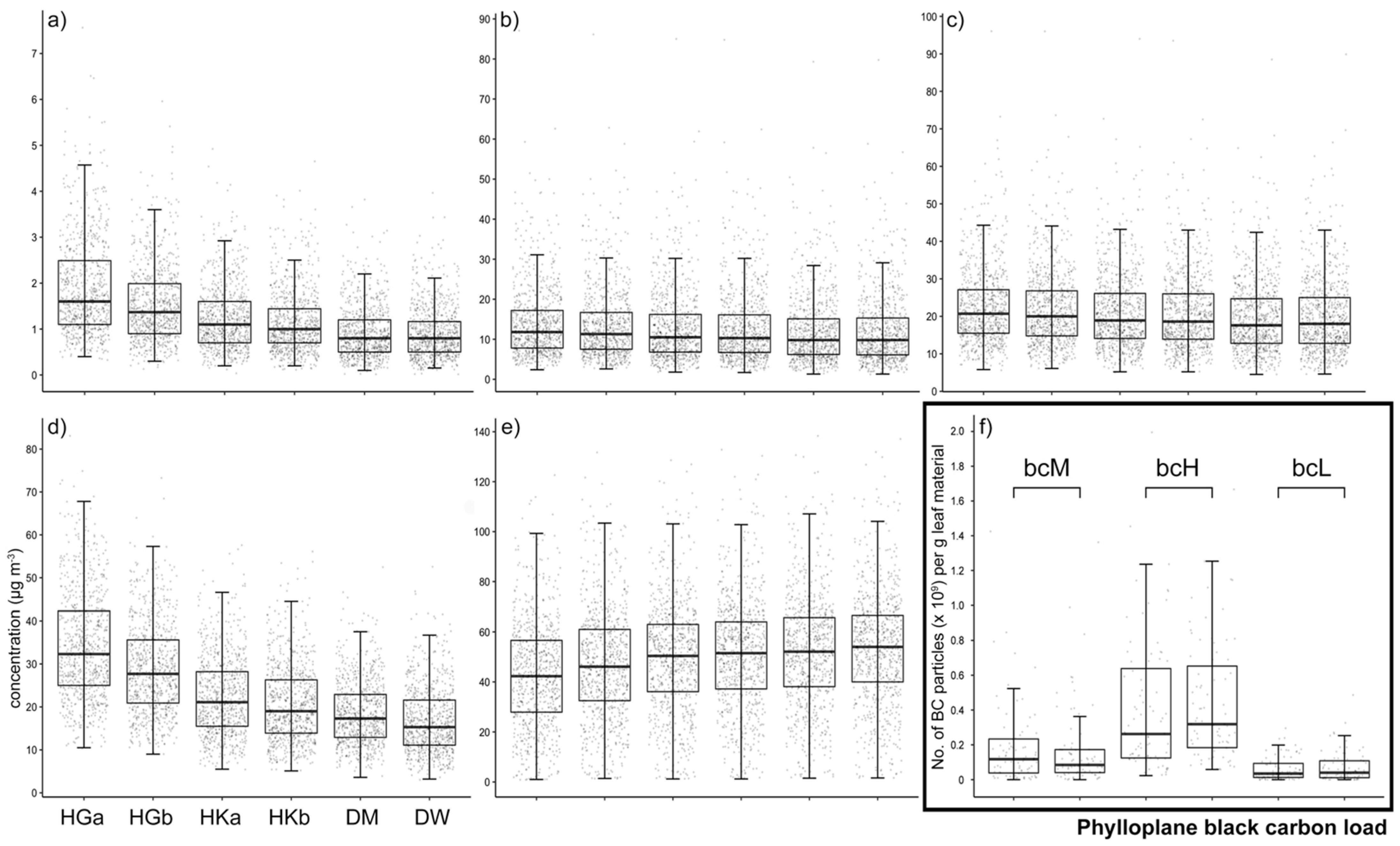
| Sampling Site | BC (µg·m−3) | PM2.5 (µg·m−3) | PM10 (µg·m−3) | NO2 (µg·m−3) | O3 (µg·m−3) | Phylloplane BC Load * | |
|---|---|---|---|---|---|---|---|
| HGa | bcM | 1.9 ± 1.0 vwxyz | 14.1 ± 9.2 yz | 22.7 ± 10.4 xyz | 34.2 ± 12.5 vwxyz | 42.9 ± 21.5 vwxyz | 1.9 ± 2.4 wxyz |
| HGb | 1.5 ± 0.8 uwxyz | 13.7 ± 9.2 yz | 22.2 ± 10.5 yz | 28.9 ± 10.6 uwxyz | 46.9 ± 22.1 uxyz | 1.7 ± 2.3 wxyz | |
| HKa | bcH | 1.3 ± 0.7 uvyz | 13.0 ± 9.2 | 21.3 ± 10.4 | 22.4 ± 9.2 uvxyz | 49.9 ± 22.1 uz | 4.1 ± 3.8 uvyz |
| HKb | 1.1 ± 0.7 uvyz | 12.9 ± 9.2 | 21.1 ± 10.5 u | 20.7 ± 8.9 uvwyz | 51.0 ± 22.2 uv | 4.4 ± 3.4 uvyz | |
| DM | bcL | 0.9 ± 0.6 uvwx | 12.1 ± 8.7 uv | 19.8 ± 10.0 uv | 18.5 ± 7.4 uvwxz | 52.1 ± 22.5 uv | 0.6 ± 0.7 uvwx |
| DW | 0.9 ± 0.5 uvwx | 12.2 ± 8.9 uv | 20.1 ± 10.3 uv | 16.8 ± 7.7 uvwxy | 53.6 ± 22.5 uvw | 0.7 ± 0.9 uvwx |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevens, V.; Thijs, S.; Bongaerts, E.; Nawrot, T.; Marchal, W.; Van Hamme, J.; Vangronsveld, J. Ambient Air Pollution Shapes Bacterial and Fungal Ivy Leaf Communities. Microorganisms 2021, 9, 2088. https://doi.org/10.3390/microorganisms9102088
Stevens V, Thijs S, Bongaerts E, Nawrot T, Marchal W, Van Hamme J, Vangronsveld J. Ambient Air Pollution Shapes Bacterial and Fungal Ivy Leaf Communities. Microorganisms. 2021; 9(10):2088. https://doi.org/10.3390/microorganisms9102088
Chicago/Turabian StyleStevens, Vincent, Sofie Thijs, Eva Bongaerts, Tim Nawrot, Wouter Marchal, Jonathan Van Hamme, and Jaco Vangronsveld. 2021. "Ambient Air Pollution Shapes Bacterial and Fungal Ivy Leaf Communities" Microorganisms 9, no. 10: 2088. https://doi.org/10.3390/microorganisms9102088
APA StyleStevens, V., Thijs, S., Bongaerts, E., Nawrot, T., Marchal, W., Van Hamme, J., & Vangronsveld, J. (2021). Ambient Air Pollution Shapes Bacterial and Fungal Ivy Leaf Communities. Microorganisms, 9(10), 2088. https://doi.org/10.3390/microorganisms9102088







