Coastal Bacterial Community Response to Glacier Melting in the Western Antarctic Peninsula
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection along the Salinity Transect
2.2. Dilution Experiments and Experimental Setup
2.3. Microbial Biomass Concentration and Nucleic Acid Extractions
2.4. 16S rRNA Gene Amplification, Sequencing and Taxonomic Annotation
2.5. Metatranscriptomic Taxonomy and Metabolic Annotation Analyses
2.5.1. Marine Microbial Protein Database Construction
2.5.2. Metatranscriptomic Transcriptional Level Analysis
3. Results
3.1. Environmental Conditions
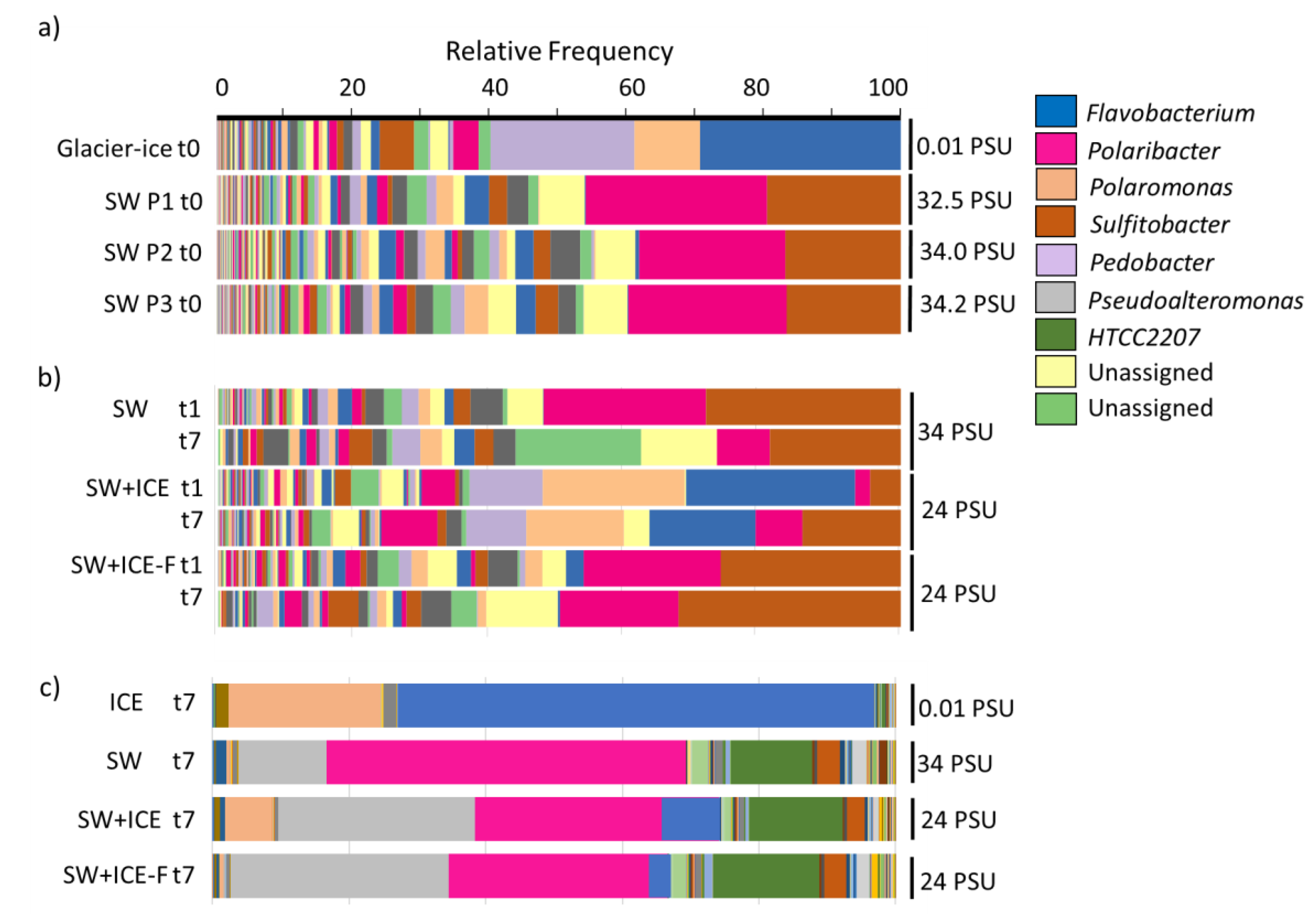
3.2. Changes in Bacterial Composition
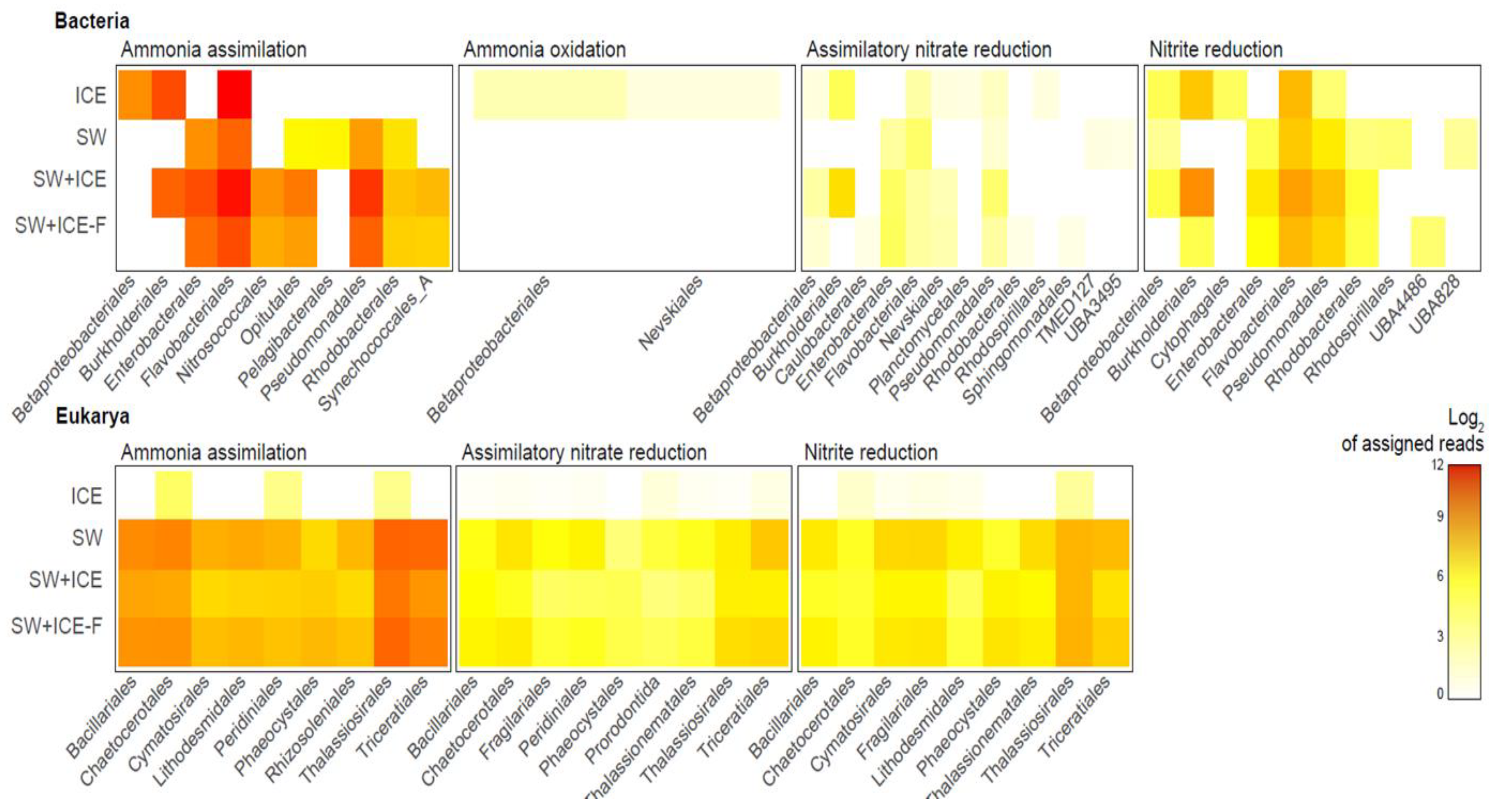
3.3. Active Response of Microbial Assimilatory Carbon and Nitrogen Pathways to Freshening
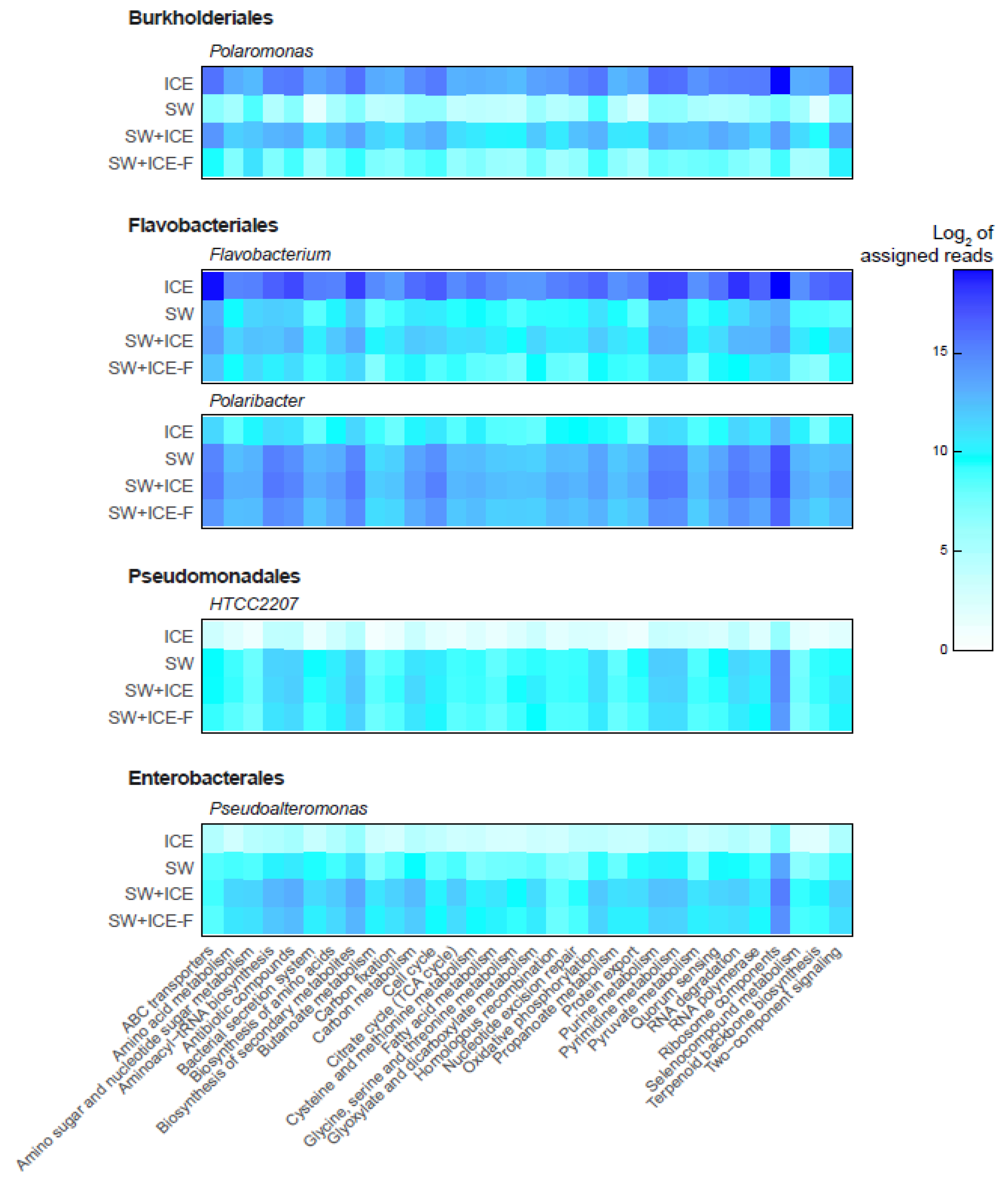
3.4. Active Microbial Sentinels in Response to Freshening
4. Discussion
4.1. Chile Bay: A Coastal WAP Scenario to Test Microbial Community Response to Freshening
4.2. Bacterial Sentinels in Response to Manipulated Freshening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steig, E.J.; Schneider, D.P.; Rutherford, S.D.; Mann, M.E.; Comiso, J.C.; Shindell, D.T. Warming of the Antarctic ice-sheet surface since the 1957 International Geophysical Year. Nature 2009, 457, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Hoegh-Guldberg, O.; Jacob, D.; Taylor, M.; Bindi, M.; Brown, S.; Camilloni, I.; Diedhiou, A.; Djalante, R.; Ebi, K.L.; Engelbrecht, F.; et al. Impacts of 1.5 °C Global Warming on Natural and Human Systems. In Global Warming of 1.5 °C. An IPCC Special Report on the Impacts of Global Warming of 1.5 °C above Pre-Industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty; Masson-Delmotte, V., Zhai, P., Pörtner, H.O., Roberts, D., Skea, J., Shukla, P.R., Pirani, A., Moufouma-Okia, W., Péan, C., Pidcock, R., et al., Eds.; 2018; Available online: https://www.ipcc.ch/sr15/chapter/chapter-3/ (accessed on 25 December 2020).
- Turner, J.; Colwell, S.R.; Marshall, G.J.; Lachlan-Cope, T.A.; Carleton, A.M.; Jones, P.D.; Lagun, V.; Reid, P.A.; Iagovinka, S. Antarctic climate change during the last 50 years. Int. J. Climatol. 2005, 25, 279–294. [Google Scholar] [CrossRef]
- Meredith, M.P.; King, J.C. Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- DeConto, R.M.; Pollard, D. Contribution of Antarctica to past and future sea-level rise. Nature 2006, 531, 591–597. [Google Scholar] [CrossRef]
- Turner, J.; Barrand, N.E.; Bracegirdle, T.J.; Convey, P.; Hodgson, D.A.; Jarvis, M.; Jenkins, A.; Marshall, G.; Meredith, M.P.; Roscoe, H.; et al. Antarctic climate change and the environment: An update. Polar Record. 2014, 50, 237–259. [Google Scholar] [CrossRef]
- Turner, J.; Phillips, T.; Marshall, G.J.; Hosking, J.S.; Pope, J.O.; Bracegirdle, T.J.; Deb, P. Unprecedented springtime retreat of Antarctic sea ice in 2016. Geophys. Res. Lett. 2017, 44, 6868–6875. [Google Scholar] [CrossRef]
- Steinberg, D.K.; Ruck, K.E.; Gleiber, M.R.; Garzio, L.M.; Cope, J.S.; Bernard, K.S.; Stammerjohn, S.E.; Schofield, O.M.; Quetin, L.B.; Ross, R.M. Long-term (1993–2013) changes in macrozooplankton off the Western Antarctic Peninsula. Deep Sea Res. Part I Oceanogr. Res. Pap. 2015, 101, 54–70. [Google Scholar] [CrossRef]
- Piñones, A.; Fedorov, A. Projected changes of Antarctic krill habitat by the end of the 21st century. Geophys. Res. Lett. 2016, 43, 8580–8589. [Google Scholar] [CrossRef]
- Schloss, I.R.; Abele, D.; Moreau, S.; Demers, S.; Bers, A.V.; González, O.; Ferreyra, G.A. Response of phytoplankton dynamics to 19-year (1991–2009) climate trends in Potter Cove (Antarctica). J. Mar. Syst. 2012, 92, 53–66. [Google Scholar] [CrossRef]
- Bers, A.V.; Momo, F.; Schloss, I.R.; Abele, D. Analysis of trends and sudden changes in long-term environmental data from King George Island (Antarctica): Relationships between global climatic oscillations and local system response. Clim. Chang. 2013, 116, 789–803. [Google Scholar] [CrossRef][Green Version]
- Dierssen, H.M.; Smith, R.C.; Vernet, M. Glacial meltwater dynamics in coastal waters west of the Antarctic peninsula. Proc. Natl. Acad. Sci. USA 2002, 99, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Vernet, M.; Martinson, D.; Iannuzzi, R.; Stammerjohn, S.; Kozlowski, W.; Sines, K.; Smith, R.; Garibotti, I. Primary production within the sea-ice zone west of the Antarctic Peninsula: I—Sea ice, summer mixed layer, and irradiance. Deep Sea Res. Part II 2008, 55, 2068–2085. [Google Scholar] [CrossRef]
- Gonçalves-Araujo, R.; Silva de Souza, M.; Tavano, V.M.; Garcia, C.A. Influence of oceanographic features on spatial and interannual variability of phytoplankton in the Bransfeld Strait, Antarctica. J. Mar. Syst. 2015, 142, 1–15. [Google Scholar] [CrossRef]
- Hopwood, M.J.; Carroll, D.; Browning, T.J.; Meire, L.; Mortensen, J.; Krisch, S.; Achterberg, E.P. Non-linear response of summertime marine productivity to increased meltwater discharge around Greenland. Nat. Commun. 2018, 9, 3256. [Google Scholar] [CrossRef]
- Borges-Mendes, C.R.; Silva de Souza, M.; Garcia, V.M. Dynamics of phytoplankton communities during late summer around the tip of the Antarctic Peninsula. Deep Sea Res. Part I Oceanogr. Res. Pap. 2012, 65, 1–14. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Vergin, K.L. Seasonality in Ocean Microbial Communities. Science 2012, 335, 671–676. [Google Scholar] [CrossRef]
- Kang, S.-H.; Jae-Shin, K.; Sanghoon, L.; Kyung, H.; Dongseon, K.; Myung, G.P. Antarctic Phytoplankton Assemblages in the Marginal Ice Zone of the Northwestern Weddell Sea. J. Plankton Res. 2001, 23, 333–352. [Google Scholar] [CrossRef]
- Chown, S.L.; Huiskes, A.; Gremmen, N.; Lee, T.A.; Crosbie, K.; Frenot, Y.; Hughes, K.; Imura, S.; Kiefer, K.; Lebouvier, M.; et al. Continent-wide risk assessment for the establishment of nonindigenous species in Antarctica. Proc. Natl. Acad. Sci. USA 2012, 109, 4938–4943. [Google Scholar] [CrossRef]
- Ghiglione, J.F.; Murray, A.E. Pronounced summer to winter differences and higher wintertime richness in coastal Antarctic marine bacterioplankton. Environ. Microbiol. 2012, 14, 617–629. [Google Scholar] [CrossRef]
- Grzymski, J.; Riesenfeld, C.; Williams, T.; Dussaq, A.; Ducklow, H.; Erickson, M.; Cavicchioli, R.; Murray, A.E. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012, 6, 1901–1915. [Google Scholar] [CrossRef]
- Giudice, A.L.; Caruso, C.; Mangano, S.; Bruni, V.; De Domenico, M.; Michaud, L. Marine Bacterioplankton Diversity and Community Composition in an Antarctic Coastal Environment. Microb. Ecol. 2012, 63, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yu, Y.; Qiao, Z.; Jin, H.Y.; Li, H.R. Diversity of bacterioplankton in coastal seawaters of Fildes Peninsula, King George Island, Antarctica. Arch. Microbiol. 2012, 196, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Alcamán-Arias, M.E.; Farıas, L.; Verdugo, J.; Alarcon-Schumacher, T.; Díez, B. Microbial activity during a coastal phytoplankton bloom on the Western Antarctic Peninsula in late summer. FEMS Microbiol. Lett. 2018, 365, fny090. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Arroyo, J.; Rodríguez Marconi, S.; Masotti, I.; Alarcón Schumacher, T.; Polz, M.; Trefault, N.; De la Iglesia, R.; Díez, B. Summer phyto and bacterioplankton communities during low and high productivity scenarios in the Western Antarctic Peninsula. Polar Biol. 2019, 42, 159–169. [Google Scholar] [CrossRef]
- Ducklow, H.; Fraser, W.; Meredith, M.; Stammerjohn, S.; Doney, S.; Martinson, D.; Sailley, S.F.; Schofield, O.M.; Steinberg, D.K.; Venables, H.J.; et al. West Antarctic Peninsula: An ice-dependent coastal marine ecosystem in transition. Oceanography 2013, 26, 190–203. [Google Scholar] [CrossRef]
- Aracena, C.; González, H.E.; Garces-Vargas, J.; Lange, C.B.; Pantoja, S.; Muñoz, F.; Teca, E.; Tejos, E. Influence of summer conditions on surface water properties and phytoplankton productivity in embayments of the South Shetland Islands. Polar Biol. 2018, 41, 2135–2155. [Google Scholar] [CrossRef]
- Manganelli, M.; Malfatti, F.; Samo, T.J.; Mitchell, B.G.; Wang, H.; Azam, F. Major Role of Microbes in Carbon Fluxes during Austral Winter in the Southern Drake Passage. PLoS ONE 2009, 4, e6941. [Google Scholar] [CrossRef]
- Williams, T.J.; Long, E.; Evans, F.; DeMaere, M.Z.; Lauro, F.M.; Raftery, M.J.; Ducklow, H.; Grzymski, J.J.; Murray, A.E.; Cavicchioli, R. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012, 6, 1883–1900. [Google Scholar] [CrossRef]
- Piquet, A.M.; Bolhuis, H.; Meredith, M.P.; Buma, A.G.J. Shifts in coastal Antarctic marine microbial communities during and after meltwater-related surface stratification. FEMS Microbiol. Ecol. 2011, 76, 413–427. [Google Scholar] [CrossRef][Green Version]
- Hofer, J.; Giesecke, R.; Hopwood, M.J.; Carrera, V.; Alarcón, E.; González, H.E. The role of water column stability and wind mixing in the production/export dynamics of two bays in the Western Antarctic Peninsula. Prog. Oceanogr. 2019, 174, 105–116. [Google Scholar] [CrossRef]
- Montes-Hugo, M.A.; Vernet, M.; Martinson, D.; Smith, R.; Iannuzzi, R. Variability on phytoplankton size structure in the western Antarctic Peninsula (1997–2006). Deep Sea Res. II Top. Stud. Oceanogr. 2008, 55, 2106–2117. [Google Scholar] [CrossRef]
- Montes-Hugo, M.; Martinson, D.; Stammerjohn, S.E.; Schofield, O. Recent changes in phytoplankton western Antarctic Peninsula. Science 2009, 323, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Grasshoff, K.; Kremling, K.; Ehrhardt, M. (Eds.) Methods of Seawater Analysis; Wiley-VCH: Weinheim, Germany, 1999; p. 600. [Google Scholar]
- Strickland, J.D.H.; Parsons, T.R. A Practical Handbook of Seawater Analysis, 2nd ed.; No. 167; Fisheries Research Board of Canada: Ottawa, ON, Canada, 1972; p. 310. [Google Scholar]
- Walters, W.; Hyde, E.R.; Lyons, D.B.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2015, 1, e00009–e00015. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.; McMurdie, P.; Rosen, M.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 1–10. [Google Scholar] [CrossRef]
- Kopylova, E.; Noe, L. Sequence analysis SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Tully, B.J.; Graham, E.D.; Heidelberg, J.F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 2018, 5, 170203. [Google Scholar] [CrossRef]
- Pierre-Alain, C.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 6. [Google Scholar]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B. Preprocesscore: A Collection of Pre-Processing Functions. R Package Version 1.50.0. 2020. Available online: https://github.com/bmbolstad/preprocessCore (accessed on 23 December 2020).
- Prestat, E.; David, M.M.; Hultman, J.; Taş, N.; Lamendella, R.; Dvornik, J.; Mackelprang, R.; Myrold, D.D.; Jumpponen, A.; Tringe, S.G.; et al. FOAM (Functional Ontology Assignments for Metagenomes): A Hidden Markov Model (HMM) database with environmental focus. Nucleic Acids Res 2014, 42, e14. [Google Scholar] [CrossRef]
- Wessel, P.; Luis, J.F. The GMT/MATLAB Toolbox. Geochem. Geophys. Geosyst. 2017, 18, 811–823. [Google Scholar] [CrossRef]
- Vaughan, D.G.; Marshall, G.J.; Connolley, W.M.; Parkinson, C.; Mulvaney, R.; Hodgson, D.A.; King, J.C.; Pudsey, C.J.; Turner, J. Recent Rapid Regional Climate Warming on the Antarctic Peninsula. Clim. Chang. 2003, 60, 243–274. [Google Scholar] [CrossRef]
- Mitchell, B.G.; Holm-Hansen, O. Observations of modeling of the Antartic phytoplankton crop in relation to mixing depth. Deep Sea Res. 1991, 38, 961–1007. [Google Scholar] [CrossRef]
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.; Amann, R.; Helmke, E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ducklow, H.W.; Abele, D.; Ruiz, E.; Buma, A.; Meredoth, M.M.; Rozema, P.D.; Schofield, O.M.; Venables, H.; Schloss, I. Correction to ‘Inter-decadal variability of phytoplankton biomass along the coastal West Antarctic Peninsula’. Philos. Trans. R. Soc. A 2018, 376, 20180170. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; He, J.; Zhang, F.; Lin, L.; Gao, Y.; Zhou, Q. Diversity and community structure of bacterioplankton in surface waters off the northern tip of the Antarctic Peninsula. Polar Res. 2019, 38, 3491. [Google Scholar] [CrossRef]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico Beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Math, R.K.; Jin, H.M.; Kim, J.M.; Hahn, Y.; Park, W.; Madsen, E.L.; Jeon, C.O. Comparative genomics reveals adaptation by Alteromonas sp. SN2 to marine tidal-flat conditions: Cold tolerance and aromatic hydrocarbon metabolism. PLoS ONE 2012, 7, e35784. [Google Scholar] [CrossRef] [PubMed]
- Mas-Lladó, M.; Piña-Villalonga, J.M.; Brunet-Galmés, I.; Nogales, B.; Bosch, R. Draft genome sequences of two isolates of the Roseobacter group, Sulfitobacter sp. strains 3SOLIMAR09 and 1FIGIMAR09, from har¬bors of Mallorca Island (Mediterranean Sea). Genome Announc. 2014, 2, e00350-14. [Google Scholar] [CrossRef]
- Williams, T.J.; Wilkins, D.; Long, E.; Evans, F.; DeMaere, M.Z.; Raftery, M.J.; Cavicchioli, R. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 2013, 15, 1302–1317. [Google Scholar] [CrossRef]
- Garcia-Lopez, E.; Rodriguez-Lorente, I.; Alcazar, P.; Cid, C. Microbial Communities in Coastal Glaciers and Tidewater Tongues of Svalbard Archipelago, Norway. Front. Mar. Sci. 2019, 5, 512. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Si, J.; Wu, G.; Tian, L.; Xiang, S. Changes of the Bacterial Abundance and Communities in Shallow Ice Cores from Dunde and Muztagata Glaciers, Western China. Front. Microbiol. 2016, 7, 1716. [Google Scholar] [CrossRef]
- Paun, V.I.; Icaza, G.; Lavin, P.; Marin, C.; Tudorache, A.; Perşoiu, A.; Dorador, C.; Purcarea, C. Total and Potentially Active Bacterial Communities Entrapped in a Late Glacial Through Holocene Ice Core From Scarisoara Ice Cave, Romania. Front. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- McCammon, S.A.; Bowman, J.P. Taxonomy of Antarctic Flavobacterium species: Description of Flavobacterium gillisiae sp. nov., Flavobacterium tegetincola sp. nov., and Flavobacterium xanthum sp. nov., nom. rev. and reclassification of [Flavobacterium] salegens as Salegentibacter salegens gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2020, 50, 1055–1063. [Google Scholar] [CrossRef]
- Clarke, A.; Murphy, E.J.; Meredith, M.P.; King, J.C.; Peck, L.S.; Barnes, D.; Smith, R.C. Climate change and the marine ecosystem of the western Antarctic Peninsula. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, V.; Alurralde, G.; Meyer, B.; Aguirre, G.; Canepa, A.; Wölfl, A.C.; Hass, H.C.; Williams, G.; Scholss, I. Glacial melting: An overlooked threat to Antarctic krill. Sci. Rep. 2016, 6, 27234. [Google Scholar] [CrossRef]
- Schneider, W.; Pérez-Santos, I.; Ross, L.; Bravo, L.; Seguel, R.; Hernández, F. On the hydrography of Puyuhuapi Channel, Chilean Patagonia. Prog. Oceanogr. 2014, 128, 8–18. [Google Scholar] [CrossRef]
- Gutierrez, M.H.; Galand, P.E.; Moffat, C.; Pantoja, S. Melting glacier impacts community structure of Bcateria, Archaea and Fungi in a Chilean Patagonia fjord. Environ. Mirobiol. 2015. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Kukhlevskiy, A.D.; Zhukova, N.V. Polaribacter reichenbachii sp. nov.: A new marine bacterium associated with the green alga Ulva fenestrata. Curr. Microbiol. 2013, 66, 16–21. [Google Scholar] [CrossRef]
- Boos, W.; Eppler, T.; Winkelmann, G. (Eds.) Prokaryotic binding protein-dependent ABC transporters. In Microbial Transport Systems; Wiley-VCH: New York, NY, USA, 2001; pp. 77–114. [Google Scholar]
- Jiao, N.Z.; Zheng, Q. The microbial carbon pump: From genes to ecosystems. Appl. Environ. Microbiol. 2011, 77, 7439–7444. [Google Scholar] [CrossRef]
- Luria, C.M.; Amaral-Zettler, L.A.; Ducklow, H.W.; Repeta, D.J.; Rhyne, A.L.; Rich, J.J. Seasonal Shifts in Bacterial Community Responses to Phytoplankton-Derived Dissolved Organic Matter in the Western Antarctic Peninsula. Front. Microbiol. 2019. [Google Scholar] [CrossRef]
- Kameyama, S.; Otomaru, M. Ice melting can change DMSP production and photosynthetic activity of the Haptophyte Phaeocystis antarctica. J. Phycol. 2020, 56, 761–774. [Google Scholar] [CrossRef]
- Kennedy, F.; McMinn, A.; Martin, A. Effect of temperature on the photosynthetic efficiency and morphotype of Phaeocystis antarctica. J. Exp. Mar. Biol. Ecol. 2012, 429, 7–14. [Google Scholar] [CrossRef]
- Kropuenske, L.R.; Mills, M.M.; van Dijken, G.L.; Bailey, S.R.; Welschmeyer, D.H.; Arrigoa, N.; Kevin, R. Photophysiology in two major Southern Ocean phytoplankton taxa: Photoprotection in Phaeocystis antarctica and Fragilariopsis cylindrus. Limnol. Oceanogr. 2019, 54. [Google Scholar] [CrossRef]
- Alderkamp, A.C.; Mills, M.M.; van Dijken, G.L.; Arrigo, K.R. Photoacclimation and non-photochemical quenching under in situ irradiance in natural phytoplankton assemblages from the Amundsen Sea, Antarctica. Mar. Ecol. Prog. Ser. 2013, 475, 15–34. [Google Scholar] [CrossRef]
- Wietz, M.; Gram, L.; Jørgensen, B.; Schramm, A. Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 2010, 61, 179–189. [Google Scholar] [CrossRef]
- Fontanez, K.M.; Eppley, J.M.; Samo, T.J.; Karl, D.M.; DeLong, E.F. Microbial community structure and function on sinking particles in the north Pacific subtropical gyre. Front. Microbiol. 2015, 6, 469. [Google Scholar] [CrossRef]
- Holmström, C.; Kjelleberg, S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 1999, 30, 285–293. [Google Scholar] [CrossRef]
- Nichols, C.M.; Bowman, J.P.; Guezennec, J. Effects of incubation temperature on growth and production of exopolysaccharides by an Antarctic sea-ice bacterium grown in batch culture. Appl. Environ. Microbiol. 2005, 71, 3519–3523. [Google Scholar] [CrossRef]
- Collins, R.E.; Carpenter, S.D.; Deming, J.W. Spatial heterogeneity and temporal dynamics of particles, bacteria, and pEPS in Arctic winter sea ice. J. Mar. Syst. 2008, 74, 902–917. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcamán-Arias, M.E.; Fuentes-Alburquenque, S.; Vergara-Barros, P.; Cifuentes-Anticevic, J.; Verdugo, J.; Polz, M.; Farías, L.; Pedrós-Alió, C.; Díez, B. Coastal Bacterial Community Response to Glacier Melting in the Western Antarctic Peninsula. Microorganisms 2021, 9, 88. https://doi.org/10.3390/microorganisms9010088
Alcamán-Arias ME, Fuentes-Alburquenque S, Vergara-Barros P, Cifuentes-Anticevic J, Verdugo J, Polz M, Farías L, Pedrós-Alió C, Díez B. Coastal Bacterial Community Response to Glacier Melting in the Western Antarctic Peninsula. Microorganisms. 2021; 9(1):88. https://doi.org/10.3390/microorganisms9010088
Chicago/Turabian StyleAlcamán-Arias, María Estrella, Sebastián Fuentes-Alburquenque, Pablo Vergara-Barros, Jerónimo Cifuentes-Anticevic, Josefa Verdugo, Martin Polz, Laura Farías, Carlos Pedrós-Alió, and Beatriz Díez. 2021. "Coastal Bacterial Community Response to Glacier Melting in the Western Antarctic Peninsula" Microorganisms 9, no. 1: 88. https://doi.org/10.3390/microorganisms9010088
APA StyleAlcamán-Arias, M. E., Fuentes-Alburquenque, S., Vergara-Barros, P., Cifuentes-Anticevic, J., Verdugo, J., Polz, M., Farías, L., Pedrós-Alió, C., & Díez, B. (2021). Coastal Bacterial Community Response to Glacier Melting in the Western Antarctic Peninsula. Microorganisms, 9(1), 88. https://doi.org/10.3390/microorganisms9010088





