Comparative Secretomics Analysis Reveals the Major Components of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzyme Production and Extraction
2.3. Determination of Enzyme Activity and Protein Content
2.4. Pretreatment of Corncob Powder (Pr-CP), RS (Pr-RS), and MCC (Pr-MCC)
2.5. Enzymatic Hydrolysis of Pr-CP, Pr-RS, and Pr-MCC
2.6. SDS-PAGE of Secretome, and In-Gel Digestion
2.7. LC-MS/MS Analysis
2.8. Data Processing
3. Results
3.1. Enzymatic Activities of Major Glycoside Hydrolases (GHs)
3.2. Hydrolytic Ability of GHs
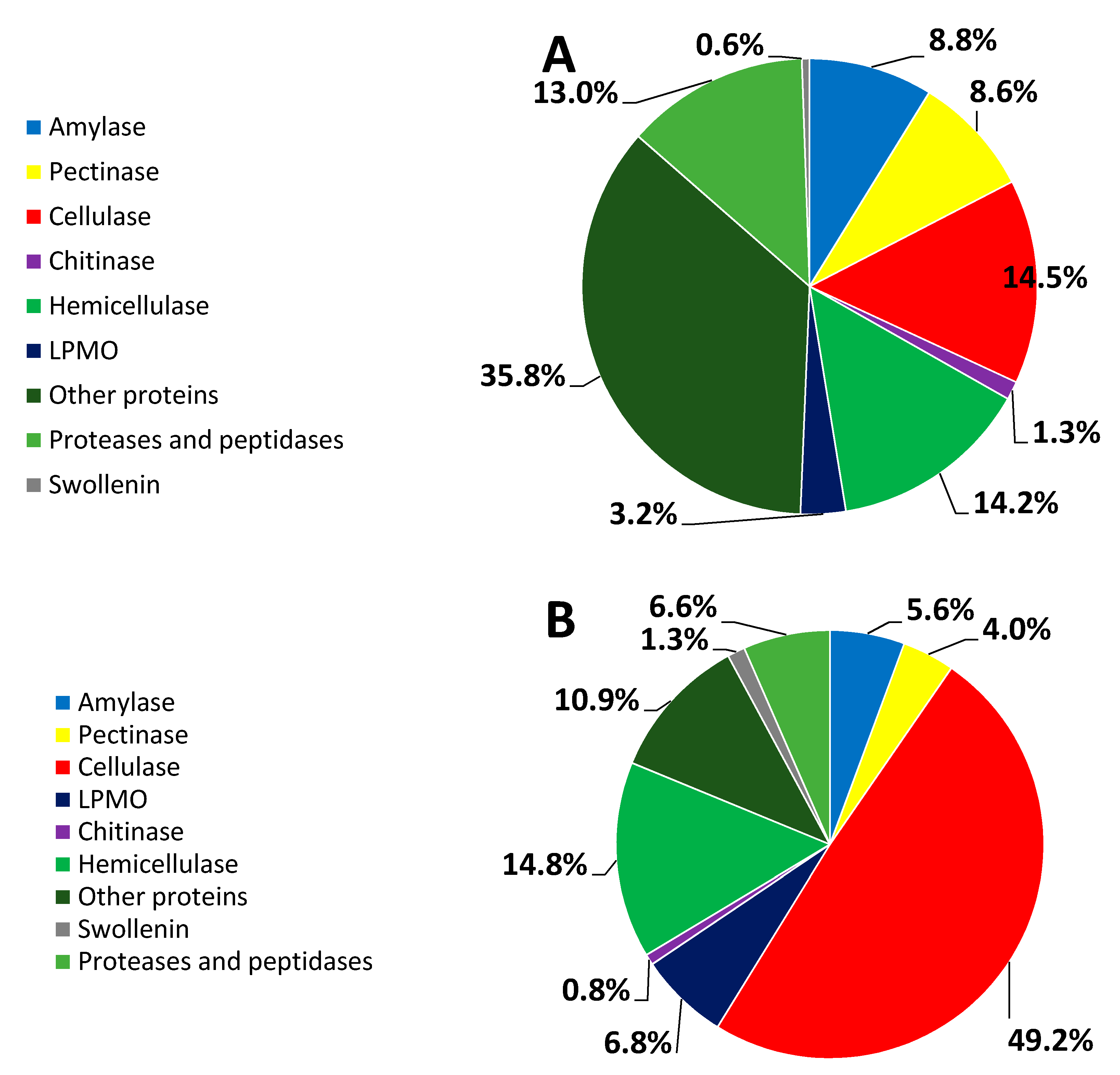
3.3. Percent Abundance of the Identified Proteins
3.4. Revealing Up-Regulated and Down-Regulated Proteins
3.5. Ascertaining Identities with a High Percentage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Watt, G.D. A new future for carbohydrate fuel cells. Renew. Energy 2014, 72, 99–104. [Google Scholar] [CrossRef]
- Zhao, X.; Yi, S.; Li, H. The optimized co-cultivation system of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30 achieved a high yield of hydrolase applied in second-generation bioethanol production. Renew. Energy 2019, 136, 1028–1035. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, L.; Xu, S.; Zhang, H.; Ren, B.; Li, T.; Zhang, S. Ionic liquid functionalized electrospun gel polymer electrolyte for use in a high-performance lithium metal battery. J. Mater. Chem. A 2018, 6, 18479–18487. [Google Scholar] [CrossRef]
- Wang, K.; Huang, Q.; Li, H.; Zhao, X. Co-evolution of β-glucosidase activity and product tolerance for increasing cellulosic ethanol yield. Biotechnol. Lett. 2020, 42, 2239–2250. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, K.; Li, H.; Yi, S.; Zhao, X. Enhancing cellulosic ethanol production through coevolution of multiple enzymatic characteristics of β-glucosidase from Penicillium oxalicum 16. Appl. Microbiol. Biotechnol. 2020, 104, 8299–8308. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.; Wu, C.; Liu, P.; Wang, W.; Wei, D. The transcription factor ACE3 controls cellulase activities and lactose metabolism via two additional regulators in the fungus Trichoderma reesei. J. Biol. Chem. 2019, 294, 18435–18450. [Google Scholar] [CrossRef]
- Duchesne, L.C.; Larson, D.W. Cellulose and the evolution of plant life. Bioscience 1989, 39, 238–241. [Google Scholar] [CrossRef]
- Saha, B.C. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 2003, 30, 279–291. [Google Scholar] [CrossRef]
- Zhang, Y.H.P.; Himmel, M.E.; Mielenz, J.R. Outlook for cellulase improvement: Screening and selection strategies. Biotechnol. Adv. 2006, 24, 452–481. [Google Scholar] [CrossRef]
- Gong, W.; Zhang, H.; Liu, S.; Zhang, L.; Gao, P.; Chen, G.; Wang, L. Comparative secretome analysis of Aspergillus niger, Trichoderma reesei, and Penicillium oxalicum during solid-state fermentation. Appl. Biochem. Biotechnol. 2015, 177, 1252–1271. [Google Scholar] [CrossRef]
- Han, X.; Wen, H.; Luo, Y.; Yang, J.; Xiao, W.; Ji, X.; Xie, J. Effects of α-amylase and glucoamylase on the characterization and function of maize porous starches. Food Hydrocoll. 2021, 116, 106661. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G.H. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Limsakul, P.; Phitsuwan, P.; Waeonukul, R.; Pason, P.; Tachaapaikoon, C.; Poomputsa, K.; Kosugi, A.; Sakka, M.; Ratanakhanokchai, K. A novel AA10 from Paenibacillus curdlanolyticus and its synergistic action on crystalline and complex polysaccharides. Appl. Microbiol. Biotechnol. 2020, 104, 7533–7550. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, T.L.R.; Dos Santos, L.V.; Pereira, G.A.G. AA9 and AA10: From enigmatic to essential enzymes. Appl. Microbiol. Biotechnol. 2016, 100, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Brunecky, R.; Yao, B.; Xie, X.; Zheng, F.; Luo, H. A Swollenin from Talaromyces leycettanus JCM12802 enhances cellulase hydrolysis toward various substrates. Front. Microbiol. 2021, 12, 715. [Google Scholar]
- Zhao, X.H.; Wang, W.; Tong, B.; Zhang, S.P.; Wei, D.Z. A newly isolated Penicillium oxalicum 16 cellulase with high efficient synergism and high tolerance of monosaccharide. Appl. Biochem. Biotechnol. 2016, 178, 173–183. [Google Scholar] [CrossRef]
- Liu, P.; Lin, A.; Zhang, G.; Zhang, J.; Chen, Y.; Shen, T.; Zhao, J.; Wei, D.; Wang, W. Enhancement of cellulase production in Trichoderma reesei RUT-C30 by comparative genomic screening. Microb. Cell Fact. 2019, 18, 81. [Google Scholar] [CrossRef] [PubMed]
- Herpoël-Gimbert, I.; Margeot, A.; Dolla, A.; Jan, G.; Mollé, D.; Lignon, S.; Mathis, H.; Sigoillot, J.C.; Monot, F.; Asther, M. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 2008, 1, 18. [Google Scholar] [CrossRef]
- Marx, I.J.; Van Wyk, N.; Smit, S.; Jacobson, D.; Viljoen-Bloom, M.; Volschenk, H. Comparative secretome analysis of Trichoderma asperellum S4F8 and Trichoderma reesei Rut C30 during solid-state fermentation on sugarcane bagasse. Biotechnol. Biofuels 2013, 6, 172. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhang, L.; Wei, X.; Zou, G.; Qin, Y.; Ma, L.; Li, J.; Zheng, H.; Wang, S.; Wang, C.; et al. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 2013, 8, e55185. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Han, X.; Qian, Y.; Liu, G.; Yao, G.; Zhong, Y.; Qu, Y. Proteomic analysis of the biomass hydrolytic potentials of Penicillium oxalicum lignocellulolytic enzyme system. Biotechnol. Biofuels 2016, 9, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandels, M.; Reese, E.T. Induction of cellulase in fungi by cellobiose. J. Bacteriol. 1960, 79, 816. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L.; Blum, R.; Glennon, W.E.; Burton, A.L. Measurement of carboxymethylcellulase activity. Anal. Biochem. 1960, 1, 127–132. [Google Scholar] [CrossRef]
- Deshpande, M.V.; Eriksson, K.E.; Pettersson, L.G. An assay for selective determination of exo-1, 4,-β-glucanases in a mixture of cellulolytic enzymes. Anal. Biochem. 1984, 138, 481–487. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 2009; pp. 17–24. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.T.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Biochem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Cao, R.; He, Q.; Zhou, J.; He, Q.; Liu, Z.; Wang, X.; Chen, P.; Xie, J.; Liang, S. High-throughput analysis of rat liver plasma membrane proteome by a nonelectrophoretic in-gel tryptic digestion coupled with mass spectrometry identification. J. Proteome Res. 2008, 7, 535–545. [Google Scholar] [CrossRef]
- Wei, J.; Sun, J.; Yu, W.; Jones, A.; Oeller, P.; Keller, M.; Woodnutt, G.; Short, J.M. Global proteome discovery using an online three-dimensional LC-MS/MS. J. Proteome Res. 2005, 4, 801–808. [Google Scholar] [CrossRef]
- Brownell, H.H.; Saddler, J.N. Steam pretreatment of lignocellulosic material for enhanced enzymatic hydrolysis. Biotechnol. Bioeng. 1987, 29, 228–235. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Q.; Cheng, G. Deconstruction of corncob by steam explosion pretreatment: Correlations between sugar conversion and recalcitrant structures. Carbohydr. Polym. 2017, 156, 351–356. [Google Scholar] [CrossRef]
- Qi, W.; He, C.; Wang, Q.; Liu, S.; Yu, Q.; Wang, W.; Leksawasdi, N.; Wang, C.; Yuan, Z. Carbon-based solid acid pretreatment in corncob saccharification: Specific xylose production and efficient enzymatic hydrolysis. ACS Sustain. Chem. Eng. 2018, 6, 3640–3648. [Google Scholar] [CrossRef]
- Liao, H.; Li, S.; Wei, Z.; Shen, Q.; Xu, Y. Insights into high-efficiency lignocellulolytic enzyme production by Penicillium oxalicum GZ-2 induced by a complex substrate. Biotechnol. Biofuels 2014, 7, 162. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Li, J.; Zhao, S.; Zhang, R.; Wang, M.; Miao, Y.; Shen, Y.; Shen, Q. Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol. Biofuels 2013, 6, 149. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.T.; Wages, J.M. New-to-nature sophorose analog: A potent inducer for gene expression in Trichoderma reesei. Enzym. Microb. Technol. 2016, 85, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, X.; Zhang, X.; Bao, J. Inhibition performance of lignocellulose degradation products on industrial cellulase enzymes during cellulose hydrolysis. Appl. Biochem. Biotechnol. 2009, 159, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Driguez, H.; Viet, C.; Schülein, M. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Nat. Biotechnol. 1985, 3, 722–726. [Google Scholar] [CrossRef]
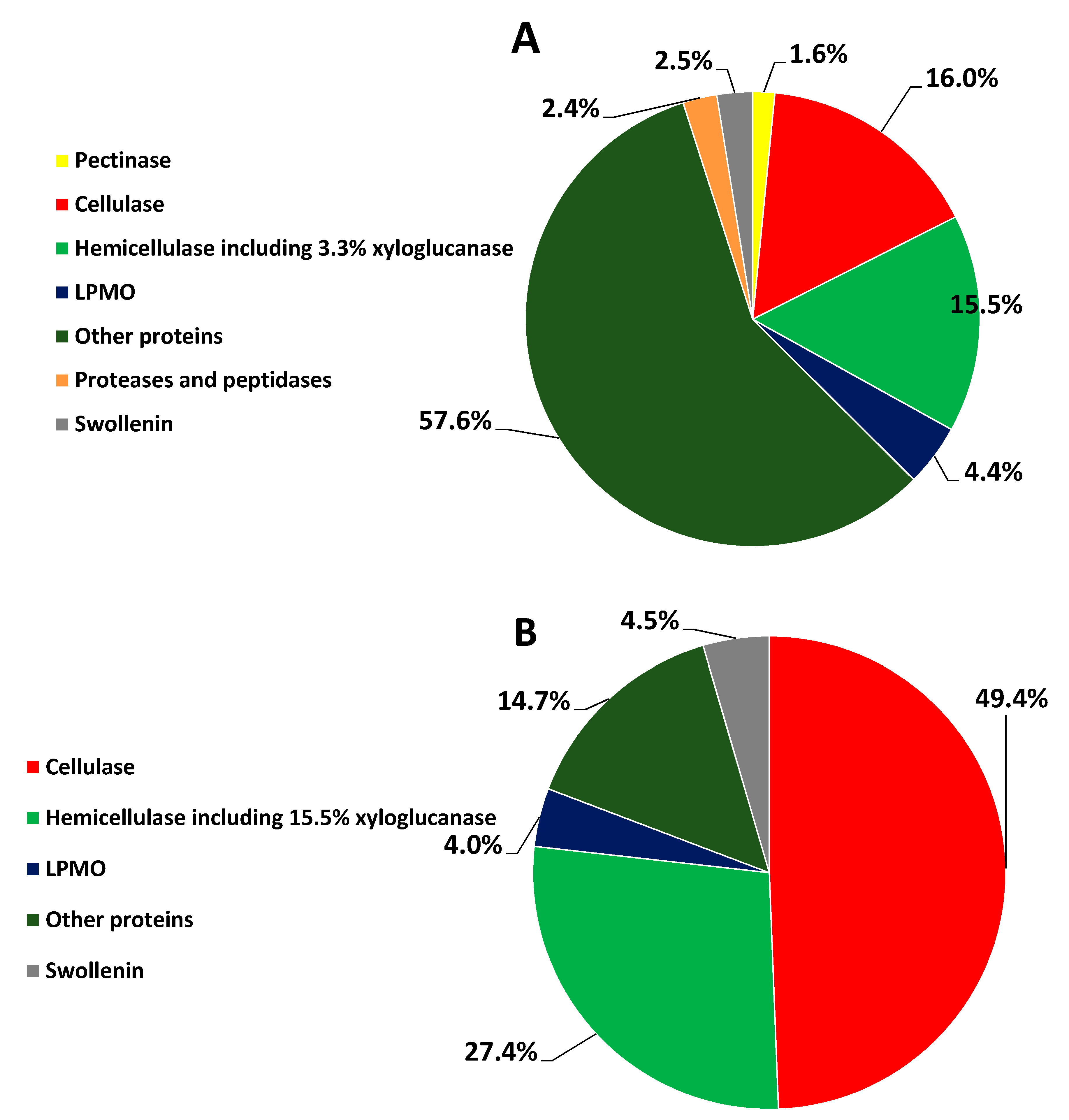
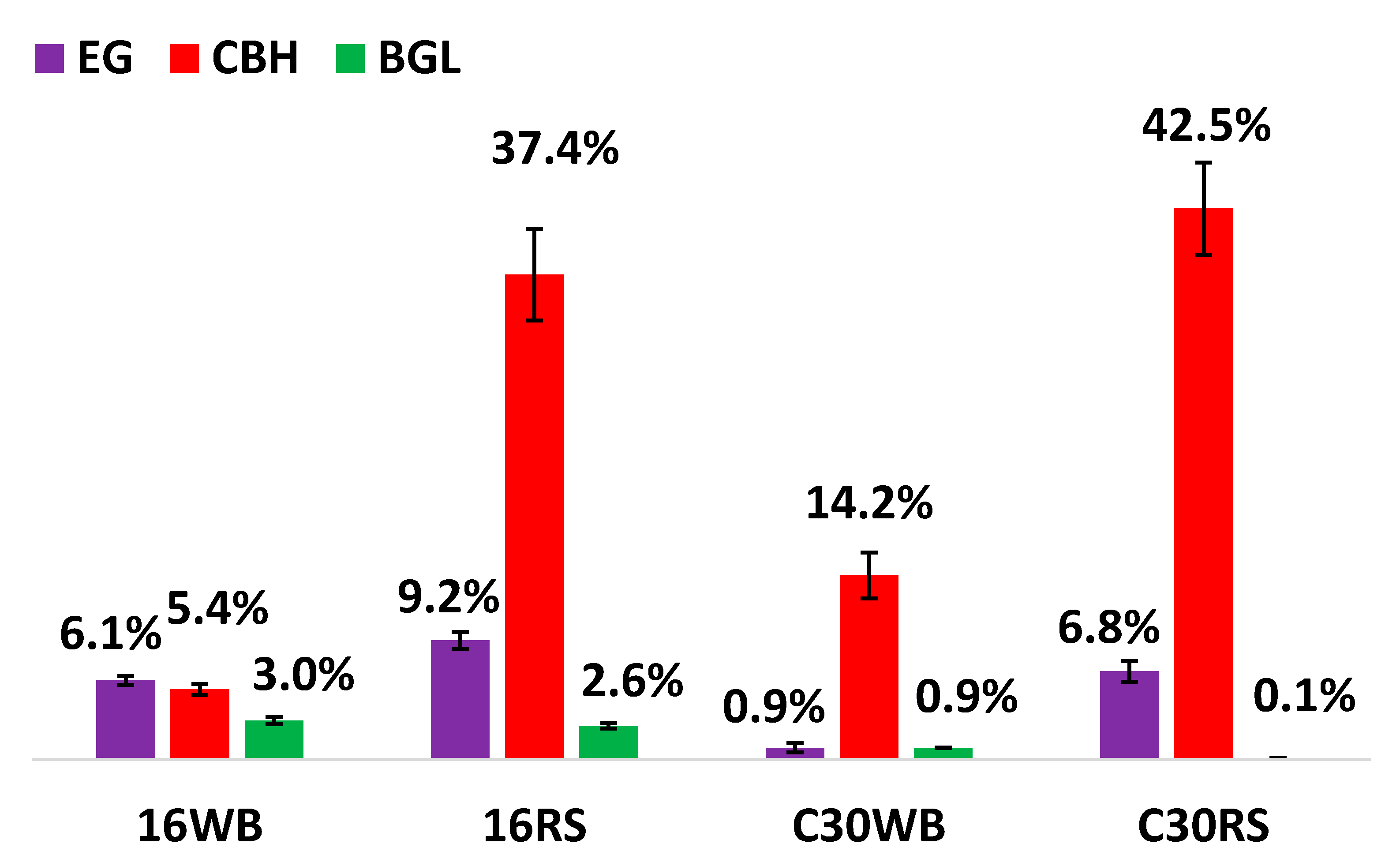
| Origins | EG (IU/gds) | CBH (IU/gds) | BGL (IU/gds) | Amylase (IU/gds) | Xylanase (IU/gds) |
|---|---|---|---|---|---|
| 16WB | 209 ± 2 | 0.02 ± 0.0 | 42 ± 1.3 | 998 ± 8 | 283 ± 2 |
| 16RS | 211 ± 2 | 0.31 ± 0.01 | 32.1 ± 1.2 | 373 ± 1 | 150 ± 3 |
| C30WB | 259 ± 1 | 0.09 ± 0.00 | 9.2 ± 1.0 | 24.8 ± 1.1 | 73 ± 0 |
| C30RS | 628 ± 4 | 0.41 ± 0.02 | 11.0 ± 0.8 | 10.2 ± 0.3 | 79 ± 6 |
| Origins | Pr-RS (μg/mL) | WB (μg/mL) | Pr-MCC (μg/mL) | Pr-CP (μg/mL) |
|---|---|---|---|---|
| 16WB | 1821 ± 72 | 10,571 ± 478 | 3475 ± 63 | 4101 ± 63 |
| 16RS | 3488 ± 81 | 6322 ± 441 | 4625 ± 370 | 8278 ± 171 |
| C30WB | 3209 ± 480 | 1948 ± 424 | 6107 ± 117 | 7978 ± 111 |
| C30RS | 3650 ± 153 | 1509 ± 43 | 7222 ± 73 | 8674 ± 189 |
| UniProt ID | Protein Description | Substrate | Classification | 16WB/16RS Ratio | Regulated Type |
|---|---|---|---|---|---|
| S8AHA8 | Putative β-xylosidase | xylooligosaccharide | GH3 | 168 ± 8 | Up |
| S8BFI1 | Putative β-mannosidase | β-mannose-oligosaccharide | GH2 | 71 ± 7 | Up |
| S7ZIW0 | Glucoamylase | dextrin | GH15 | 38 ± 1 | Up |
| S7Z5H6 | β-galactosidase | β-lactose | GH35 | 29 ± 1 | Up |
| S7ZA57 | β-xylanase | xylan | GH10 | 10 ± 0 | Up |
| S8B2R2 | Putative α-mannosidase | α-mannose-oligosaccharide | GH92 | 10 ± 1 | Up |
| S8AH74 | Putative endo-β-1,4-xylanase | xylan | GH30 | 10 ± 1 | Up |
| S7ZW00 | Putative α-L-arabinofuranosidase | arabinogalactan, arabinoglycan, etc. | GH62 | 9.4 ± 0.4 | Up |
| S8AXM3 | Putative endo-β-1,6-galactanase | β-1,6-galactan | GH30 | 9.2 ± 0.8 | Up |
| S7ZUD9 | Putative exo-β-1,3-galactanase | β-1,3-galactan | GH43 | 7.3 ± 0.6 | Up |
| S7ZBM4 | Arabinogalactan endo-β-1,4-galactanase | β-galactosidic linkages in type I arabinogalactans | GH53 | 7.1 ± 0.4 | Up |
| S8B8M7 | Putative α-L-arabinofuranosidase | arabinogalactan, arabinoglycan, etc. | GH43 | 6.6 ± 0.2 | Up |
| S7ZMB4 | Putative EG | cellulose | GH12 | 5.1 ± 0.1 | Up |
| S8B2H7 | Putative endo-β-1,4-xylanase | xylan | GH30 | 4.5 ± 0.3 | Up |
| S7ZJL3 | CBH I | MCC | GH7 | 3.0 ± 0.1 | Up |
| S8B0N0 | Putative β-glucuronidase | proteoglycan | GH2 | 2.8 ± 0.2 | Up |
| S7ZMU5 | Putative β-glucanase | β-1,3(4)-glucan | GH16 | 2.4 ± 0.1 | Up |
| S7ZFY8 | α-galactosidase | α-lactose | GH27 | 2.2 ± 0.1 | Up |
| S8AWH6 | Putative chitinase | chitin | GH18 | 1.7 ± 0.1 | Up |
| S7Z6T2 | α-amylase Amy13A | α-1,4-starch | GH13 | 1.7 ± 0.1 | Up |
| S7ZZQ8 | Putative α-L-rhamnopyranohydrolase | R-α-L-rhamnopyranoside | GH28 | 1.7 ± 0.1 | Up |
| S7ZD03 | Endo-polygalacturonase | pectin or pectinic acid | GH28 | 1.6 ± 0.1 | Up |
| S7Z4P2 | Putative α-L-arabinofuranosidase | arabinogalactan, arabinoglycan, etc. | GH43 | 1.6 ± 0.1 | Up |
| S7Z4H1 | α-1,2-mannosidase | α-1,2-mannose-oligosaccharide | GH47 | 1.6 ± 0.1 | Up |
| S7ZAV8 | β-xylanase | xylan | GH10 | 1.5 ± 0.0 | Up |
| S8B7P9 | Putative α-L-arabinofuranosidase | arabinogalactan, arabinoglycan, etc. | GH54 | 1.5 ± 0.1 | Up |
| S7ZWC7 | Putative exo-α-L-1,5-arabinanase | α-L-1,5-arabinoglycan | GH93 | 1.5 ± 0.1 | Up |
| S7ZDN1 | Putative endo-β-1,4-glucanase | cellulose | GH5 | 1.4 ± 0.1 | Up |
| S7ZCP1 | Putative β-1,3-1,4-glucanase | β-1,3-1,4-glucan | GH16 | 1.4 ± 0.1 | Up |
| S8AUX2 | Putative α-mannosidase | α-mannose-oligosaccharide | GH92 | 1.3 ± 0.1 | Up |
| S7ZR03 | Putative chitinase | chitin | GH18 | 0.76 ± 0.04 | Down |
| S8B6D7 | Glucoamylase | dextrin | GH15 | 0.70 ± 0.03 | Down |
| S8B0F3 | BGL | cellooligosaccharide | GH3 | 0.67 ± 0.03 | Down |
| S7Z8G1 | Putative chitinase | chitin | GH18 | 0.54 ± 0.03 | Down |
| S8AMF6 | Putative β-1,6-glucanase | β-1,6-glucan | GH30 | 0.53 ± 0.01 | Down |
| S8B6N1 | Putative chitinase | chitin | GH18 | 0.51 ± 0.01 | Down |
| S8AXN0 | Putative pectate lyase | pectinic acid | polysaccharide lyase 1 family | 0.43 ± 0.00 | Down |
| S7Z3I8 | Putative α-L-arabinofuranosidase | arabinogalactan, arabinoglycan, etc. | GH62 | 0.40 ± 0.02 | Down |
| S7ZPW1 | LPMO | polysaccharide | AA9 | 0.33 ± 0.00 | Down |
| S7ZAS9 | Putative endo-β-1,3-glucanase | β-1,3-glucan | Pectate lyase superfamily | 0.32 ± 0.00 | Down |
| S7ZAB6 | Putative swollenin | solid cellulose | Expansin_EG45 | 0.24 ± 0.01 | Down |
| S7ZP52 | CBH II | MCC | GH6 | 0.09 ± 0.00 | Down |
| S8B2B2 | EG1 | cellulose | GH7 | 0.09 ± 0.00 | Down |
| S7ZL65 | Putative β-1,4-mannanase | mannan | GH5 | 0.08 ± 0.00 | Down |
| S8AMN0 | Endo-1,4-β-xylanase | xylan | GH11 | 0.08 ± 0.00 | Down |
| S7ZRD6 | CBH I | MCC | GH7 | 0.08 ± 0.00 | Down |
| S8BGM3 | EG | cellulose | GH5 | 0.06 ± 0.00 | Down |
| S8BDN2 | β-xylanase | xylan | GH10 | 0.04 ± 0.00 | Down |
| S7ZX22 | EG | cellulose | GH5 | 0.04 ± 0.00 | Down |
| S8AIJ2 | EG | cellulose | GH5 | 0.02 ± 0.00 | Down |
| UniProt ID | Protein Description | Substrate | Classification | C30WB/C30RS Ratio | Regulated Type |
|---|---|---|---|---|---|
| A0A024SGF7 | α-galactosidase | α-lactose | GH27 | 12 ± 0 | Up |
| A0A024S1T5 | Chitinase | chitin | GH18 | 7.7 ± 0.7 | Up |
| A0A024SDM6 | β-xylosidase | xylooligosaccharide | GH3 | 5.9 ± 0.4 | Up |
| A0A024RWW9 | xylanase | xylan | GH30 | 5.8 ± 0.3 | Up |
| A0A024SAF4 | β-1,3-endoglucanase | β-1,3-glucan | GH17 | 4.0 ± 0.1 | Up |
| A0A024S166 | α-glucuronidase | xylan | GH67 | 3.3 ± 0.1 | Up |
| A0A024S732 | β-glucanase | β-1,3(4)-glucan | GH16 | 2.5 ± 0.2 | Up |
| A0A024S1W9 | β-1,3-endoglucanase | β-1,3-glucan | GH17 | 2.3 ± 0.1 | Up |
| A0A024S0A7 | β-1,4-endoxylanase | xylan | GH43 | 1.9 ± 0.1 | Up |
| A0A024S2Y7 | α-N-arabinofuranosidase | α-L-arabinoside | GH54 | 1.9 ± 0.1 | Up |
| A0A024S1V1 | Endopolygalacturonase | pectin, pectinic acid | GH28 | 1.8 ± 0.1 | Up |
| A0A024RUF8 | β-mannosidase A | β-mannose-oligosaccharide | GH2 | 1.5 ± 0.1 | Up |
| A0A024S0G1 | Endo-β-1,6-galactanase | β-1,6-galactan | GH30 | 1.2 ± 0.1 | Up |
| A0A024SIJ3 | β-mannase (Fragment) | mannan | GH5 | 0.62 ± 0.02 | Down |
| A0A024RZP7 | Swollenin | solid cellulose | Expansin_EG45 | 0.50 ± 0.03 | Down |
| A0A024SNB7 | EG | cellulose | GH7 | 0.49 ± 0.02 | Down |
| P36217 | Endo-1,4-β-xylanase 2 | xylan | GH11 | 0.46 ± 0.01 | Down |
| A0A024RXP8 | CBH I | MCC | GH7 | 0.33 ± 0.01 | Down |
| A0A024S0K1 | Chitnase | chitin | GH18 | 0.33 ± 0.02 | Down |
| A0A024SGE7 | α-L-arabinofuranosidase | α-L-arabinoside | GH62 | 0.30 ± 0.00 | Down |
| A0A024SH76 | CBH II | MCC | GH6 | 0.26 ± 0.01 | Down |
| A0A024S9Z6 | Xyloglucanase | xyloglucan | GH74 | 0.22 ± 0.00 | Down |
| A0A024SFJ2 | LPMO | polysaccharide | AA9 | 0.21 ± 0.00 | Down |
| A0A024RV01 | β-1,4-endoxylanase | xylan | GH30 | 0.16 ± 0.01 | Down |
| A0A024SH20 | EG | cellulose | GH5 | 0.15 ± 0.00 | Down |
| A0A024SCX9 | BGL | cellooligosaccharide | GH3 | 0.10 ± 0.00 | Down |
| A0A024SIB3 | Endo-1,4-β-xylanase 3 | xylan | GH10 | 0.09 ± 0.00 | Down |
| A0A024S2H5 | EG | cellulose | GH12 | 0.05 ± 0.00 | Down |
| Origins | UniProt ID | Description | Relative Abundance (%) | Substrate | Classification |
|---|---|---|---|---|---|
| 16WB | S7ZRD6 | CBH I | 3.7 ± 0.2 | MCC | GH7 |
| S7ZP52 | CBH II | 1.5 ± 0.0 | MCC | GH6 | |
| S7ZA57 | β-xylanase | 6.5 ± 0.3 | xylan | GH10 | |
| S8AH74 | endo-β-1,4-xylanase | 1.1 ± 0.0 | xylan | GH30 | |
| S8B6D7 | Glucoamylase | 5.7 ± 0.3 | dextrin | GH15 | |
| S7Z6T2 | α-amylase my13A | 1.8 ± 0.1 | starch | GH13 | |
| S7ZDN1 | Endo-β-1,4-mannanase F | 1.7 ± 0.0 | mannan | GH5 | |
| S7ZMB4 | EG | 1.5 ± 0.0 | cellulose | GH12 | |
| S8B0F3 | BGL | 3.0 ± 0.1 | cellooligosaccharide | GH3 | |
| S7ZPW1 | LPMO | 3.2 ± 0.1 | polysaccharide | AA9 | |
| S7ZAB6 | Swollenin | 0.6 ± 0.0 | solid cellulose | Expansin_EG45 | |
| 16RS | S7ZRD6 | CBH I | 26 ± 3 | MCC | GH7 |
| S7ZP52 | CBH II | 9.6 ± 0.6 | MCC | GH6 | |
| S8BDN2 | β-xylanase | 14 ± 1 | xylan | GH10 | |
| S8B6D7 | Glucoamylase | 5.2 ± 0.3 | dextrin | GH15 | |
| S7Z6T2 | α-amylase my13A | 0.7 ± 0.0 | starch | GH13 | |
| S8BGM3 | EG | 3.7 ± 0.1 | cellulose | GH5 | |
| S7ZX22 | EG | 3.3 ± 0.1 | cellulose | GH5 | |
| S8AIJ2 | EG | 1.2 ± 0.0 | cellulose | GH5 | |
| S8B0F3 | BGL | 2.5 ± 0.1 | cellooligosaccharide | GH3 | |
| S7Z716 | LPMO | 1.2 ± 0.0 | polysaccharide | AA9 | |
| S7ZPW1 | LPMO | 5.6 ± 0.2 | polysaccharide | AA9 | |
| S7ZAB6 | Swollenin | 1.3 ± 0.1 | solid cellulose | Expansin_EG45 | |
| C30WB | A0A024RXP8 | CBH I | 7.8 ± 0.4 | MCC | GH7 |
| A0A024SH76 | CBH II | 6.2 ± 0.3 | MCC | GH6 | |
| A0A024S9Z6 | Xyloglucanase | 3.3 ± 0.0 | xyloglucan | GH74 | |
| A0A024RWA5 | BGL | 0.5 ± 0.2 | cellooligosaccharide | GH3 | |
| A0A024SM10 | LPMO | 3.4 ± 0.2 | polysaccharide | AA9 | |
| A0A024SFJ2 | LPMO | 1.0 ± 0.1 | polysaccharide | AA9 | |
| A0A024RZP7 | Swollenin | 2.5 ± 0.1 | solid cellulose | Expansin_EG45 | |
| C30RS | A0A024RXP8 | CBH I | 21 ± 2 | MCC | GH7 |
| A0A024SH76 | CBH II | 21± 1 | MCC | GH6 | |
| A0A024S9Z6 | Xyloglucanase | 16 ± 1 | xyloglucan | GH74 | |
| A0A024SH20 | EG | 5.5 ± 0.4 | cellulose | GH5 | |
| A0A024S2H5 | EG | 1.2 ± 0.0 | cellulose | GH12 | |
| A0A024SFJ2 | LPMO | 4.0 ± 0.1 | polysaccharide | AA9 | |
| A0A024RZP7 | Swollenin | 4.5 ± 0.2 | solid cellulose | Expansin_EG45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Zhang, N.; Pearce, R.; Yi, S.; Zhao, X. Comparative Secretomics Analysis Reveals the Major Components of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30. Microorganisms 2021, 9, 2042. https://doi.org/10.3390/microorganisms9102042
Wang K, Zhang N, Pearce R, Yi S, Zhao X. Comparative Secretomics Analysis Reveals the Major Components of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30. Microorganisms. 2021; 9(10):2042. https://doi.org/10.3390/microorganisms9102042
Chicago/Turabian StyleWang, Kexin, Nian Zhang, Robin Pearce, Shi Yi, and Xihua Zhao. 2021. "Comparative Secretomics Analysis Reveals the Major Components of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30" Microorganisms 9, no. 10: 2042. https://doi.org/10.3390/microorganisms9102042
APA StyleWang, K., Zhang, N., Pearce, R., Yi, S., & Zhao, X. (2021). Comparative Secretomics Analysis Reveals the Major Components of Penicillium oxalicum 16 and Trichoderma reesei RUT-C30. Microorganisms, 9(10), 2042. https://doi.org/10.3390/microorganisms9102042





