Abstract
Plant biomass offers great potential as a sustainable resource, and microbial consortia are primordial in its bioconversion. The wheat-straw-biodegradative bacterial strain w15 has drawn much attention as a result of its biodegradative potential and superior degradation performance in bacterial-fungal consortia. Strain w15 was originally assigned to the species Sphingobacterium multivorum based on its 16S ribosomal RNA (rRNA) gene sequence. A closer examination of this taxonomic placement revealed that the sequence used has 98.9% identity with the ‘divergent’ 16S rRNA gene sequence of S. multivorum NCTC 11343T, yet lower relatedness with the canonical 16S rRNA sequence. A specific region of the gene, located between positions 186 and 210, was found to be highly variable and determinative for the divergence. To solve the identity of strain w15, genome metrics and analyses of ecophysiological niches were undertaken on a selection of strains assigned to S. multivorum and related species. These analyses separated all strains into three clusters, with strain w15, together with strain BIGb0170, constituting a separate radiation, next to S. multivorum and S. siyangense. Moreover, the strains denoted FDAARGOS 1141 and 1142 were placed inside S. siyangense. We propose the renaming of strains w15 and BIGb0170 as members of the novel species, coined Sphingobacterium paramultivorum.
1. Introduction
In our studies on bacterial-fungal consortia that are able to degrade, and thrive on, wheat straw (WS), consistent evidence was obtained for the tenet that organisms identified as Sphingobacterium multivorum are consistently present and have defined roles in WS degradation processes [1,2]. A key facet of these Sphingobacterium-like organisms is their potential to produce a broad range of carbohydrate-active proteins, of which particular endo-1,4-beta-xylanases, alpha-l-arabinofuranosidases and alpha-l-fucosidases (belonging to CAZy families GH3, GH10, GH43, GH51, GH67 and GH95) stand out as having likely roles in consortial degradation processes [2]. In the course of our studies, a strain (denoted w15), preliminarily identified as S. multivorum based on its 16S rRNA sequence, was selected for further work, as it revealed a remarkable behavior when grown in combination with the fungus Coniochaeta ligniaria and the bacterium Citrobacter freundii so4 [3]. Other studies have also pointed to a role of S. multivorum (and alike) strains in the degradation of organic polymers from plant remains [4,5], keratan (endo-/β-galactosidase activity) [6], arabinoxylan [7], hexaconazole triazole fungicide [8] as well as petroleum and polycyclic aromatic hydrocarbon [9,10,11]. Moreover, a role in drought tolerance and growth promotion in tomato has been suggested [12]. On the other hand, S. multivorum strains have been isolated from clinical specimens, often as organisms involved in diverse kinds of opportunistic infections [13], causing bacteremia and acute meningitis in young immunocompetent adults [14]; and being involved in multiple myeloma [15] and cystic fibrosis [16].
This ecological versatility of S. multivorum strains is another clear motif to further investigate the identity of strain w15. Although its full genome sequence has been produced [17], it has not been fully explored, and hence an examination of its characteristics regarding identity was deemed necessary. We thus examined the available data regarding the origin, ecological niche, nature and diversity of, next to strain w15, selected S. multivorum and related strains, including two type strains present in different strain collections (NCTC 11343T, DSM 11691T).
The bacterial species S. multivorum derives from an early analysis of a group of 28 strains that originally fell within the genus Flavobacterium as a well-defined cluster (group IIk, biotype 2) [13]. Based on their coherent characteristics, these 28 strains were classified as forming one taxonomically concise, new species, coined Flavobacterium multivorum. The type strain, deposited in the NCTC culture collection, is the aforementioned NCTC 11343T. Briefly, the species, then considered to be typically clinical, was described as an aerobic, Gram-negative, nonmotile and oxidative bacterium, forming rod-shaped cells and growing at mesophilic temperatures. It was consistently able to produce acid from a range of carbohydrates. Strikingly, considerable resistance to a broad range of antimicrobial agents was found across the strains [13]. Later, based on the finding of high levels of sphingophospholipids as cellular lipid components, Yabuuchi et al. [18] founded the novel genus Sphingobacterium and transferred F. multivorum to it, yielding Sphingobacterium multivorum [13,18]. Currently, the genus Sphingobacterium encompasses at least 58 other species, including the close relatives of S. multivorum, namely S. siyangense and S. spiritivorum (type strain of the genus) [19,20,21,22]. Moreover, since its original description, several ‘environmental’ S. multivorum strains have been added to the species [4,6], and—following a search for strains related to our strain w15—we recently came across a highly interesting strain, BIGb0170 [23].
In recent years, bioinformatics-based genome metrics have developed into the standard for delineating bacterial species [24]. In particular, average nucleotide identity based on the BLASTn algorithm (ANIb) and genome BLAST distance phylogeny (GBDP)-based digital DNA: DNA hybridization (dDDH) methods have become the methods of choice to determine species boundaries and confirm identities [25]. In addition, tetranucleotide frequency (TETRA) assessments can yield supporting information on bacterial relatedness [26]. However, in spite of the fuzziness of the current species designations around S. multivorum, such advanced genome-based metrics have not yet been applied to the respective species.
Based on the emerging information gathered so far, we hypothesized that strain w15 might either represent a new subclass within the species S. multivorum (potentially adding a novel ecological type to this species) or belong to a group that is closely associated with S. multivorum. Hence, we examine the available evidence (based on analyses of both molecular and experimental data) with respect to the taxonomic and ecophysiological position of strain w15 and comparator strains, including the S. multivorum type strain. The results of these analyses clearly demonstrate that strain w15 belongs to a novel clade that is associated with S. multivorum and S. siyangense, for which we propose the name S. paramultivorum.
2. Materials and Methods
2.1. Strains and Genomes
Strain w15 was used as the ‘start’ organism. Moreover, data on diverse strains of S. multivorum, including the type strains, were selected across strain collections: NCTC 11343T; DSM 11691T (FDAARGOS 1203), FDAARGOS 1141 (DSM 11689, a.k.a. IFO 14087; NBRC 14087; F-179); FDAARGOS 1142 (DSM 11703), FDAARGOS 1140 (DSM 11688, KS 0433), FDAARGOS 1143 (DSM 15469, NCTC 11034), BIGb0170. In addition, the following close relatives of S. multivorum were also used: S. siyangense PDNC006, S. siyangense SY1T, Sphingobacterium sp. G1-14.
2.2. Cloning of Strain w15 16S rRNA Genes and Sequencing
Cells of strain w15 were recovered from −80 °C stocks and placed on tryptic soy agar (TSA, Sigma–Aldrich, Darmstadt, Germany) plates. After 24h incubation at 28 °C, a single colony was picked up and streaked to a new TSA plate, which was incubated at 28 °C for another 24 h. Afterwards, a single colony was introduced into 20 mL Luria-Bertani (LB, Sigma–Aldrich, Darmstadt, Germany) broth in a 100 mL Erlenmeyer flask. After 16h incubation at 28 °C, 1.8 mL of culture was harvested for DNA extraction using the DNeasy UltraClean Microbial Kit (Qiagen, GmbH, Hilden, Germany). Next, two sets of primers (B8F/515R and B8F/1492R) were used to generate 16S rRNA gene based amplicons by standard PCR [27]. The amplicons were then cleaned using the QIAquick PCR Purification Kit (Qiagen, GmbH, Hilden, Germany), and used for cloning in the pGEM-T Easy Vector Systems Kit (Promega, GmbH, Walldorf, Germany). The cloned amplicons were then subjected to sequencing at BaseClear (BaseClear B.V., Leiden, The Netherlands) using Sanger technology.
2.3. Detection of 16S rRNA Gene Types—And Their Ratio—In Transcriptomes
Three different sequence variants were found in the region between the 16S rRNA positions 186–210. These sequences (and their reverse complementary sequences) were used as proxies for (at least) three different 16S rRNA genes (Supplementary Material, Table S1) in order to assess their prevalence in RNA samples taken from a three-partner (strain w15, Citrobacter freundii so4, Coniochaeta sp. 2T2.1) wheat straw degradation experiment (Wang et al., in preparation). The sequences were not found in either C. freundii so4 or in Coniochaeta sp. 2T2.1. Other differences were found in the 16S rRNA gene region 463–476 (AAATACGTGTATTT and TTCTACGTGTAGGG). Different sequences found in this region were also used as proxies in a dichotomic scheme. The numbers of reads of each sequence type were summed and used for the ratio calculations, as given below. Three variants (denoted as C_GA, T_GA and T_CA) were found at positions 186–210, and two variants (denoted as AAA_TTT and TTC_GGG) were found in region 463–476. The ratio of a variant was calculated as, for example, ratio of variant C_GA = read count of variant C_GA/(sum of read counts of C_GA, T_GA and T_CA), ratio of AAA_TTT = read count of AAA_TTT/(sum of read counts of AAA_TTT and TTC_GGG).
2.4. Phylogenetic Analyses
Multiple sequence alignments were performed using the ‘Muscle’ default setting in the Mega 7 software [28]. The best DNA/Protein models (ML) were determined within this platform using partial deletion of gaps/missing data with 95% site coverage cutoff. Then, phylogenetic trees were inferred using maximum likelihood analyses [29] under the K2+G+I mode of evolution [28]. Bootstrap analyses were performed using 1000 resamplings to estimate the confidence values of the tree topologies [30].
To clarify the taxonomic position of w15, sequences of five conserved marker genes (rplC, groL, ftsA, dnaE, gyrB) were retrieved from NCBI (whole genome assemblies), and phylogenetic analyses were performed using the concatenated sequences in Mega 7 [28]. Each gene was first aligned independently using the ‘Muscle’ default setting. The best DNA/Protein models (ML) were determined using partial deletion of gaps/missing data with 95% site coverage cutoff [28]. The genes were then concatenated and aligned, and maximum likelihood phylogeny was determined under the LG + α + γ model of evolution and with 1000 bootstrap replicates.
2.5. Comparative Genome Analyses
ANI values and TETRA correlation coefficients were obtained using SpeciesWS [31]. The ANI analyses relied on the algorithms BLAST+ (ANIb) and MUMmer-Maximum Unique Matches (ANIm). Additionally, Tetra Correlation Search (TCS) analyses (shown as Z-values) were also done to provide a hit list for insights into the relationships with genomes of the reference genome database [31]. Typically, the ANI values between genomes of the same species are above ~95%, and so we used this threshold for species designations.
2.6. Genome-Based Taxonomic Analyses
Whole genome-based taxonomic analyses and dDDH were performed using the type strain genome server (TYGS, https://tygs.dsmz.de, accessed on 20 August 2021) [32] with the following methods.
Determination of closely-related type strains—Determination of the relatedness of type strain genomes was done in two complementary ways. First, ‘user’ genomes were compared against all type strain genomes available in the TYGS database via the rapid MASH algorithm [33], and then ten type strains with the smallest MASH distances were chosen per genome. Secondly, an additional set of ten closely-related type strains was determined via the 16S rRNA gene sequences. These were extracted from the user genomes using RNAmmer [34] and each sequence was subsequently BLASTed [35] against the 16S rRNA gene sequence of each of the currently 14,723 type strains available in the TYGS database. Distances calculated by formula d5 [25] were finally used to determine the ten closest type strain genomes for each of the user genomes.
Pairwise comparison of genome sequences—For the phylogenomic inference, pairwise comparisons across selected genomes were conducted using GBDP and accurate intergenomic distances inferred under the algorithm ‘trimming’ with distance formula d5 [25]. One hundred distance replicates were calculated for each. dDDH values and confidence intervals were calculated using the recommended settings of the GGDC 2.1 [25].
Phylogenetic inference—The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.6.1 including SPR postprocessing [36]. Branch support was inferred from one hundred pseudo bootstrap replicates each. The trees were rooted at the midpoint [37] and visualized with PhyD3 [38].
Type species-based species and subspecies clustering—Type species-based species clustering using a 70% dDDH radius around 28 selected Sphingobacterium type strains was done as previously described [32]. Subspecies clustering was done using a 79% dDDH threshold as previously recommended [39].
3. Results
Although initial analyses—based on the then-available almost-full 16S rRNA gene sequence—had placed strain w15 within the species S. multivorum [17], questions arose with respect to its precise affiliation. Major issues were the (potential) microheterogeneity across the ribosomal operons, and close association only with the so-called ‘divergent’ 16S rRNA gene of the type strain. We thus first set out to examine the overall genomic features of a selection of relevant strains associated with S. multivorum. We then made an in-depth analysis of the strain w15 16S rRNA genes, based on detailed analyses of the microheterogeneity across such operons.
3.1. General Properties of the Genomes of Strains Assigned or Related to S. multivorum and S. siyangense
To shed light on the nature of the association of strain w15 with the species S. multivorum, key genome features were first examined. As comparators, the genomes of two S. siyangense strains (SY1T; PDNC006) and one Sphingobacterium sp., GI-14, were also examined (Table 1). All S. multivorum strain genomes (NCTC 11343T; NCTC 11034; DSM 11691T; FDAARGOS 1140; FDAARGOS 1143) showed G+C% of 40.0–40.1%, genome sizes of 5.7–6.0 Mbp, and had totals of 4879–5310 identifiable genes. The S. siyangense strains (SY1T; PDNC006) showed G+C% of 39.8%, genome sizes of 6.3–6.8 Mbp, and had totals of 5482–5741 identifiable genes. Regarding the genome features, strain w15 had a G+C% of 39.8%, a genome size of 6.68 Mbp, with a total of 5541 identifiable genes. Interestingly, it was very akin to a recently described strain, BIGb0170, which revealed a G+C% of 39.9%, a genome size of 6.43 Mbp, and a total of 5348 genes.

Table 1.
Genome information of strains used in this study.
Across the board, and assuming an average bacterial gene size of about 1000 bp, relatively high gene loads and, consequently, relatively low levels of non-coding or non-assigned DNA were found. These were estimated to be around 83–89% (strains w15 and BIGb0170 83%, S. siyangense SY1T 87% and S. multivorum NCTC 11343T 89%) (Table 1). This indicated a moderate to strong control of genome quality via deletion bias in these organisms.
Interestingly, from the first analyses, we initially found evidence for the existence of 5–7 5S rRNA operons across all genomes, and 6–7 (all S. multivorum strains) versus just 1 (w15, and SY1T) 16S rRNA operon(s). This puzzling finding constituted the basis for our further investigations.
3.2. Unraveling the 16S rRNA Gene Features of Strain w15 and Other Strains
Here, we examine the 16S rRNA operons found across all S. multivorum strains, including the type strains NCTC 11343T and DSM 11691T, with implications for the identity of strain w15. First, the available 16S rRNA gene sequence data (not shown) indeed indicated strain DSM 11691T to be identical to NCTC 11343T, suggesting these are interchangeable (also including strain NBRC 14947T, a copy of NCTC 11343T). Hence, for further work, we used information gathered from strain NCTC 11343T, as the ‘canonical’ S. multivorum type strain. Remarkably, detailed analyses of this strain’s genome revealed the presence of seven ribosomal RNA operons (Table 1). Of these, six were largely identical (internal differences < 0.5%), with one being rather divergent. For the purpose of our study, the first group of six operons was coined the canonical group, and the outlier was coined the divergent operon.
3.2.1. Small Regions in the 16S rRNA Gene V2 Region Drive the Microheterogeneity
Major differences between the canonical and the divergent 16S rRNA gene of S. multivorum NCTC 11343T were found to be located in a small region (part of variable region 2 – V2) between positions 186 and 210 (13 differences/gap in a total of 25 base positions), as listed in Table 2. The remainder of the about 1500-bp sequence was rather similar (not shown). Thus, the divergent 16S rRNA gene sequence (UAUU01000005.1_rrna_60) had 98.89% similarity with the six canonical ones, while the similarities among the latter were 99.54–99.93% (Table 2). In this study, a random sequence from the canonical group was chosen to represent the canonical 16S rRNA gene (UAUU01000011.1_rrna_80) of S. multivorum NCTC 11343T.

Table 2.
16S rRNA gene sequence similarity within Sphingobacterium multivorum NCTC 11343T.
Remarkably, the genome-derived 16S rRNA gene sequence of strain w15 turned out to be most similar (98.89%) to the divergent sequence present in S. multivorum strain NCTC 11343T, and lower similarities were found with the canonical ones (98.48–98.76%, data not shown). Expectedly, the differences across the w15 and NCTC 11343T 16S rRNA gene sequences were—to a high extent–also driven by divergence in the 186–210 region (13 differences/gap in a total of 25 base positions), with one-base differences at just 8–9 positions over the remaining length (~1500 bases).
3.2.2. Exploring the 16S rRNA Operons in Strain w15 and Related Strains
For strain w15, we PCR-generated 40 clones, all of which were fully sequenced (see Figures S1–S3, Table S1). In the 40 clones, differences were found at three positions (in different combinations): position 186: C or T (24 C, 14 T); positions 207–209: G-A or A-C (35 G-A, 4 A-C); positions 463–465/474–476: 23 AAA-TTT, 15 TTC-GGG. The 40 sequences were sorted into two major groups based on the differences at positions 463–465/474–476: AAA-TTT and TTC-GGG. We then examined how the differences at the other positions were distributed within these two groups. Results are summarized in Table 3. A total of six sequence types was found, with differences among these being < 1% (Table 4). We hypothesized these represent (partial) sequences of at least six 16S rRNA gene operons. Of these, five almost-full sequences from each type of 16S rRNA gene operon were used in later analyses (w15, seq 1 to seq 5, Table 4).

Table 3.
Different 16S rRNA gene sequences in strain w15.

Table 4.
16S rRNA sequence similarities across Sphingobacterium strains, referring to w15 rrna_66.
To examine whether the different 16S rRNA gene copies play different roles in gene expression in the biodegradative consortia, the ratios of different 16S rRNA types of strain w15 growing in a three-partner consortium (Wang et al. in prep) were evaluated; these were calculated separately in the V2 and V3 regions (due to the technical limitation). Samples from three time points (early-24h, mid-5 days, and late-13 days) obtained from two shaking speeds (60 and 180 rpm) were tested. The ratio of variants T_AC/T_GA/C_GA (V2 region) and variants AAA-TTT/TTC-GGG (V3 region) were found to be stable over time and shaking speed, being around 1:2.1:4.0 and 1.6:1 respectively (Figure 1), similar to the ratios detected by the cloning and sequencing effort (Table 3).
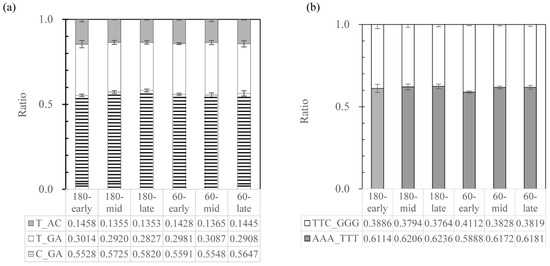
Figure 1.
16S rRNA types of strain w15 (a) ratio of T_AC (grey)/T_GA (blank)/C_GA (zebra lines) at V2 region, and (b) ratio of TTC_GGG (blank)/AAA_TTT (grey) at V3 region. 180: samples from 180 rpm shake cultures; 60: samples from 60 rpm cultures; early: 24 h; mid: 5 days; late: 13 days.
Analysis of the 16S rRNA sequences within each strain with the different types of 16S rRNA sequences (labelled seq 1, seq 2…; Table 4) revealed all seven 16S rRNA gene sequences of strain BIGb0170 to diverge < 0.2% across each other. Moreover, none of these seven sequences had >99% similarity with either the divergent or canonical 16S rRNA gene sequences of S. multivorum NCTC 11343T or S. siyangense SY1T. In contrast, all seven were >99% similar to sequences of strain w15, with the main differences being found at positions 186, 207–209, and 463–465/474–476. This resembled the variances found within strain w15 (Table 4), and indicated that strains BIGb0170 and w15 are very closely related.
We further found all seven 16S rRNA gene operons of strain FDAARGOS 1141 to have differences < 1%, with none of them having >99% similarity with either the divergent or the canonical 16S rRNA genes of S. multivorum NCTC 11343T. Instead, these had >99% similarity with the strain w15 16S rRNA gene sequences and one had 99.4% similarity with S. siyangense SY1T (Table 4).
However, the situation was complex for S. siyangense (strain PDNC006), as it showed <1% difference among all seven 16S rRNA gene operons, with two operons being similar to the canonical 16S rRNA gene of strain NCTC 11343T (99.48% similarity), and the other five having <1% difference with the similar ones of S. siyangense SY1T. An overview of these differences and similarities can be found in Table 4.
3.3. Phylogenetic Analyses
The analyses based on alignment of the 16S rRNA genes thus showed that the divergent and canonical 16S rRNA gene sequences of S. multivorum NCTC 11343T clustered into two groups (Figure 2a). Clearly, strains w15, BIGb0170, FDAARGOS 1141 and FDAARGOS 1142 were separate from these S. multivorum clusters. Whereas the sequences of the latter strains showed potential intermixing with S. siyangense SY1T, those of strains w15 and BIGb0170 revealed grouping within a separate cluster.
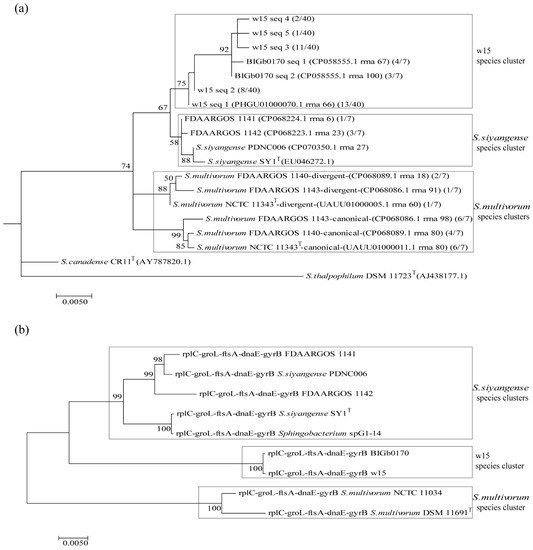
Figure 2.
Phylogenetic tree of strain w15, BIGb0170, S. multivorum NCTC 11343T, S. siyangense SY1T and related species, based on (a) 16S rRNA and (b) five concatenated core genes (rplC, groL, ftsA, dnaE, gyrB).
To further examine these uncertain placements, we constructed a five-gene concatenate based gene tree (Figure 2b). Analysis of the similarities of the core genes rplC, groL, ftsA, dnaE, gyrB showed that the w15 genes had high (>98.7%) amino acid similarities with the relevant counterpart sequences of all tested strains (Table S2), with the highest (100%) similarities with the respective genes of strain BIGb0170 (Table S2). However, at the DNA level, the five-gene concatenate trees clearly showed the existence of three groups among the selected strains: group 1—S. multivorum (DSM 11691T/NCTC 11343T, NCTC 11034); group 2—strains w15 and BIGb0170; group 3—S. siyangense (SY1T, PDN006). Interestingly, strains FDAARGOS 1141 and FDAARGOS 1142 both clustered tightly into group 3 (S. siyangense).
3.4. Average Nucleotide Identity (ANI) and Tetranucleotide Frequency (TETRA) Analyses
To further clarify the identities of strains w15, BIGb0170, FDAARGOS 1141 and FDAARGOS 1142, we calculated the ANI and TETRA values between the genomes of these four strains, next to 11 other ones, together belonging to six related Sphingobacterium species. The data indicated that the type strains of S. multivorum (NCTC 11343T and DSM 11691T) were virtually identical, with ANIb, ANIm and TETRA values being 99.97%, 99.97% and 1.0000, respectively (Figure 3). Furthermore, the only other strains denoted as S. multivorum that passed both the ANIb and ANIm species thresholds (>95%), were NCTC 11034, FDAARGOS 1140 and FDAARGOS 1143 (a.k.a. NCTC 11034, DSM 15469) (Figure 3). However, these did not pass the TETRA species thresholds. With respect to strain w15, it appeared as part of a small group including strain BIGb0170 (and FDAARGOS 1141), with >95% ANIb relatedness between them (Figure 3a), versus < 95% with the other strains including S. multivorum. This grouping was confirmed by the ANIm values (Figure 3b), albeit at higher relatedness levels. The TETRA values (Figure 3c) were largely in line with this contention, with strain w15 having 0.9994 similarity with BIGb0170 (but lower with FDAARGOS 1141, examined further in the next section), versus TETRA levels of <~0.9981 with both the S. multivorum and S siyangense (type) strains.

Figure 3.
Heat maps of average nucleotide identities; (a) ANIb, (b) ANIm; and tetranucleotide frequencies (c) TETRA analyses of strains w15, BIGb0170, S. multivorum NCTC 11343T, S. siyangense SY1T and related species. Selected ANIb (> 95%) and TETRA (> 0.999) values were used as species thresholds.
3.4.1. Taxonomic Placement of FDAARGOS 1141 and FDAARGOS 1142
Surprisingly, the genomes of strains w15, BIGb0170, FDAARGOS 1141 and FDAARGOS 1142 only showed 91–92% ANIb and ANIm values to S. multivorum NCTC 11343T. This provided strong evidence for the tenet that these four strains fall outside of the species S. multivorum. Furthermore, only strains FDAARGOS 1141 and FDAARGOS 1142 showed ANIb (and ANIm) values > 95% with S. siyangense SY1T, whereas strains w15 and BIGb0170 had ANIb values of only 94.57% and 94.78, respectively (Figure 3). In contrast, FDAARGOS 1141 and FDAARGOS 1142 both failed the TETRA species threshold with S. siyangense SY1T, as both revealed values of 0.9984; this was also found above (S. multivorum species). Based on these data, we consider the placement of FDAARGOS 1141 and FDAARGOS 1142 into a (sub)group related to S. siyangense to be commendable; however, this is to be examined further.
3.4.2. Proposal of a New Species Based on Strains w15 and BIGb0170
Clearly, the genomes of w15 and BIGb0170 were highly similar, with ANIb and ANIm values (98.3% and 98.9% between them) exceeding the respective species thresholds (>95%). Remarkably, as discussed in the foregoing, the two genomes also passed the TETRA species cutoff value (>0.999), being 0.9994 (Figure 3). Finally, the core gene concatenate comparisons (Figure 2b) were consistent with the high relatedness of the genomes of these two strains. Thus, the collective evidence points to the conclusion that strains w15 and BIGb0170 form a taxonomically tight group, which differs substantially from S. multivorum and S. siyangense, and establishes a novel species, for which the name Sphingobacterium paramultivorum is proposed.
3.5. Genome-Based Taxonomic Analyses
Using the cutoff value of dDDH (>70%), the aforementioned genomes were analyzed by digital hybridization analyses. The data showed that the genomes of strains w15 and BIGb0170 both had dDDH values of <70% with those of the S. multivorum and S. siyangense type strains (being 46% and 66%, respectively). Overall, the genomes clustered into three groups (Figure 4), i.e., the predicted S. multivorum and S. siyangense, and the newly proposed S. paramultivorum (w15 and BIGb0170) groups. Similarly, the genomes of both FDAARGOS 1141 and FDAARGOS 1142 had dDDH values of 46–47% with that of S. multivorum NCTC 11343T (below the species boundary) and 71–72% with that of S. siyangense SY1T. The clustering was further consistent with the five-gene phylogenetic tree and the ANI analyses. A 79% dDDH threshold was used by us for further (subspecies) clustering, which resulted in two subspecies identified under S. multivorum and S. siyangense (Table 5, Figure 5); this was in line with the five-gene phylogenetic tree and the TETRA analysis.
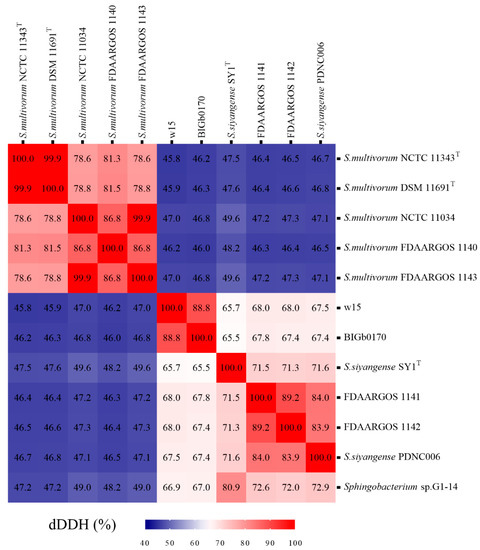
Figure 4.
Heat map of dDDH analyses of strain w15, BIGb0170, S. multivorum NCTC 11343T, S. siyangense SY1T and closely related species.

Table 5.
Taxonomic identification based on whole genome sequences.
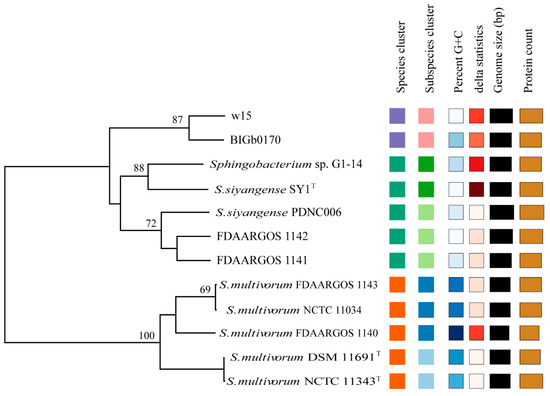
Figure 5.
Tree inferred with FastME 2.1.6.1 [36] from GBDP distances calculated from the genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above the branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 65.2%. The tree was rooted at the midpoint [37]. Colored squares indicate relatedness (same color, same parameter value class, on the basis of standard FastME settings).
Furthermore, the whole genome-based taxonomic analyses using the type strain database (TYGS) confirmed that strains FDAARGOS 1140, FDAARGOS 1143 and NCTC 11034 all belong to the species S. multivorum, and strain PDNC006 to S. siyangense (Table 5). The analysis also confirmed the conclusion that strains w15 and BIGb0170, together, form a new species, and FDAARGOS 1141 and FDAARGOS 1142 are highly related to S. siyangense (Table 5).
3.6. Examination of Strain Origins and Ecophysiological Data
In an attempt to determine the ecological ‘niches’ of the selected strains, all strain descriptions were examined with respect to the strain (ecophysiological) metadata. For strain w15, we used data generated in our own laboratory [17]. Table 6 shows a summary of these analyses. Clearly, two major types of strain origin were found: clinical or environmental (soil, nematode-associated or rotting plant matter) settings. Moreover, one may discern the glimpses of two types of ecological behavior, as it is likely the strains first found in clinical settings are often involved in infectious processes (interactions with hosts leading to infection), whereas those found in environmental settings often revealed involvement in biodegradative processes. Strain w15 is strongly involved in WS degradation, whereas the sister strain BIGb0170 is a context-dependent colonizer of Caenorhabditis elegans, in which host biodegradative processes also play roles. Clearly, the evidence further shows that strains of all three clusters are able to utilize a plethora of carbon sources, such as D-xylose, maltose, D-melibiose, D-fructose, D-glucose, sucrose, D-galactose, trehalose, lactose, cellobiose, melezitose, salicin, and N-acetyl-D-glucosamine.

Table 6.
Strain origins and ecophysiological data.
Other traits differentiate the three clusters, as follows. Only S. siyangense SY1T was able to grow at 4 and 42 °C, and survive at low pH (<4.0); it performs hydrolysis of L-arginine, D-sorbitol, L-sorbose, D-mannitol, xylitol, adonitol and glycerol. In contrast, neither w15 nor S. multivorum NCTC 11343T were able to grow at 4 °C, 42 °C, pH 4.0 or utilize these carbon sources. Remarkably, all strains from the clinic (S. multivorum NCTC 11343T, NCTC 11034) were able to hydrolyse Tween 80, while w15 and BIGb0170 and S. siyangense SY1T were not. S. multivorum NCTC 11343T was not able to utilize D-mannose or L-rhamnose, but w15 and S. siyangense SY1T were.
4. Discussion
In this study, we examined the available data on the genomic, phylogenetic and ecophysiological traits of selected relevant S. multivorum and related Sphingobacterium strains, including the biodegradative strain w15 (Table 1). Following this data gathering exercise, we placed a focus on key data that assist us in the elucidation of the taxonomic placement of the relevant strains w15, BIGb0170 and several FDAARGOS ones, which so far have been associated loosely with S. multivorum. Indeed, following the coining of the genus Sphingobacterium by Yabuuchi et al. in 1983 [18], there have been many additions to this genus, including the species S. multivorum. However, the exact placement of S. multivorum has been under debate and the position of several related organisms has also remained unclear.
Unraveling the taxonomic identity of selected strains associated with S. multivorum—16S rRNA gene features—Clearly, the fact that multiple 16S rRNA operons occur within and across the selected Sphingobacterium- strains, including the S. multivorum type strain, poses key problems for the taxonomic consistency in these organisms. This facet also has strong implications for the taxonomic identity of strain w15, which was previously assigned to S. multivorum based on just a single 16S rRNA gene sequence [17]. In addition, previous studies revealed the species S. multivorum to be very close to S. siyangense [20], and these two species were often noticed to cluster together in 16S rRNA sequence based phylogenetic trees [19,20,44]. Thus we surmised that a “species cluster” (<1% divergence at the 16S rRNA gene sequence level) might be present. However, this tenet appeared to be insufficient to delineate species, as in species containing multiple 16S rRNA gene copies, sequences with up to 1% divergence or more can occur (Table 4). The current way of categorizing bacteria into species by 16S rRNA gene sequence identity would unavoidably classify a great diversity of phylogenetically different bacteria into the same species [45]. To the best of our knowledge, this is the first report on multiple 16S rRNA sequences with such high (1% or more) divergence within one single strain. The data obtained in this study indicate that a complex 16S rRNA gene sequence picture is present across all strains. In fact, we present evidence for the contention that strain w15 has at least six 16S rRNA genes, with microheterogeneity between them. Moreover, this situation was akin to that in strain BIGb0170 and also to that of the species S. multivorum, although it was different by sequence. In detail, a highly variable 16S rRNA gene region—position 186–210—was identified as being mainly responsible for the differences, whereas another region—position 463–465/474–476—was also important. The use of the sequence divergences in these two regions in rRNA expressed under different conditions and over time in degrader consortia, however, did not reveal differences in the ratio of the different rRNA types (Figure 1). This debunked our hypothesis of an ecophysiological driver for the existence of the multiple diverse 16S rRNA gene copies, at least within the limits of the experimental contexts.
The differentiation of the selected strains into three groups is robust and supported by several lines of evidence—Although the 16S rRNA gene analyses were blurred by multiple sequences and sequence microheterogeneities, at least three lines of evidence provided solid evidence for the separation of strains w15 and BIGb0170 from S. multivorum and S. siyangense, and their placement together into a novel species. These were, first, the solid evidence yielded by ANIb (and ANIm), in which the w15/BIGb0170 group stood out as clearly separate from the other groups, passing the species threshold set at 95% (Figure 3a,b). This was supported by the following TETRA analyses, in which a threshold of 0.999 was used (Figure 3c). The second thread was provided by the dDDH and whole genome based taxonomic analyses; the <70% dDDH values with the S. multivorum and S. siyangense comparator genomes point to a new species formed by w15 and BIGb0170 (Figure 4 and Figure 5, Table 5). The third thread came from the five gene-concatenate analyses, which clearly showed the existence of three clusters, with strains w15 and BIGb0170 clustering closely together. Overall, we consider these analyses to provide very convincing evidence for the contention that strains w15 and BIGb0170 together constitute a novel species, coined Sphingobacterium paramultivorum, which is related to S. siyangense and S. multivorum.
Habitats, niche and ecological versatility of the selected Sphingobacterium strains: among all selected strains, several, including w15, BIGb0170, FDAARGOS 1141 (from soil) and FDAARGOS 1142 (from activated sludge) came from environmental samples, whereas others, including S. multivorum NCTC 11343T and NCTC 11034 were from clinical specimens. This shows the divergent environmental habitats in which the organisms that were loosely associated with S. multivorum can be found. It is logical to posit that the organisms in this broad group can thrive in environments in which pulses of nutrients become available at times, establishing conditions for rapid growth provided a quick start (kick-off) can be made. Previous studies have shown that S. multivorum strains can thrive in various environments, from plant polymers [4,5] to hexaconazole triazole/petroleum hydrocarbon contaminated soil [8,9,10,11]. One can postulate that such ecological situations can and will arise in both the environmental and clinical settings, establishing suitable niches and driving the growth and evolution of the niche occupants. Given that the strains studied were obtained from soils, plant waste, plastics, and clinical settings (Table 6), we posit that their primary ‘habitat’ might be broad. What does this mean for the ecological niche they occupy, and can we discern differences in this niche occupancy across the three groups we identified? Defining niche as the combination of conditions, energy generation and nutritional parameters, yielding a multiple-axis ecological space area, it appears difficult to affirm the precise niche (and habitat) space occupied by the S. multivorum associated organisms. Clearly, based on their (shared) genome traits, it appears that all organisms can thrive in carbon- and oxygen-rich settings, where growth depends on oxidative processes. In the respective habitats, it is thought that pH and temperature are drivers that possibly define the (sub)niches, with obvious differences between environmental and clinical settings (Table 6).
Given the above arguments, the situation with respect to the ecophysiological characteristics of the w15/BIGb0170 group, and its differentiation from the other two groups, is not clear and subject to speculation. Whereas strain w15 was selected by WS into a WS degradation process and so may find its niche there; BIGb0170 is adapted to life within nematodes (where conditions of biopolymer degradation may reign as well). A commonality is the soil origin of both organisms, in which conditions of fluctuating temperature, pH and nutrient levels commonly reign. Furthermore, from the genomic analyses plus some of the physiological tests, it appears that strains w15 and BIGb0170 are both quick responders to the appearance of carbonaceous substrate [3,23]. Together, such commonalities are consistent with the genome-based arguments for the placement of w15 and BIGb0170 into a new species, in which they might even represent different ecotypes. In contrast, we lack evidence for a thorough consideration of the niches and ecophysiological traits of the members of the other two groups.
Can the nature of the genomes and the 16S rRNA genes across the selected sphingobacteria be linked to niche occupancy? All strains analyzed had relatively large genomes, with those of the w15/BIGb0170 group, together with those of the S. siyangense strains, having larger sizes than those of the S. multivorum group (Table 1). This could be explained by the contention that clinical S. multivorum group strains to some extent adapt to a host setting, having lost some functions that are important for survival in harsh (soil) settings [46]. Moreover, all genomes had relatively high gene loads (83–89%)—as is usual for bacterial genomes—which is usually taken as evidence for high gene conservation pressures due to regular events of selection, in the light of the commonly high deletion bias [46,47]. The genomes had two additional salient features, i.e., the ability to furnish the (secreted) enzymes for utilization of a plethora of different substrates, often derived from organic polymers, as carbon and energy sources, and the high number (up to seven) of 16S rRNA operons. The presence of multiple such operons, and the potential importance in the growth kick-off, was already reported for Escherichia coli [48,49] and some bacilli [50] but it was new for strain w15. In detail, strain w15 had at least six, slightly divergent, 16S rRNA gene operons, mainly driven by the highly variable 186–210 region. It indeed was a highly efficient grower in aerobic sequential batches with WS as the substrate, in which short growth periods, interspersed with periods of slow growth or growth arrest, were stimulated at the expense of WS as the carbon and energy source [3,17]. Indeed, in WS-driven consortia, strain w15 is—upon each introduction—confronted with a sudden flush of abundant resources, like the sugars (or organic acids) coming off the WS polymers hemicellulose and cellulose. Therefore, w15 will have to rapidly scale the transcriptional and translational apparatus up and down, and potentially this is reflected in ribosome numbers and also types of ribosomes (loaded with either the canonical or the divergent 16S rRNA). The fact that no changes in the ratios between different types were found (Figure 1) indicated the absence of strong selective pressures favoring one or another rRNA type, across the conditions (Wang et al., in prep). Thus, one may surmise that such operons (within the constraints of the study) are selected to a rather similar extent.
The importance of multiple 16S rRNA operons for the ecology of strain w15—As brought forward in the literature and the foregoing text, the resource availability in an environmental setting, being either high, moderate or low, is a key driver of the population dynamics of bacteria like strain w15. Thus, fluctuating resource concentrations result in particular temporal resource dynamics which likely selected—over evolutionary time—strains like w15 for optimal fitness [51]. Previous work has already shown that the number of rRNA gene copies may predict an organism’s growth rate as well as growth efficiency as affected by resource availability [51,52]. Thus, in light of the tenet that multiple rRNA operons go with possibilities for high (initial) growth rates, the rRNA gene copy numbers may dictate not only the growth rate, but also the speed of response to growth condition changes, in particular upshifts [48,52,53]. The rapid bacterial growth observed by us for strain w15 (Wang et al. in prep), may require a substantial instantaneous increase in cellular ribosome numbers (as compared to slow growth or growth arrest). Clearly, such multiple rRNA gene copies may support the rapid build-up of sufficient ribosome numbers, simply by facilitating rRNA transcriptions. To what extent the microheterogeneities across the different gene copies in w15 have effects is so far unclear, as the data reported here (Figure 1) appear to show relative constancy of the type ratios. Similar reasoning may apply to the other multi rRNA copy organisms studied here.
5. Conclusions
Altogether, it can be concluded from this study that there is consistency in the data which unequivocally show that strains w15 and BIGb0170 represent a new species, for which the name Sphingobacterium paramultivorum is proposed. The type strain will be S. paramultivorum w15, deposited at DSMZ under number DSM 106342. Briefly, it is characterized as: Gram-negative, non-motile, non-spore forming, rod-shaped, forming yellowish colonies; growth in aerobic conditions, in a narrow pH range across neutral pH (5.0–9.0, optimal at pH 7.0), mesophilic temperature range (20–30 °C, optimal at 28 °C), and moderate NaCl levels (0–1%); able to grow on multiple sugar compounds (α-methyl-D-glucoside, α-D-lactose, D-cellobiose, α-methyl-D-glucoside, N-acetyl-D-glucosamine, pectin, dextrin, inulin), but not on acid compounds (L-phenylalanine, i-erythritol, D-galacturonic acid, D-galactonic acid lactone, putrescine, L-serine, D-mannitol, L-asparagine, L-threonine, glycogen, itaconic acid).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9102057/s1; Figure S1: Difference could be due to B8F primer (AGAGTTTGATCMTGGCTCAG), Figure S2: Random differences, Figure S3: Differences with a clear ratio/frequency, Table S1: sequences used as probes for 16S rRNA ratio of w15, and Table S2: similarity of 9 strains to w15 of 5 protein genes (amino acid).
Author Contributions
Conceptualization, Y.W. and J.D.v.E.; methodology, Y.W. and J.K.B.; software, Y.W.; investigation, Y.W.; data curation, Y.W.; writing—original draft preparation, Y.W. and J.D.v.E.; writing—review and editing, J.K.B. and J.D.v.E.; visualization, Y.W.; supervision, J.D.v.E.; project administration, J.D.v.E.; funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
Y.W. was supported by the China Scholarship Council (CSC) though the Ph.D. scholarship program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank Akbar Adjie Pratama, Xiu Jia, Xipeng Liu, and Zhenhua Zhang for their advice on the analysis method.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed potential conflicts of interest.
References
- Cortes-Tolalpa, L.; Jiménez, D.J.; de Lima Brossi, M.J.; Salles, J.F.; van Elsas, J.D. Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl. Microbiol. Biotechnol. 2016, 100, 7713–7725. [Google Scholar] [CrossRef]
- Jiménez, D.J.; Maruthamuthu, M.; van Elsas, J.D. Metasecretome analysis of a lignocellulolytic microbial consortium grown on wheat straw, xylan and xylose. Biotechnol. Biofuels 2015, 8, 199. [Google Scholar] [CrossRef]
- Cortes-Tolalpa, L.; Salles, J.F.; van Elsas, J.D. Bacterial synergism in lignocellulose biomass degradation–complementary roles of degraders as influenced by complexity of the carbon source. Front. Microbiol. 2017, 8, 1628. [Google Scholar] [CrossRef]
- Ventorino, V.; Aliberti, A.; Faraco, V.; Robertiello, A.; Giacobbe, S.; Ercolini, D.; Amore, A.; Fagnano, M.; Pepe, O. Exploring the microbiota dynamics related to vegetable biomasses degradation and study of lignocellulose-degrading bacteria for industrial biotechnological application. Sci. Rep. 2015, 5, 8161. [Google Scholar] [CrossRef] [PubMed]
- Photphisutthiphong, Y.; Vatanyoopaisarn, S. Dyadobacter and Sphingobacterium isolated from herbivore manure in Thailand and their cellulolytic activity in various organic waste substrates. Agric. Nat. Resour. 2019, 53, 89–98. [Google Scholar] [CrossRef]
- Kitamikado, M.; Ito, M. Isolation of keratanase-producing bacteria from natural habitats. J. Fact. Agric. Kyushu Univ. 1979, 24, 101–112. [Google Scholar] [CrossRef]
- Malfliet, S.; Justé, A.; Crauwels, S.; Willems, K.; De Cooman, L.; Lievens, B.; Aerts, G. Assessing the xylanolytic bacterial diversity during the malting process. Food Microbiol. 2013, 36, 406–415. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Tian, C.; Xu, J.; Dong, F.; Liu, X.; Wu, X.; Zheng, Y. Characterization of hexaconazole-degrading strain Sphingobacterium multivorum and analysis of transcriptome for biodegradation mechanism. Sci. Total Environ. 2020, 722, 137171. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.A.; Mohamedelhassan, E.E.; Yanful, E.K.; Weselowski, B.; Yuan, Z.C. Isolation and characterization of novel bacterial strains for integrated solar-bioelectrokinetic of soil contaminated with heavy petroleum hydrocarbons. Chemosphere 2019, 237, 124514. [Google Scholar] [CrossRef]
- Cheng, J.; Sun, Z.; Yu, Y.; Li, X.; Li, T. Effects of modified carbon black nanoparticles on plant-microbe remediation of petroleum and heavy metal co-contaminated soils. Int. J. Phytoremed. 2019, 21, 634–642. [Google Scholar] [CrossRef]
- Balu, S.; Bhunia, S.; Gachhui, R.; Mukherjee, J. Assessment of polycyclic aromatic hydrocarbon contamination in the Sundarbans, the world’s largest tidal mangrove forest and indigenous microbial mixed biofilm-based removal of the contaminants. Environ. Pollut. 2020, 266, 115270. [Google Scholar] [CrossRef]
- Eke, P.; Kumar, A.; Sahu, K.P.; Wakam, L.N.; Sheoran, N.; Ashajyothi, M.; Patel, A.; Fekam, F.B. Endophytic bacteria of desert cactus (Euphorbia trigonas Mill) confer drought tolerance and induce growth promotion in tomato (Solanum lycopersicum L.). Microbiol. Res. 2019, 228, 126302. [Google Scholar] [CrossRef]
- Holmes, B.; Owen, R.J.; Weaver, R.E. Flavobacterium multivorum, a new species isolated from human clinical specimens and previously known as group IIk, biotype 2. Int. J. Syst. Bacteriol. 1981, 31, 21–34. [Google Scholar] [CrossRef]
- Abro, A.H.; Rahimi Shahmirzadi, M.R.; Jasim, L.M.; Badreddine, S.; Al Deesi, Z. Sphingobacterium multivorum Bacteremia and Acute Meningitis in an immunocompetent adult patient: A case report. Iran. Red Crescent Med. J. 2016, 18, e38750. [Google Scholar] [CrossRef][Green Version]
- Konala, V.M.; Naramala, S.; Bose, S.; Gayam, V.; Madhira, B.R.; Adapa, S. Bacteremia secondary to uncommon gram-negative Bacilli transmitted from the canine in a patient with multiple Myeloma. J. Investig. Med. High Impact Case Rep. 2020, 8. [Google Scholar] [CrossRef]
- Lambiase, A.; Rossano, F.; Del Pezzo, M.; Raia, V.; Sepe, A.; de Gregorio, F.; Catania, M.R. Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res. Notes 2009, 2, 262. [Google Scholar] [CrossRef]
- Cortes-Tolalpa, L.; Wang, Y.; Salles, J.F.; van Elsas, J.D. Comparative genome analysis of the lignocellulose degrading bacteria Citrobacter freundii so4 and Sphingobacterium multivorum w15. Front. Microbiol. 2020, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, E.; Kaneko, T.; Yano, I.; Moss, C.W.; Miyoshi, N. Sphingobacterium gen. nov., Sphingobacterium spiritivorum comb. nov., Sphingobacterium multivorum comb. nov., Sphingobacterium mizutae sp. nov., and Flavobacterium indologenes sp. nov.: Glucose-Nonfermenting Gram-Negative Rods in CDC Groups IIK-2 and IIb. Int. J. Syst. Evol. Microbiol. 1983, 33, 580–598. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Marayati, B.F.; Wada-Katsumata, A.; Wasserberg, G.; Schal, C.; Apperson, C.S.; Ponnusamy, L. Sphingobacterium phlebotomi sp. nov., a new member of family Sphingobacteriaceae isolated from sand fly rearing substrate. Int. J. Syst. Evol. Microbiol. 2021, 71, 004809. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, H.; Zhang, C.X.; Yang, S.Y.; Liu, X.H.; Zhang, K.Y.; Lai, R. Sphingobacterium siyangense sp. nov., isolated from farm soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 1458–1462. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar Chaudhary, D.; Kim, J. Sphingobacterium terrae sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.; Owen, R.J.; Hollis, D.G. Flavobacterium spiritittorum, a new species isolated from human clinical specimens. Int. J. Syst. Bacteriol. 1982, 32, 157–165. [Google Scholar] [CrossRef]
- Dirksen, P.; Assié, A.; Zimmermann, J.; Zhang, F.; Tietje, A.M.; Marsh, S.A.; Félix, M.A.; Shapira, M.; Kaleta, C.; Schulenburg, H.; et al. CeMbio—The Caenorhabditis elegans Microbiome Resource. G3 Genes Genomes Genet. 2020, 10, 3025–3039. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Pride, D.T.; Meinersmann, R.J.; Wassenaar, T.M.; Blaser, M.J. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003, 13, 145–158. [Google Scholar] [CrossRef]
- Grizard, S.; Dini-Andreote, F.; Tieleman, B.I.; Salles, J.F. Dynamics of bacterial and fungal communities associated with eggshells during incubation. Ecol. Evol. 2014, 4, 1140–1157. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Gen. Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Gen. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Takeuchi, M.; Yokota, A. Proposals of Sphingobacterium faecium sp. nov., Sphingobacterium piscium sp. nov., Sphingobacterium heparinum comb. nov., Sphingobacterium thalpophilum comb. nov., and two genospecies of the genus Sphingobacterium, and synonymy of Flavobacterium yabuuchiae and Sphingobacterium spiritivorum. J. Gen. Appl. Microbiol. 1992, 38, 465–482. [Google Scholar] [CrossRef]
- Steyn, P.L.; Segers, P.; Vancanneyt, M.; Sandra, P.; Kersters, K.; Joubert, J.J. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int. J. Sys. Bact. 1998, 48, 165–177. [Google Scholar] [CrossRef]
- Oyaizu, H.; Komagata, K. Chemotaxanomic and phenotypic characterization of the strains of species in the Flavobacterium-cytophaga complex. J. Gen. Appl. Microbiol. 1981, 27, 57–107. [Google Scholar] [CrossRef]
- Yin, P.; Liu, S.; Wu, X. Whole genome sequence of Sphingobacterium sp. G1-14, a strain with effective paichongding biodegradation. J. Biol. Life Sci. 2019, 10, 58. [Google Scholar] [CrossRef]
- Lee, D.; Hur, J.S.; Kahng, H.Y. Sphingobacterium cladoniae sp. nov., isolated from lichen, Cladonia sp., and emended description of Sphingobacterium siyangense. Int. J. Syst. Evol. Microbiol. 2013, 63, 755–760. [Google Scholar] [CrossRef]
- Tang, L.; Li, Y.; Deng, X.; Johnston, R.N.; Liu, G.R.; Liu, S.L. Defining natural species of bacteria: Clear-cut genomic boundaries revealed by a turning point in nucleotide sequence divergence. BMC Genom. 2013, 14, 489. [Google Scholar] [CrossRef]
- Bobay, L.M.; Ochman, H. The Evolution of Bacterial Genome Architecture. Front Genet. 2017, 30, 72. [Google Scholar] [CrossRef]
- Mira, A.; Ochman, H.; Moran, N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001, 17, 589–596. [Google Scholar] [CrossRef]
- Condon, C.; Liveris, D.; Squires, C.; Schwartz, I.; Squires, C.L. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 1995, 177, 4152–4156. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.S.; Schmidt, T.M. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 2004, 70, 6670–6677. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.M.; Noll, M.; Liesack, W. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 2007, 9, 2464–2474. [Google Scholar] [CrossRef]
- Roller, B.R.K.; Schmidt, T.M. The physiology and ecological implications of efficient growth. ISME J. 2015, 9, 1481–1487. [Google Scholar] [CrossRef]
- Klappenbach, J.A.; Dunbar, J.M.; Schmidt, T.M. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 2000, 66, 1328–1333. [Google Scholar] [CrossRef]
- Stevenson, B.S.; Schmidt, T.M. Growth rate-dependent expression of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 1998, 180, 1970–1972. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).