Yeast Biodiversity in Fermented Doughs and Raw Cereal Matrices and the Study of Technological Traits of Selected Strains Isolated in Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Fermented and Raw Matrices Prospected for Yeast Isolation
2.1.1. Type I Mother Doughs
2.1.2. Leavened Doughs from Bakeries
2.1.3. Cereal Grains and Flours
2.2. Yeast Isolation, Diversity Analysis, Species Identification, and S. cerevisiae Strain Distinction
2.2.1. Yeast Isolation
2.2.2. DNA Extraction
2.2.3. RAPD Pattern Analysis
2.2.4. Genus and Species Identification
2.2.5. Distinguishing S. cerevisiae Strains
2.3. Metataxonomic Analysis of Selected MDs
2.4. Phenotypic Analysis
2.4.1. Yeast Strains and Culture Conditions
2.4.2. Leavening Ability of Yeasts in Doughs
2.4.3. ANKOM-Gas Production
2.4.4. Hydrolytic Enzyme Assays
2.4.5. Assays for Vitamin Requirement and Riboflavin Production
2.4.6. Assays for Extracellular Enzyme Activities
3. Results
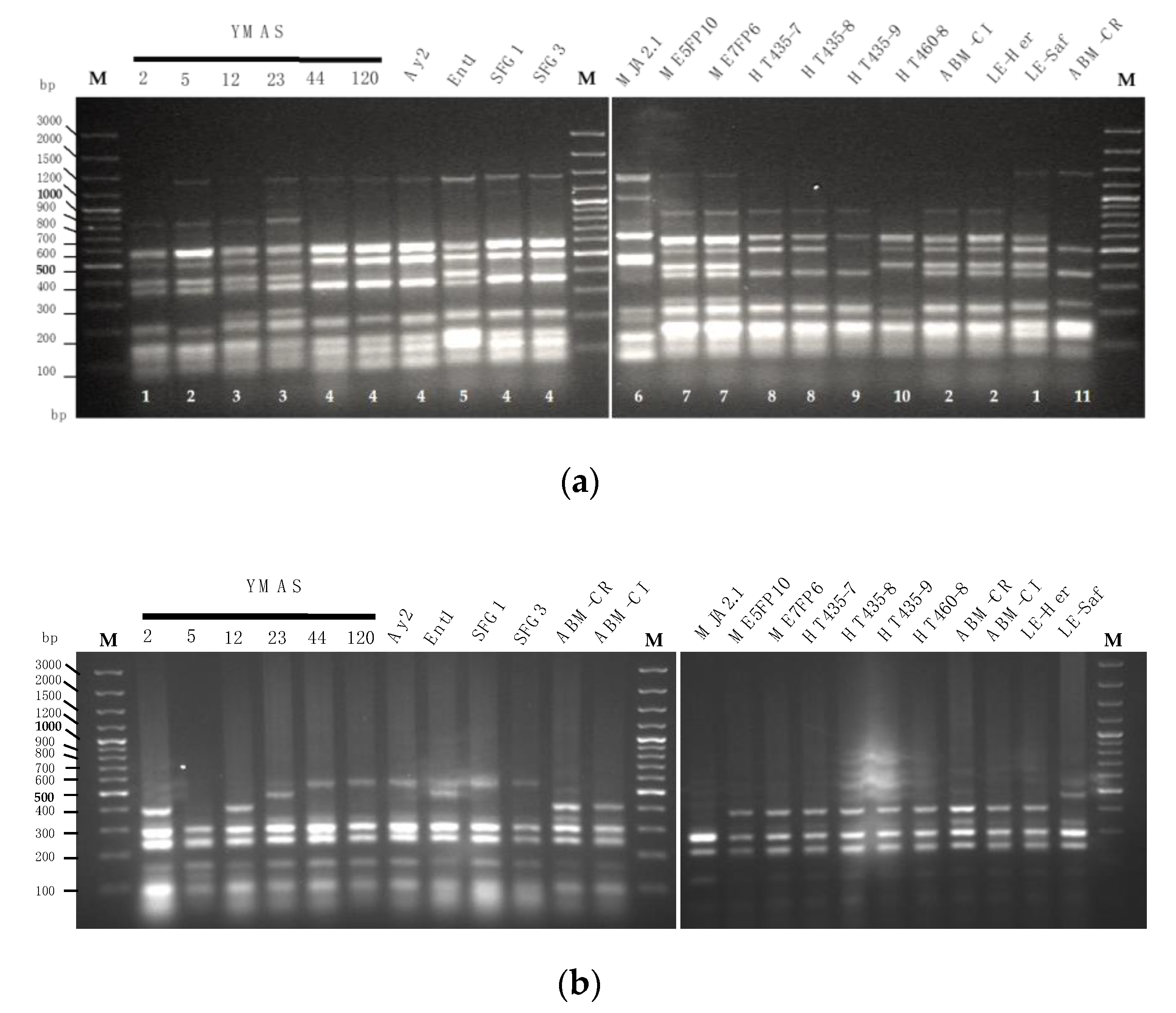
3.1. Genotyping of the Yeast Isolates
3.1.1. Analysis of the 5.8S-ITS and RAPD Patterns
3.1.2. Analysis of the 5.8S-ITS and D1/D2 Regions
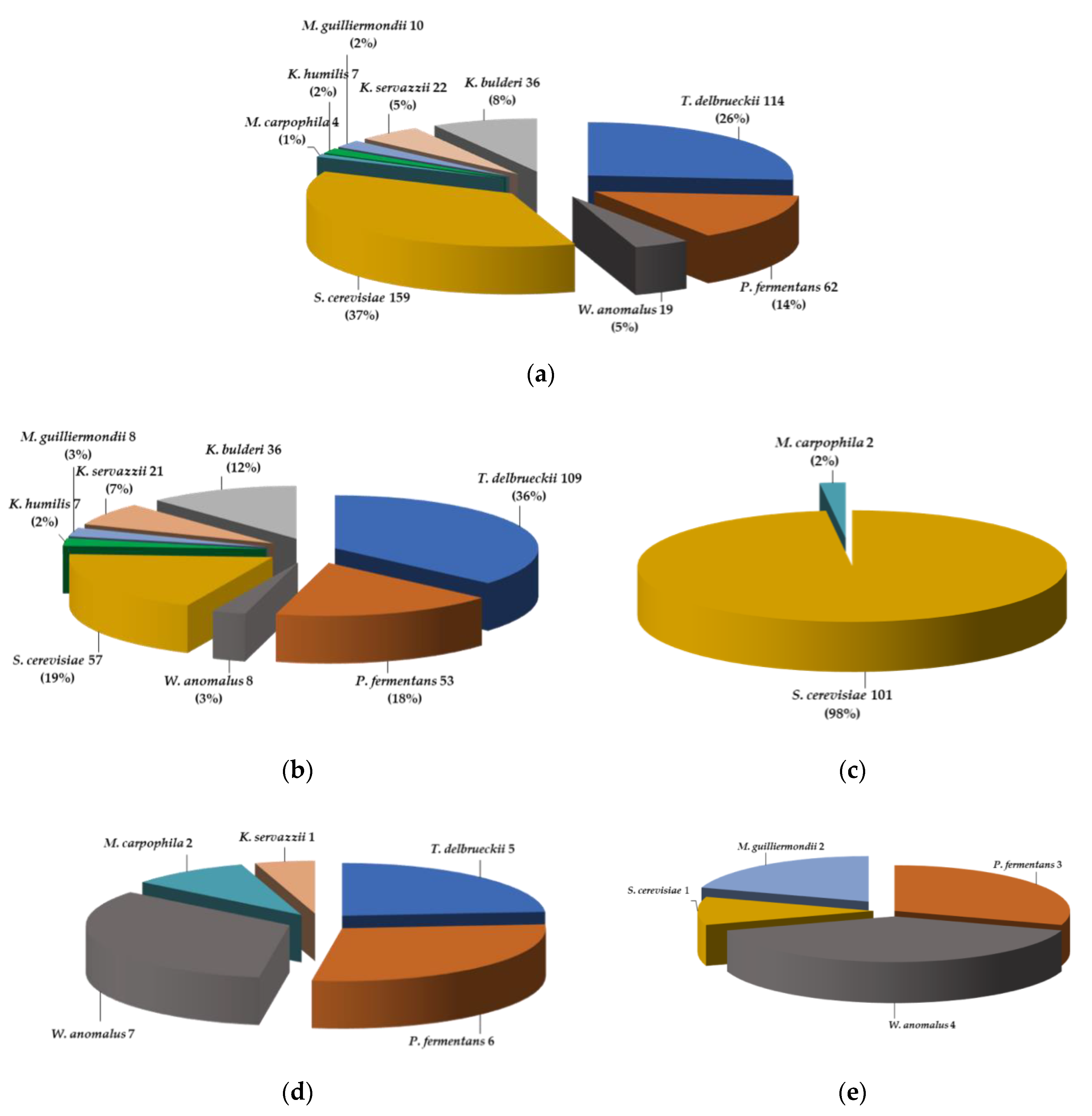
3.2. Distribution of the Yeast Species and Isolates
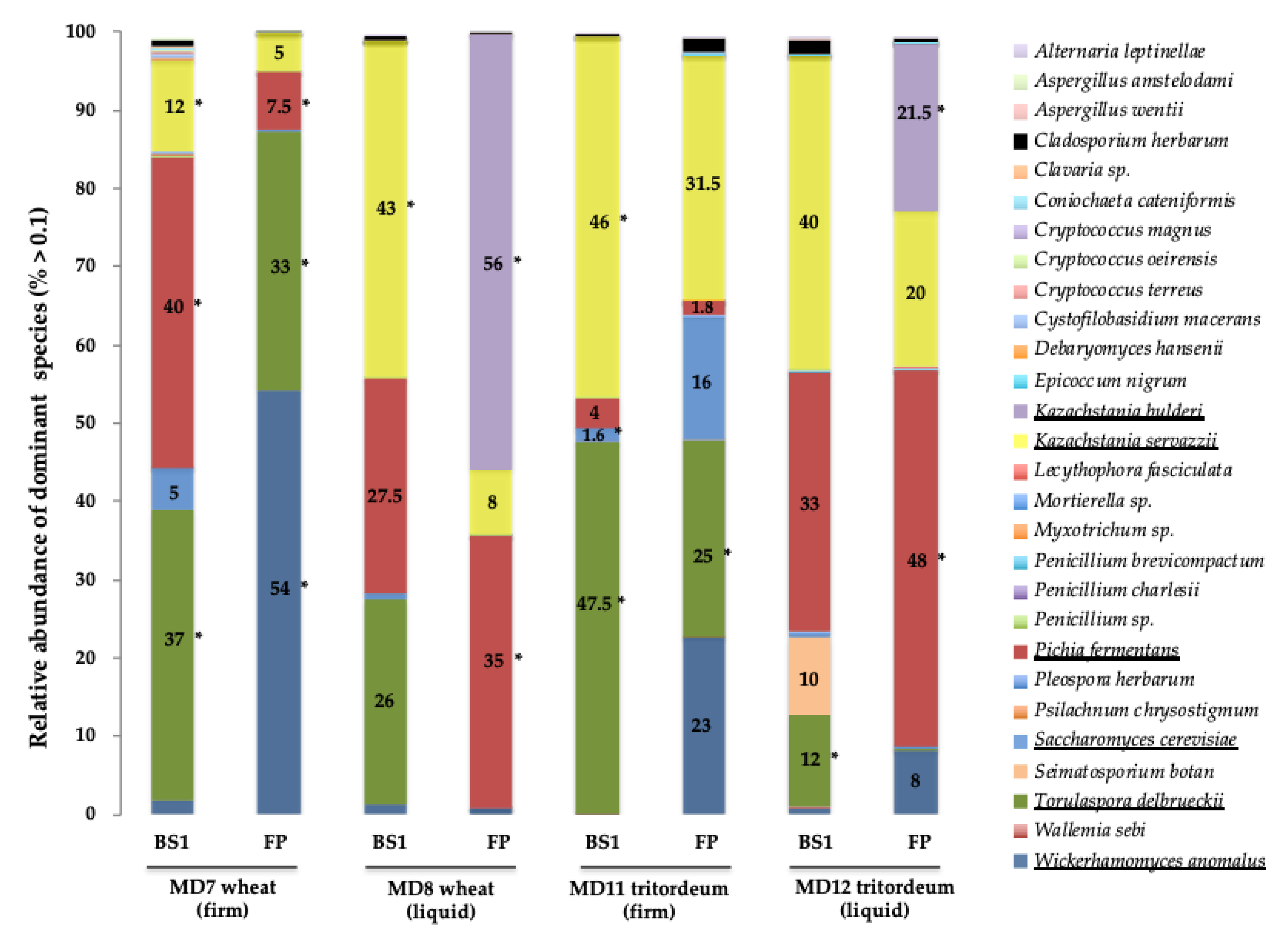
3.3. Metataxonomic Analysis of the Fungal Microbiome in Four Selected Mother Doughs
3.4. Analysis of Traits of Technological Interest in Selected Yeast Strains
3.4.1. Saccharomyces cerevisiae Strain Discrimination
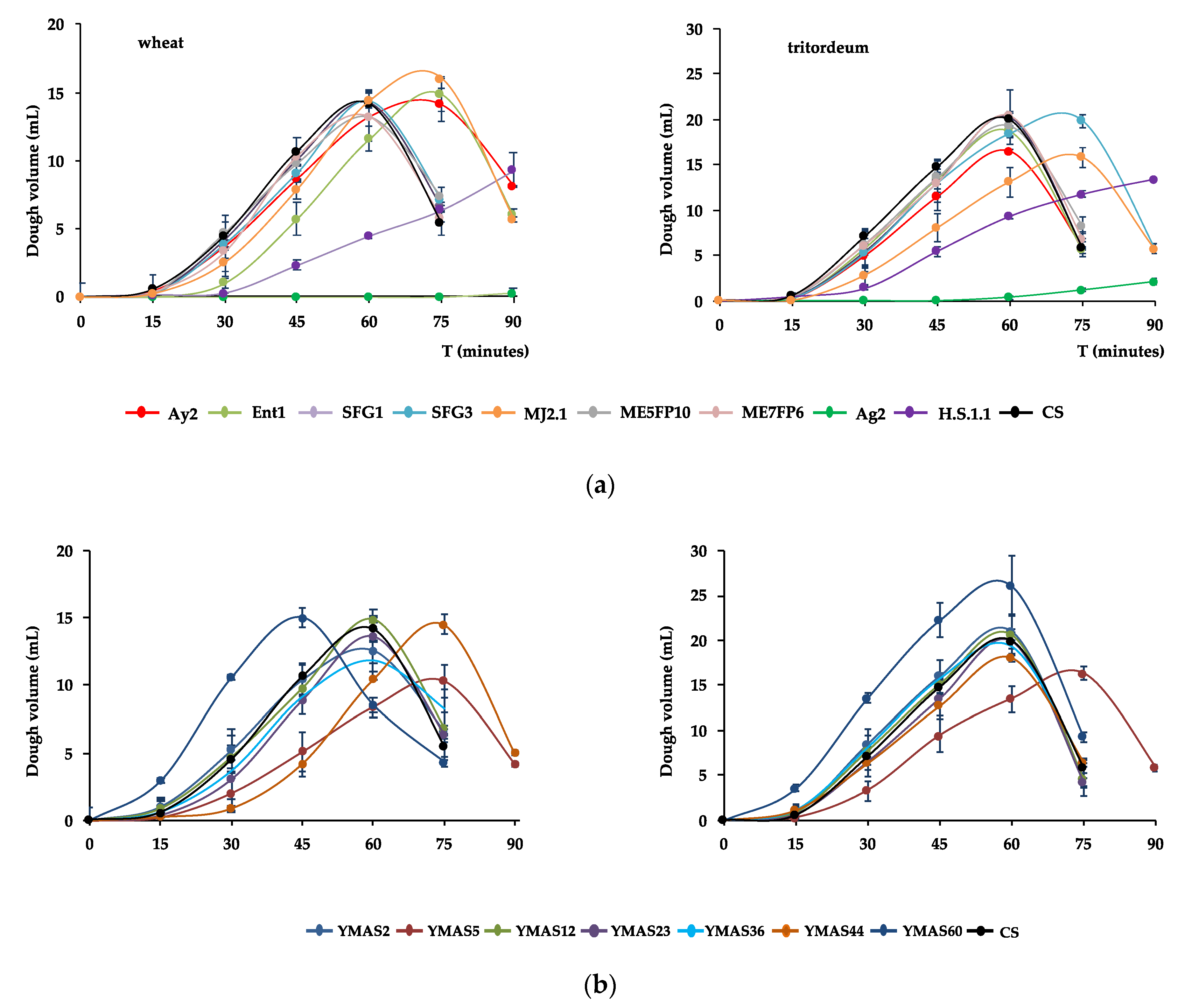
3.4.2. Yeast Capacity to Leaven Doughs
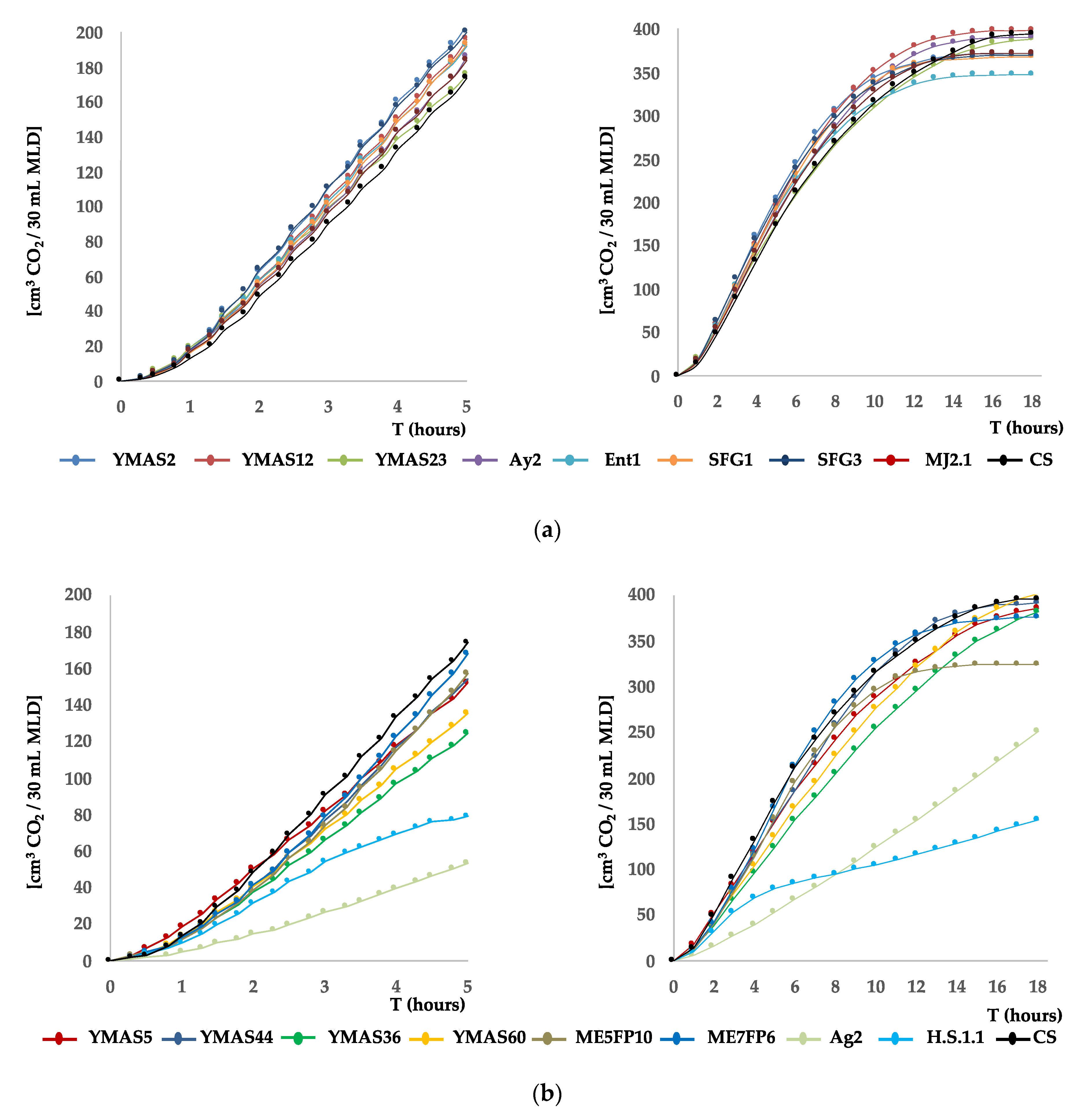
3.4.3. Yeast CO2 Production
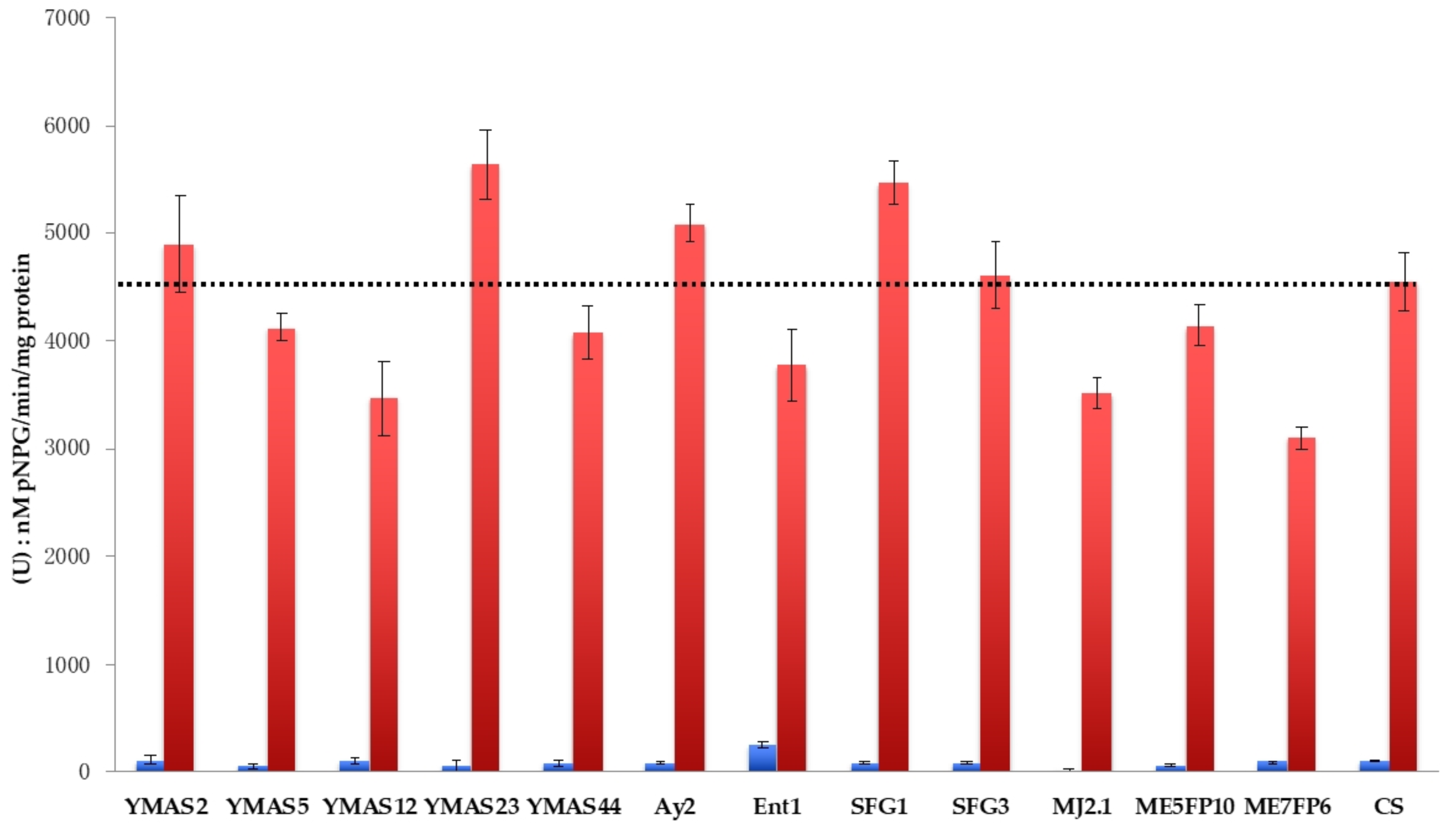
3.4.4. Sugar Hydrolytic Activities of Saccharomyces cerevisiae Strains
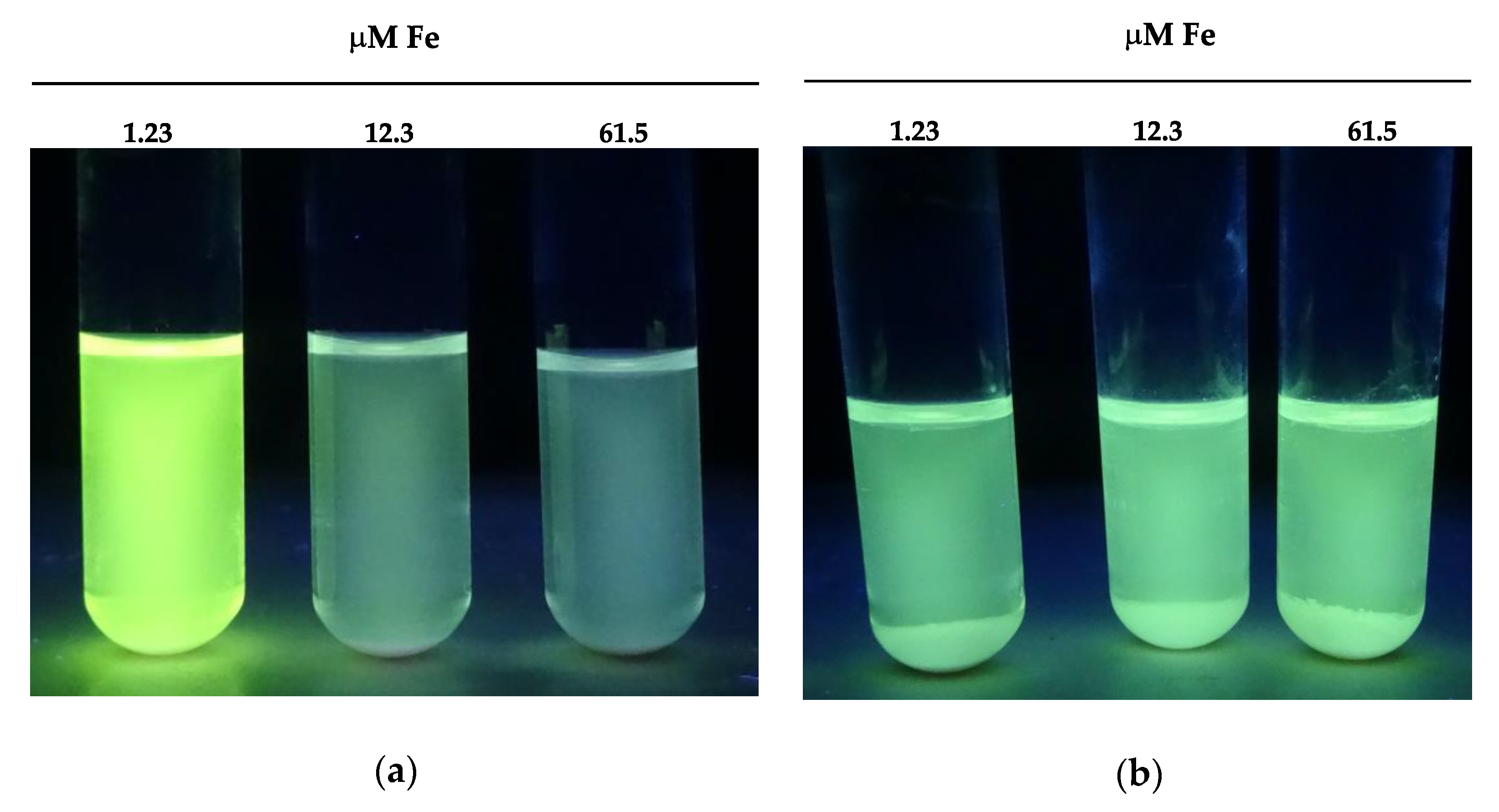
3.4.5. Screening for Yeast Vitamin Requirement and Riboflavin Production
3.4.6. Exocellular Enzymatic Activities of Non-Saccharomyces Yeasts
4. Discussion
4.1. Species Biodiversity in 433 Yeasts Isolated from 67 Cereal Matrices
4.2. Influence of the Flour, Consistency, and Age on the Yeast Species Found in MDs
4.3. Metataxonomic Analysis of Yeast Communities in Four Selected MDs
4.4. Phenotypic Analysis of Selected Yeast Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Randez-Gil, F.; Córcoles-Sáez, I.; Prieto, J.A. Genetic and phenotypic characteristics of baker’s yeast: Relevance to baking. Annu. Rev. Food. Sci. Technol. 2013, 4, 191–214. [Google Scholar] [CrossRef] [PubMed]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Carbonetto, B.; Ramsayer, J.; Nidelet, T.; Legrand, J.; Sicard, D. Bakery yeasts, a new model for studies in ecology and evolution. Yeast 2018, 35, 591–603. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Leroy, F. Microbial Ecology and Process Technology of Sourdough Fermentation. Adv. Appl. Microbiol. 2017, 100, 49–160. [Google Scholar]
- Carbonetto, B.; Nidelet, T.; Guezenec, S.; Perez, M.; Segond, D.; Sicard, D. Interactions between Kazachstania humilis Yeast Species and Lactic Acid Bacteria in Sourdough. Microorganisms 2020, 8, 240. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, X.; Bi, Y.; Zhao, R.; Nie, Y.; Yuan, W. Dynamics of microbial community and changes of metabolites during production of type Ι sourdough steamed bread made by retarded sponge-dough. Food Chem. 2020, 330, 127316. [Google Scholar] [CrossRef]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelflife extension. Crit. Rev. Food Sci. Nutr. 2017, 57, 3528–3542. [Google Scholar] [CrossRef]
- Quattrini, M.; Liang, N.; Fortina, M.G.; Xiang, S.; Curtis, J.M.; Gänzle, M. Exploiting synergies of sourdough and antifungal organic acids to delay fungal spoilage of bread. Int. J. Food Microbiol. 2019, 302, 8–14. [Google Scholar] [CrossRef]
- Gobbetti, M.; Rizzello, C.G.; Di Cagno, R.; De Angelis, M. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol. 2014, 37, 30–40. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Calasso, M.; Archetti, G.; Rizzello, C.G. Novel insights on the functional/nutritional features of the sourdough fermentation. Int. J. Food Microbiol. 2019, 302, 103–113. [Google Scholar] [CrossRef]
- Laatikainen, R.; Koskenpato, J.; Hongisto, S.M.; Loponen, J.; Poussa, T.; Huang, X.; Sontag-Strohm, T.; Salmenkari, H.; Korpela, R. Pilot Study: Comparison of Sourdough Wheat Bread and Yeast-Fermented Wheat Bread in Individuals with Wheat Sensitivity and Irritable Bowel Syndrome. Nutrients 2017, 9, 1215. [Google Scholar] [CrossRef] [PubMed]
- Loponen, J.; Gänzle, M.G. Use of Sourdough in Low FODMAP Baking. Foods 2018, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Capurso, A.; Capurso, C. The Mediterranean way: Why elderly people should eat wholewheat sourdough bread—A little known component of the Mediterranean diet and healthy food for elderly adults. Aging Clin. Exp. Res. 2020, 32, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Portincasa, P.; Montemurro, M.; Di Palo, D.M.; Lorusso, M.P.; De Angelis, M.; Bonfrate, L.; Genot, B.; Gobbetti, M. Sourdough Fermented Breads are More Digestible than Those Started with Baker’s Yeast Alone: An In Vivo Challenge Dissecting Distinct Gastrointestinal Responses. Nutrients 2019, 11, 2954. [Google Scholar] [CrossRef]
- Menezes, L.A.A.; Minervini, F.; Filannino, P.; Sardaro, M.L.S.; Gatti, M.; Lindner, J.D. Effects of Sourdough on FODMAPs in Bread and Potential Outcomes on Irritable Bowel Syndrome Patients and Healthy Subjects. Front. Microbiol. 2018, 9, 1972. [Google Scholar] [CrossRef]
- Zannini, E.; Gobbetti, M. Sourdough for health. In The 7th International Symposium on Sourdough. Int. J. Food Microbiol. 2019, 302, 1–2. [Google Scholar] [CrossRef]
- Struyf, N.; Laurent, J.; Lefevere, B.; Verspreet, J.; Verstrepen, K.J.; Courtin, C.M. Establishing the relative importance of damaged starch and fructan as sources of fermentable sugars in wheat flour and whole meal bread dough fermentations. Food Chem. 2017, 218, 89–98. [Google Scholar] [CrossRef]
- Reese, A.T.; Madden, A.A.; Joossens, M.; Lacaze, G.; Dunn, R.R. Influences of Ingredients and Bakers on the Bacteria and Fungi in Sourdough Starters and Bread. mSphere 2020, 5, e00950-19. [Google Scholar] [CrossRef]
- Koistinen, V.M.; Mattila, O.; Katina, K.; Poutanen, K.; Aura, A.M.; Hanhineva, K. Metabolic profiling of sourdough fermented wheat and rye bread. Sci. Rep. 2018, 8, 5684. [Google Scholar] [CrossRef]
- Korcari, D.; Ricci, G.; Quattrini, M.; Fortina, M.G. Microbial consortia involved in fermented spelt sourdoughs: Dynamics and characterization of yeasts and lactic acid bacteria. Lett. Appl. Microbiol. 2020, 70, 48–54. [Google Scholar] [CrossRef]
- Nionelli, L.; Rizzello, C.G. Sourdough-Based Biotechnologies for the Production of Gluten-Free Foods. Foods 2016, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Polo, A.; Rizzello, C.G. The sourdough fermentation is the powerful process to exploit the potential of legumes, pseudo-cereals and milling by-products in baking industry. Crit. Rev. Food Sci. Nutr. 2019, 60, 2158–2173. [Google Scholar] [CrossRef] [PubMed]
- Carbó, R.; Gordún, E.; Fernández, A.; Ginovart, M. Elaboration of a spontaneous gluten-free sourdough with a mixture of amaranth, buckwheat, and quinoa flours analyzing microbial load, acidity, and pH. Food Sci. Technol. Int. 2020, 26, 344–352. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lhomme, E.; Lattanzi, A.; Dousset, X.; Minervini, F.; De Angelis, M.; Lacaze, G.; Onno, B.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of sixteen French traditional sourdoughs. Int. J. Food Microbiol. 2015, 215, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Lhomme, E.; Urien, C.; Legrand, J.; Dousset, X.; Onno, B.; Sicard, D. Sourdough microbial community dynamics: An analysis during French organic bread-making processes. Food Microbiol. 2016, 53, 41–50. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Sadiq, F.A.; Yang, H.; Gu, J.; Yuan, L.; Lee, Y.J.; He, G. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chem. 2018, 242, 404–411. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Celano, G.; Gobbetti, M. House microbiotas as sources of lactic acid bacteria and yeasts in traditional italian sourdoughs. Food Microbiol. 2015, 52, 66–76. [Google Scholar] [CrossRef]
- Urien, C.; Legrand, J.; Montalent, P.; Casaregola, S.; Sicard, D. Fungal Species Diversity in French Bread Sourdoughs Made of Organic Wheat Flour. Front. Microbiol. 2019, 10, 201. [Google Scholar] [CrossRef]
- Fraberger, V.; Unger, C.; Kummer, C.; Domig, K.J. Insights into microbial diversity of traditional Austrian sourdough. LWT-Food Sci. Technol. 2020, 127, 109358. [Google Scholar] [CrossRef]
- Barber, S.; Baguena, R.; Martínez-Anaya, M.A.; Torner, M.J. Microflora of the sour dough of wheat flour bread. I. Identification and functional properties of microorganisms of industrial sour doughs. Rev. Agroquim. Tecnol. Alimentos. 1983, 23, 552–562. [Google Scholar]
- Barber, S.; Baguena, R. Microflora of the sour dough of wheat flour bread. V. Isolation, identification and evaluation of functional properties of sour dough’s microorganisms. Rev. Agroquim. Tecnol. Alimentos. 1988, 28, 67–78. [Google Scholar]
- Martínez-Anaya, M.A.; Pitarch, B.; Bayarri, P.; de Barber, C.B. Microflora of the sour dough of wheat flour bread. X, Interactions between yeast and lactic acid bacteria in wheat doughs and their effects in bread quality. Cereal Chem. 1990, 67, 85. [Google Scholar]
- Gordún, E.; del Valle, L.J.; Ginovart, M.; Carbó, R. Comparison of the microbial dynamics and biochemistry of laboratory sourdoughs prepared with grape, apple and yogurt. Food Sci. Technol. Int. 2015, 21, 428–439. [Google Scholar] [CrossRef]
- Coda, R.; Cagno, R.D.; Gobbetti, M.; Rizzello, C.G. Sourdough lactic acid bacteria: Exploration of non-wheat cereal-based fermentation. Food Microbiol. 2014, 37, 51–58. [Google Scholar] [CrossRef]
- Katina, K.; Juvonen, R.; Laitila, A.; Flander, L.; Nordlund, E.; Kariluoto, S.; Piironen, V.; Poutanen, K. Fermented wheat bran as a functional ingredient in baking. Cereal Chem. 2012, 89, 126–134. [Google Scholar] [CrossRef]
- Rosenkvist, H.; Hansen, A. Contamination profiles and characterization of Bacillus species in wheat bread and raw materials for bread production. Int. J. Food Microbiol. 1995, 26, 353–363. [Google Scholar] [CrossRef]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- Valmorri, S.; Tofalo, R.; Settanni, L.; Corsetti, A.; Suzzi, G. Yeast microbiota associated with spontaneous sourdough fermentations in the production of traditional wheat sourdough breads of the Abruzzo region (Italy). Antonie Van Leeuwenhoek 2010, 97, 119–129. [Google Scholar] [CrossRef]
- Succi, M.; Reale, A.; Andrighetto, C.; Lombardi, A.; Sorrentino, E.; Coppola, R. Presence of yeasts in southern Italian sourdoughs from Triticum aestivum flour. FEMS Microbiol. Lett. 2003, 225, 143–148. [Google Scholar] [CrossRef][Green Version]
- Martorana, A.; Giuffrè, A.M.; Capocasale, M.; Zappia, C.; Sidari, R. Sourdoughs as a source of lactic acid bacteria and yeasts with technological characteristics useful for improved bakery products. Eur. Food Res. Technol. 2018, 244, 1873–1885. [Google Scholar] [CrossRef]
- Gordún, E.; Puig, A.; Pujol, L.; Carbó, R. Identification of yeast isolated from laboratory sourdoughs prepared with grape, apple and yogurt. J. Microbiol. Biotechnol. Food Sci. 2018, 7, 399–403. [Google Scholar] [CrossRef]
- De Filippis, F.; Parente, E.; Ercolini, D. Metagenomics insights into food fermentations. Microb. Biotechnol. 2017, 10, 91–102. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “terroir” Concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef]
- Wecks, S.; Van Kerrebroeck, S.; De Vuyst, L. Omics approaches to understand sourdough fermentation processes. Int. J. Food Microbiol. 2019, 302, 90–102. [Google Scholar] [CrossRef]
- Comasio, A.; Verce, M.; Van Kerrebroeck, S.; De Vuyst, L. Diverse microbial composition of sourdoughs from different origins. Front. Microbiol. 2020, 11, 1212. [Google Scholar] [CrossRef]
- Brandt, M.J. Starter cultures for cereal based foods. Food Microbiol. 2014, 37, 41–43. [Google Scholar] [CrossRef]
- Winters, M.; Panayotides, D.; Bayrak, M.; Rémont, G.; Viejo, C.G.; Liu, D.; Le, B.; Liu, Y.; Luo, J.; Zhang, P.; et al. Defined co-cultures of yeast and bacteria modify the aroma, crumb and sensory properties of bread. J. Appl. Microbiol. 2019, 127, 778–793. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Georgalaki, M.; Alexandraki, V.; Kazou, M.; Anastasiou, R.; Tsakalidou, E. Chapter 6, Sourdough Bread. In Innovations in Traditional Foods, 1st ed.; Galanakisf, C.M., Ed.; Food Waste Recovery Group, ISEKI Food Association: Vienna, Austria, 2019; Volume 1, pp. 127–158. [Google Scholar]
- Steensels, J.; Snoek, T.; Meersman, E.; Picca Nicolino, M.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Zhou, N.; Schifferdecker, A.J.; Gamero, A.; Compagno, C.; Boekhout, T.; Piškur, J.; Knecht, W. Kazachstania gamospora and Wickerhamomyces subpelliculosus: Two alternative baker’s yeasts in the modern bakery. Int. J. Food Microbiol. 2017, 250, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.; Álvarez, J.; Martin, L.M.; Barro, F.; Ballesteros, J. The development of tritordeum: A novel cereal for food processing. J. Cereal Sci. 1999, 30, 85–95. [Google Scholar]
- Vaquero, L.; Comino, I.; Vivas, S.; Rodríguez-Martín, L.; Giménez, M.J.; Pastor, J.; Sousa, C.; Barro, F. Tritordeum: A novel cereal for food processing with good acceptability and significant reduction in gluten immunogenic peptides in comparison with wheat. J. Sci. Food. Agric. 2018, 98, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Lin, X.; Song, H.Y.; Xiao, D.G. Effects of MAL61 and MAL62 overexpression on maltose fermentation of baker’s yeast in lean dough. World J. Microbiol. Biotechnol. 2015, 31, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Myers, D.K.; Lawlor, D.T.; Attfield, P.V. Influence of invertase activity and glycerol synthesis and retention on fermentation of media with a high sugar concentration by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 145–150. [Google Scholar] [CrossRef]
- Nevoigt, E.; Stahl, U. Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997, 21, 231–241. [Google Scholar] [CrossRef]
- Batifoulier, F.; Verny, M.A.; Chanliaud, E.; Demigné, C. Effect of different breadmaking methods on thiamine, riboflavin and pyridoxine contents of wheat bread. J. Cereal Sci. 2005, 42, 101–108. [Google Scholar] [CrossRef]
- Jacques, N.; Sarilar, V.; Urien, C.; Lopes, M.R.; Morais, C.G.; Uetanabaro, A.P.T.; Tinsley, C.R.; Rosa, C.A.; Sicard, D.; Casaregola, S. Three novel ascomycetous yeast species of the Kazachstania clade, Kazachstania saulgeensis sp. nov., Kazachstania serrabonitensis sp. nov. and Kazachstania australis sp. nov. Reassignment of Candida humilis to Kazachstania humilis f.a. comb. nov. and Candida pseudohumilis to Kazachstania pseudohumilis f.a. comb. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5192–5200. [Google Scholar]
- Kariluoto, S.; Edelmann, M.; Nyström, L.; Sontag-Strohm, T.; Salovaara, H.; Kivelä, R.; Herranen, M.; Korhola, M.; Piironen, V. In situ enrichment of folate by microorganisms in beta-glucan rich oat and barley matrices. Int. J. Food Microbiol. 2014, 176, 38–48. [Google Scholar] [CrossRef]
- Degre, R.; Zhang, Z.; Edwards, G. Novel Vitamin D2 Yeast Preparation, a Method for Producing the Same and the Use Thereof. U.S. Patent Application 11/977,56, 12 June 2008. [Google Scholar]
- García-Estepa, R.M.; Guerra-Hernández, E.; García-Villanova, B. Phytic acid content in milled cereal products and breads. Food Res. Int. 1999, 32, 217–221. [Google Scholar] [CrossRef]
- Schlemmer, U.; Frølich, W.; Prieto, R.M.; Grases, F. Phytate in Foods and Significance for Humans: Food Sources, Intake, Processing, Bioavailability, Protective Role and Analysis. Mol. Nutr. Food Res. 2009, 53, S330–S375. [Google Scholar] [CrossRef] [PubMed]
- Nuobariene, L.; Hansen, Å.S.; Arneborg, N. Isolation and identification of phytase-active yeasts from sourdoughs. LWT-Food Sci. Technol. 2012, 48, 190–196. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek 2011, 99, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Kunze, G.; Satyanarayana, T. Yeast phytases: Present scenario and future perspectives. Crit Rev Biotechnol. 2007, 27, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 2017, 250, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Palla, M.; Agnolucci, M.; Calzone, A.; Giovannetti, M.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G.; Pontonio, E. Exploitation of autochthonous Tuscan sourdough yeasts as potential starters. Int. J. Food Microbiol. 2019, 302, 59–68. [Google Scholar] [CrossRef]
- Scherf, K.A.; Wieser, H.; Koehler, P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Int. J. Food Microbiol. 2018, 110, 62–72. [Google Scholar] [CrossRef]
- Gobbetti, M.; Pontonio, E.; Filannino, P.; Rizzello, C.G.; De Angelis, M.; Di Cagno, R. How to improve the gluten-free diet: The state of the art from a food science perspective. Food Res. Int. 2018, 110, 22–32. [Google Scholar] [CrossRef]
- Sakandar, H.A.; Kubow, S.; Azadi, B.; Faryal, A.; Ali, B.; Ghazanfar, S.; Quraishi, U.M.; Imran, M. Wheat fermentation with Enterococcus mundtii QAUSD01 and Wickerhamomyces anomalus QAUWA03 consortia induces concurrent gliadin and phytic acid degradation and inhibits gliadin toxicity in Caco-2 monolayers. Front. Microbiol. 2019, 9, 3312. [Google Scholar] [CrossRef]
- Aslankoohi, E.; Herrera-Malaver, B.; Rezaei, M.N.; Steensels, J.; Courtin, C.M.; Verstrepen, K.J. Non-conventional yeast strains increase the aroma complexity of bread. PLoS ONE 2016, 11, e0165126. [Google Scholar] [CrossRef]
- Gamero, A.; Quintilla, R.; Groenewald, M.; Alkema, W.; Boekhout, T.; Hazelwood, L. High-throughput screening of a large collection of non-conventional yeasts reveals their potential for aroma formation in food fermentation. Food Microbiol. 2016, 60, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yin, Y.; Ali, B.; Zhang, Y.; Guo, L.; Xu, X. Isolation of yeast strains from Chinese liquor Daqu and its use in the wheat sourdough bread making. Food Biosci. 2019, 31, 100443. [Google Scholar] [CrossRef]
- Coda, R.; Cassone, A.; Rizzello, C.G.; Nionelli, L.; Cardinali, G.; Gobbetti, M. Antifungal Activity of Wickerhamomyces anomalus and Lactobacillus plantarum during Sourdough Fermentation: Identification of Novel Compounds and Long-Term Effect during Storage of Wheat Bread. Appl. Environ. Microbiol. 2011, 77, 3484–3492. [Google Scholar] [CrossRef] [PubMed]
- Coda, R.; Rizzello, C.G.; Di Cagno, R.; Trani, A.; Cardinali, G.; Gobbetti, M. Antifungal activity of Meyerozyma guilliermondii: Identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiol. 2013, 33, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Foschino, R.; Gallina, S.; Andrighetto, C.; Rossetti, L.; Galli, A. Comparison of cultural methods for the identification and molecular investigation of yeasts from sourdoughs for Italian sweet baked products. FEMS Yeast Res. 2004, 4, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Vingataramin, L.; Frost, E.H. A single protocol for extraction of gDNA from bacteria and yeast. BioTechniques 2015, 58, 120–125. [Google Scholar] [CrossRef]
- Andrighetto, C.; Psomas, E.; Tzanetakis, N.; Suzzi, G.; Lombardi, A. Randomly amplified polymorphic DNA (RAPD) PCR for the identification of yeasts isolated from dairy products. Lett. Appl. Microbiol. 2000, 30, 5–9. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimization of inter-d analysis for Saccharomyces cerevisiae strain characterization. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Siesto, G.; Capece, A.; Sipiczki, M.; Csoma, H.; Romano, P. Polymorphism detection among wild Saccharomyces cerevisiae strains of different wine origin. Ann. Microbiol. 2013, 63, 661–668. [Google Scholar] [CrossRef]
- Marinangeli, P.; Angelozzi, D.; Ciani, M.; Clementi, F.; Mannazzu, I. Minisatellites in Saccharomyces cerevisiae genes encoding cell wall proteins: A new way towards wine strain characterization. FEMS Yeast Res. 2004, 4, 427–435. [Google Scholar] [CrossRef][Green Version]
- Marinangeli, P.; Clementi, F.; Ciani, M.; Mannazzu, I. SED1 polymorphism within the genus Saccharomyces. FEMS Yeast Res. 2004, 5, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Boveri, S.; Rainieri, S.; Pulvirenti, A. Method for the validation of intraspecific crosses of Saccharomyces cerevisiae strains by minisatellite analysis. Can. J. Microbiol. 2012, 58, 350–358. [Google Scholar] [CrossRef]
- Codón, A.C.; Benítez, T. Variability of the physiological features and of the nuclear and mitochondrial genomes of baker’s yeasts. Syst. Appl. Microbiol. 1995, 18, 343–352. [Google Scholar]
- Rincón, A.M.; Codón, A.C.; Castrejón, F.; Benítez, T. Improved properties of baker’s yeast mutants resistant to 2-deoxy-D-glucose. Appl. Environ. Microbiol. 2001, 67, 4279–4285. [Google Scholar] [CrossRef]
- Panadero, J.; Randez-Gil, F.; Prieto, J.A. Validation of a flour-free model dough system for throughput studies of baker’s yeast. Appl. Environ. Microbiol. 2005, 71, 1142–1147. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chaplin, M.F. Monosaccharides. In Carbohydrate Analysis: A Practical Approach; Chaplin, M.F., Kennedy, J.F., Eds.; IRL Press: Oxford, UK, 1986; pp. 1–36. [Google Scholar]
- Wickerham, L.J. A critical evaluation of the nitrogen assimilation tests as commonly used in the classification of the yeasts. J. Bacteriol. 1946, 52, 293–301. [Google Scholar] [CrossRef]
- Boretsky, Y.R.; Protchenko, O.V.; Prokopiv, T.M.; Mukalov, I.O.; Fedorovych, D.V.; Sibirny, A.A. Mutations and environmental factors affecting regulation of riboflavin synthesis and iron assimilation also cause oxidative stress in the yeast Pichia guilliermondii. J. Basic Microbiol. 2007, 47, 371–377. [Google Scholar] [CrossRef]
- Carrasco, M.; Rozas, J.M.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012, 12, 251. [Google Scholar] [CrossRef] [PubMed]
- Strauss, M.L.; Jolly, N.P.; Lambrechts, M.G.; van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Olstorpe, M.; Schnürer, J.; Passoth, V. Screening of yeast strains for phytase activity. FEMS Yeast Res. 2009, 9, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F.; Fink, G.; Hicks, J. Methods in Yeast Genetics: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1986; pp. 163–169. [Google Scholar]
- Higgins, V.J.; Braidwood, M.; Bell, P.; Bissinger, P.; Dawes, I.W.; Attfield, P.V. Genetic evidence that high noninduced maltase and maltose permease activities, governed by MALx3-encoded transcriptional regulators, determine efficiency of gas production by baker’s yeast in unsugared dough. Appl. Environ. Microbiol. 1999, 65, 680–685. [Google Scholar] [CrossRef]
- Jiang, T.; Xiao, D.; Gao, Q. Characterization of maltose metabolism in lean dough by lagging and non-lagging baker’s yeast strains. Ann. Microbiol. 2008, 58, 655. [Google Scholar] [CrossRef]
- Fedorovich, D.; Protchenko, O.; Lesuisse, E. Iron uptake by the yeast Pichia guilliermondii. Flavinogenesis and reductive iron assimilation are co-regulated processes. Biometals 1999, 12, 295–300. [Google Scholar] [CrossRef]
- Hierro, N.; González, A.; Mas, A.; Guillamón, J.M. New PCR-based methods for yeast identification. J. Appl. Microbiol. 2004, 97, 792–801. [Google Scholar] [CrossRef]
- Atkins, S.D.; Clark, I.M. Fungal molecular diagnostics: A mini review. J. Appl. Genet. 2004, 45, 3–15. [Google Scholar]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.; Crous, P.W.; Boekhout, T.; et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Ranjard, L.; Poly, F.; Lata, J.C.; Mougel, C.; Thioulouse, J.; Nazaret, S. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: Biological and methodological variability. Appl. Environ. Microbiol. 2001, 67, 4479–4487. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.F.; Hammes, W.P.; Habermeyer, M.; Engel, K.H.; Knorr, D.; Eisenbrand, G.; Senate Commission on Food Safety (SKLM) of the German Research Foundation. Microbial food cultures-opinion of the Senate Commission on Food Safety (SKLM) of the German Research Foundation (DFG). Mol. Nutr. Food Res. 2011, 55, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Brandt, M.J. Industrial production of sourdoughs for the baking branch—An overview. Int. J. Food Microbiol. 2019, 302, 3–7. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: Suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 2020, 18, e06174. [Google Scholar]
- Buzzini, P.; Di Mauro, S.; Turchetti, B. Yeasts as starter cultures. In Starter Cultures in Food Production; Speranza, B., Bevilacqua, A., Corbo, M.R., Sinigaglia, M., Eds.; Wiley Editors: New York, NY, USA, 2017; pp. 16–49. [Google Scholar]
- Pulvirenti, A.; Solieri, L.; Gullo, M.; De Vero, L.; Giudici, P. Occurrence and dominance of yeast species in sourdough. Lett. Appl. Microbiol. 2004, 38, 113–117. [Google Scholar] [CrossRef]
- Vrancken, G.; De Vuyst, L.; Van der Meulen, R.; Huys, G.; Vandamme, P.; Daniel, H.M. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 2010, 10, 471–481. [Google Scholar] [CrossRef]
- Vernocchi, P.; Valmorri, S.; Dalai, I.; Torriani, S.; Gianotti, A.; Suzzi, G.; Guerzoni, M.E.; Mastrocola, D.; Gardini, F. Characterization of the yeast population involved in the production of a typical Italian bread. J. Food Sci. 2004, 69, M182–M186. [Google Scholar] [CrossRef]
- Iacumin, L.; Cecchini, F.; Manzano, M.; Osualdini, M.; Boscolo, D.; Orlic, S.; Comi, G. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 2009, 26, 128–135. [Google Scholar] [CrossRef]
- Boyaci Gunduz, C.P.; Gaglio, R.; Franciosi, E.; Settanni, L.; Erten, H. Molecular analysis of the dominant lactic acid bacteria of chickpea liquid starters and doughs and propagation of chickpea sourdoughs with selected Weissella confusa. Food Microbiol. 2020, 91, 103490. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Influence of artisan bakery-or laboratory-propagated sourdoughs on the diversity of lactic acid bacterium and yeast microbiotas. Appl. Environ. Microbiol. 2012, 78, 5328–5340. [Google Scholar] [CrossRef] [PubMed]
- Harth, H.; Kerrebroeck, S.V.; De Vuyst, L. Impact of process conditions on the microbial community dynamics and metabolite production kinetics of teff sourdough fermentations under bakery and laboratory conditions. Food Sci. Nutr. 2018, 6, 1438–1455. [Google Scholar] [CrossRef] [PubMed]
- Taccari, M.; Aquilanti, L.; Polverigiani, S.; Osimani, A.; Garofalo, C.; Milanović, V.; Clementi, F. Microbial diversity of type I sourdoughs prepared and back-slopped with wholemeal and refined soft (Triticum aestivum) wheat flours. J. Food Sci. 2016, 81, M1996–M2005. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the lactic acid bacterium and yeast microbiota in the switch from firm-to liquid-sourdough fermentation. Appl. Environ. Microbiol. 2014, 80, 3161–3172. [Google Scholar] [CrossRef]
- Arici, M.; Ozulku, G.; Yildirim, R.M.; Sagdic, O.; Durak, M.Z. Biodiversity and technological properties of yeasts from Turkish sourdough. Food Sci. Biotechnol. 2017, 27, 499–508. [Google Scholar] [CrossRef]
- Guerzoni, M.E.; Serrazanetti, D.I.; Vernocchi, P.; Gianotti, A. Physiology and Biochemistry of Sourdough Yeasts. In Handbook on Sourdough Biotechnology; Gobbetti, M., Gänzle, M., Eds.; Springer: Boston, MA, USA, 2013; pp. 155–181. [Google Scholar]
- Van Kerrebroeck, S.; Maes, D.; De Vuyst, L. Sourdoughs as a function of their species diversity and process conditions, a meta-analysis. Trends Food Sci. Technol. 2017, 68, 152–159. [Google Scholar] [CrossRef]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2015, 16, 43–56. [Google Scholar] [CrossRef]
- Wachowska, U.; Irzykowski, W.; Jędryczka, M. Agrochemicals: Effect on genetic resistance in yeasts colonizing winter wheat kernels. Ecotoxicol. Environ. Saf. 2018, 30, 77–84. [Google Scholar] [CrossRef]
- Amorim, J.C.; Piccoli, R.H.; Duarte, W.F. Probiotic potential of yeasts isolated from pineapple and their use in the elaboration of potentially functional fermented beverages. Food Res. Int. 2018, 107, 518–527. [Google Scholar] [CrossRef]
- Celano, G.; De Angelis, M.; Minervini, F.; Gobbetti, M. Different flour microbial communities drive to sourdoughs characterized by diverse bacterial strains and free amino acid profiles. Front. Microbiol. 2016, 8, 1770. [Google Scholar] [CrossRef]
- Poutanen, K.; Flander, L.; Katina, K. Sourdough and cereal fermentation in a nutritional perspective. Food Microbiol. 2009, 26, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Nionelli, L.; Coda, R.; De Angelis, M.; Gobbetti, M. Effect of sourdough fermentation on stabilization, and chemical and nutritional characteristics of wheat germ. Food Chem. 2010, 119, 1079–1089. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Pini, N.; Guerrini, S.; Granchi, L.; Vincenzini, M. Liquid and firm sourdough fermentation: Microbial robustness and interactions during consecutive backsloppings. LWT Food Sci. Technol. 2019, 105, 9–15. [Google Scholar] [CrossRef]
- Giannone, V.; Longo, C.; Damigella, A.; Raspagliesi, D.; Spina, A.; Palumbo, M. Technological properties of bakers’ yeasts in durum wheat semolina dough. J. Ind. Microbiol. Biotechnol. 2010, 37, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Chambers, P.J.; Pretorius, I.S. The Development of Superior Yeast Strains for the Food and Beverage Industries: Challenges, Opportunities and Potential Benefits. In Yeasts in Food and Beverages; Querol, A., Fleet, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 399–444. [Google Scholar]
- Higgins, V.J.; Bell, P.J.; Dawes, I.W.; Attfield, P.V. Generation of a novel Saccharomyces cerevisiae strain that exhibits strong maltose utilization and hyperosmotic resistance using nonrecombinant techniques. Appl. Environ. Microbiol. 2001, 67, 4346–4348. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, U.; Öste, R.; Jägerstad, M. Cereal fructans: Hydrolysis by yeast invertase, in vitro and during fermentation. J. Cereal Sci. 1987, 6, 53–60. [Google Scholar] [CrossRef]
- Struyf, N.; Laurent, J.; Verspreet, K.J.; Verstrepen, K.J.; Courtin, C.M. Saccharomyces cerevisiae and Kluyveromyces marxianus co-cultures allow reduction of fermentable oligo-, di-, and monosaccharides and polyols levels in whole wheat bread. J. Agric. Food Chem. 2017, 65, 8704–8713. [Google Scholar] [CrossRef] [PubMed]
- Laurent, J.; Timmermans, E.; Struyf, N.; Verstrepen, K.J.; Courtin, C.M. Variability in yeast invertase activity determines the extent of fructan hydrolysis during wheat dough fermentation and final FODMAP levels in bread. Int. J. Food Microbiol. 2020, 326, 108648. [Google Scholar] [CrossRef]
- Agnastopoulos, D.A.; Tsaltas, D. Fermented Foods and Beverages. In Innovations in Traditional Foods; Galanakis, C.M., Ed.; Woodhead Publishing: Cambridge, UK, 2019; pp. 257–291. [Google Scholar] [CrossRef]
- Perli, T.; Wronska, A.K.; Ortiz-Merino, R.A.; Pronk, J.T.; Daran, J.M. Vitamin requirements and biosynthesis in Saccharomyces cerevisiae. Yeast 2020, 37, 283–304. [Google Scholar] [CrossRef]
- De Marco, L.; Epis, S.; Capone, A.; Martin, E.; Bozic, J.; Crotti, E.; Ricci, I.; Sassera, D. The Genomes of Four Meyerozyma caribbica Isolates and Novel Insights into the Meyerozyma guilliermondii Species Complex. G3 2018, 8, 755–759. [Google Scholar] [CrossRef]
- Kloots, W.; Op den Kamp, D.; Abrahamse, L. In vitro iron availability from iron-fortified whole-grain wheat flour. J. Agric. Food Chem. 2004, 52, 8132–8136. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Capozzi, V.; Arena, M.P.; Spadaccino, G.; Dueñas, M.T.; López, P.; Fiocco, D.; Spano, G. Riboflavin-overproducing strains of Lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 2014, 98, 3691–3700. [Google Scholar] [CrossRef] [PubMed]
- Mohedano, M.L.; Hernández-Recio, S.; Yépez, A.; Requena, T.; Martínez-Cuesta, M.C.; Peláez, C.; Russo, P.; LeBlanc, J.C.; Spano, G.; Aznar, R.; et al. Real-Time detection of riboflavin production by Lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using mCherry labeling. Front. Microbiol. 2019, 10, 1748. [Google Scholar] [CrossRef] [PubMed]
- Lomolino, G.; Zocca, F.; Spettoli, P.; Lante, A. Detection of β-Glucosidase and esterase activities in wild yeast in a distillery environment. J. Inst. Brew. 2006, 112, 97–100. [Google Scholar] [CrossRef]
- Kurita, O. Increase of acetate ester-hydrolysing esterase activity in mixed cultures of Saccharomyces cerevisiae and Pichia anomala. J. Appl. Microbiol. 2008, 104, 1051–1058. [Google Scholar] [CrossRef]
- Dank, A.; Smid, E.J.; Notebaart, R.A. CRISPR-Cas genome engineering of esterase activity in Saccharomyces cerevisiae esters aroma formation. BMC Res. Notes 2018, 11, 682. [Google Scholar] [CrossRef]
- Bonciani, T.; De Vero, L.; Giannuzzi, E.; Verspohl, A.; Giudici, P. Qualitative and quantitative screening of the β-glucosidase activity in Saccharomyces cerevisiae and Saccharomyces uvarum strains isolated from refrigerated must. Lett. Appl. Microbiol. 2018, 67, 72–78. [Google Scholar] [CrossRef]


| MD# | Consistency | Flour Type | Baker (Location) |
|---|---|---|---|
| MD1 | Firm | wheat, W180–200 (a) | EM (Zamora, Spain) |
| MD2 | Liquid | wheat, W180–200 (a) | EM (Zamora, Spain) |
| MD3 | Firm | wheat, W130–150 (a) | EM (Zamora, Spain) |
| MD4 | Liquid | wheat, W130–150 (a) | EM (Zamora, Spain) |
| MD5 | Firm | T. Zamorana W230–250 (TZM) (a) | EM (Zamora, Spain) |
| MD6 | Liquid | T. Zamorana W230–250 (TZM) (a) | EM (Zamora, Spain) |
| MD7 | Firm | wholemeal wheat (WMW) (a) | EM (Zamora, Spain) |
| MD8 | Liquid | wholemeal wheat (WMW) (a) | EM (Zamora, Spain) |
| MD9 | Firm | tritordeum W100–110 (tr) (a) | EM (Zamora, Spain) |
| MD10 | Liquid | tritordeum W100–110 (tr) (a) | EM (Zamora, Spain) |
| MD11 | Firm | wholemeal tritordeum (WMtr) (a) | EM (Zamora, Spain) |
| MD12 | Liquid | wholemeal tritordeum (WMtr) (a) | EM (Zamora, Spain) |
| MD13 | Firm | wheat “Oromas” (b) | EM (Zamora, Spain) |
| MD14 | Liquid | mixture of 6 flours (MO6F) (a) | EM (Zamora, Spain) |
| MD15 | Firm | wheat (d) | MJA (Valladolid, Spain) |
| MD16 | Firm | tritordeum W100–110 (Tr) (c) | MJA (Valladolid, Spain) |
| MD17 | Firm | wholemeal tritordeum (WMtr) (a) | MJA (Valladolid, Spain) |
| MD18 | Firm | tritordeum W100–110 (tr) (c) | JAR (Barcelona, Spain) |
| MD19 | Firm | wheat “T-80” W200–220 (c) | JAR (Barcelona, Spain) |
| MD20 | Firm | wheat (d) | TB (Boulogne-sur-Mer, France) |
| MD21 | Firm | wheat (d) | FB (Boulogne-sur-Mer, France) |
| Province | BD# | Bakery or Baker (Town) |
|---|---|---|
| Salamanca | BD1 | A. Sánchez (Cabrerizos) |
| BD2 | San Morales (Cabrerizos) | |
| BD3 | V. Hernández (San Felices de los Gallegos) | |
| BD4 | L.E. Río (Lumbrales) | |
| Ávila | BD5 | R. Hernández C. B. (Muñogalindo) |
| BD6 | La Garrosa (Solosancho) | |
| BD7 | La Candelaria (Sotalbo) | |
| BD8 | I. López Hernández (La Horcajada) | |
| Zamora | BD9 | Repostería Mateos S.A. (Entrala) |
| BD10 | Mayagus S.L. (Mayalde) | |
| BD11 | A. Rodríguez (Videmala) | |
| BD12 | Sta. María de la Vega (Bretó de la Ribera) | |
| BD13 | Lomar (Ayoó de Vidriales) | |
| BD14 | Santa Marina (Aguilar de Tera) | |
| BD15 | Montero Mezquita (Gallegos del Río) | |
| León | BD16 | Pedro González (Val de San Lorenzo) |
| Sample | Grain | Source |
|---|---|---|
| EE1 | 5 cereal mixture | Emilio Esteban (Valladolid, Spain) |
| EE4 | rye | Emilio Esteban (Valladolid, Spain) |
| EE5 | barley | Emilio Esteban (Valladolid, Spain) |
| EE6 | wheat | Emilio Esteban (Valladolid, Spain) |
| EE9 | oat | Emilio Esteban (Valladolid, Spain) |
| T1 | tritordeum—aucan | * Irnasa (CSIC) (Salamanca, Spain) |
| T4 | tritordeum—aucan | (Jaén, Spain) |
| T6 | tritordeum—aucan | (Córdoba, Spain) |
| T7 | tritordeum—bulel | (Córdoba, Spain) |
| Gtit | tritordeum | (Bari, Italy) |
| Flour | ||
| HS2 | WM rye | Stone, Rincón del Segura (Albacete, Spain) |
| HS1 | WM wheat | Stone, Rincón del Segura (Albacete, Spain) |
| H1 | wheat W180–200 | Cylinders, Molinos del Duero (Zamora, Spain) |
| H2 | WM wheat | Cylinders, La Vilafranquina (Ávila, Spain) |
| H3 | wheat W130–150 | Cylinders, Molinos del Duero (Zamora, Spain) |
| H4 | wheat “Oromas” | Cylinders, La Vilafranquina (Ávila, Spain) |
| H5 | T. Zamorana W230–250 | Stone, Molinos del Duero (Zamora, Spain) |
| H6 | WM tritordeum | (Málaga, Spain) |
| H7 | tritordeum | (Málaga, Spain) |
| T9 | tritordeum—aucan | (Barcelona, Spain) |
| ACoF | tritordeum—aucan | (Córdoba, Spain) |
| AJF | tritordeum—aucan | (Jaén, Spain) |
| BCF | tritordeum—bulel | (Sevilla, Spain) |
| BCoF | tritordeum—bulel | (Córdoba, Spain) |
| BJF | tritordeum—bulel | (Jaén, Spain) |
| ACoI | WM tritordeum—aucan | (Córdoba, Spain) |
| BCI | WM tritordeum—bulel | (Sevilla, Spain) |
| BCoI | WM tritordeum—bulel | (Córdoba, Spain) |
| THI | WM tritordeum—aucan | (Sevilla, Spain) |
| HRTit | tritordeum—bulel | (Bari, Italy) |
| Representative Yeast Strains (Laboratory Code) | ITS Type | RAPD Groups | Closest Type Strain Accession Numbers 5.8-ITS/LSU (D1/D2) | ITS/LSU (D1/D2) Similarity (%) |
|---|---|---|---|---|
| P7FP8, P6FP8, P5FP3, P4FP5, P5FP8, P5FP10, P6FP2, P1FP9, P3FP5, P1FP2 | I | A, B, C, D, E, F, G, I, J, K | Kazachstania bulderi CBS 8638T KY103628/KY107908 | 100/100 |
| MTB-1 | I | H | Kazachstania humilis CBS 5658T KY102142/KY106507 | 97.7/100 |
| H41 | I | L | Kazachstania servazzii CBS 4311T KY103668/KM454442 | 99.9/99.8 |
| P2A5 | I | M | K. servazzii CBS 4311T KY103668/KM454442 | 99.6/99.8 |
| P4A6, GTi7 | II | A, C | Meyerozyma guilliermondii CBS 2030T NR_111247/KY108542 | 100/99.8 |
| Ag2 | II | D | Meyerozyma carpophila CBS 5256T MK394110/KY106386 | 100/99.8 |
| H6.3 | II | B | M. carpophila CBS 5256T MK394110/KY106386 | 99.8/99.8 |
| ME2FP5, HRTi8, HRTi7 | III | A, H, J | Pichia fermentans CBS 187T KY104545/KY108804 | 98.4/99.5 |
| EE9B | III | L | P. fermentans CBS 187T KY104545/KY108804 | 99.1/99.5 |
| ME2FP2, ME1A5, ME3A1 | III | F, G, I | P. fermentans CBS 187T KY104545/KY108804 | 99.8/99.5 |
| H5.2, P6FP4, ME2A1, ME4A7, P3FP2, EE1A | III | B, C, D, E, K, M | P. fermentans CBS 187T KY104545/KY108804 | 100/99.5 |
| YMAS12, ME1A8 | IV | A, B | Saccharomyces cerevisiae CBS 1171T AB018043/KC881066 | 100/100 |
| GTi5 | IV | D | S. cerevisiae CBS 1171T AB018043/KC881066 | 99.6/100 |
| MJA2.1, ME5FP10 | IV | E, G | S. cerevisiae CBS 1171T AB018043/KC881066 | 99.8/100 |
| YMAS3, YMAS2 | IV | C, F | S. cerevisiae CBS 1171T AB018043/KC881066 | 99.9/100 |
| ME3FP9, ME5A5, P3D6, P6A10, P7F10, P5A7, P5A8, P5A2, ME6FP10, ME3A7, ME3A2, H.S.1.1, ME2A2 | V | A, B, C, D, E, F, G, H, I, L, M, O, Q | Torulaspora delbrueckii CBS 1146T KY105617/AJ508558 | 100/100 |
| ME2FP10, ME3FP3, ME2FP7, ME4FP1, ME5FP6 | V | J, K, N, R | T. delbrueckii CBS 1146T KY105617/AJ508558 | 99.9/100 |
| T9, GTi1, ME2FP3, ME1FP9 | VI | A, C, D, E | Wickerhamomyces anomalus CBS 5759T KY105894/EU057562 | 100/100 |
| ME5FP8 | VI | B | W. anomalus CBS 5759T KY105894/EU057562 | 98.7/100 |
| MDs: | MD7 Wheat, Firm | MD8 Wheat, Liquid | MD11 Tritordeum, Firm | MD12 Tritordeum, Liquid | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fermentation step: | BS1 FP | BS1 FP | BS1 FP | BS1 FP | ||||||||||||
| I | % | I | % | I | % | I | % | I | % | I | % | I | % | I | % | |
| Kazachstania bulderi | - | - | - | - | - | - | 4 | 56 | - | - | - | - | - | - | 9 | 21.5 |
| Kazachstania servazzii | 1 | 12 | - | 5 | 10 | 43 | - | 8 | 1 | 46 | - | 31.5 | - | 40 | - | 20 |
| Pichia fermentans | 3 | 40 | 4 | 7.5 | - | 27.5 | 6 | 35 | - | 4 | - | 1.5 | - | 33 | 1 | 48 |
| Saccharomyces cerevisiae | - | 5 | - | - | - | 0.7 | - | 0.1 | 1 | 1.6 | - | 15.7 | - | 0.2 | - | 0.1 |
| Torulaspora delbrueckii | 4 | 37 | 3 | 33 | - | 26 | - | 0.1 | 5 | 48 | 10 | 25 | 10 | 12 | - | 0.3 |
| Wickerhamomyces anomalus | - | 2 | 3 | 54 | - | 1 | - | 0.7 | - | 0.1 | - | 23 | - | 0.8 | - | 8 |
| Fermentation Rate (mL/min) | Maximal Volume, mL (min) | |||
|---|---|---|---|---|
| Strain | Wheat | Tritordeum | Wheat | Tritordeum |
| Ay2 | 331 ± 4 | 393 ± 4 | 14.1 ± 1.3 (75′) | 16.4 ± 0.2 (60′) |
| Ent1 | 352 ± 4 | 444 ± 3 | 14.9 ± 1.3 (75′) | 18.7 ± 0.5 (60′) |
| SFG1 | 321 ± 1 | 479 ± 1 | 14.3 ± 0.4 (60′) | 20.2 ± 3.1 (60′) |
| SFG3 | 324 ± 1 | 416 ± 1 | 14.4 ± 0.5 (60′) | 19.9 ± 0.7 (75′) |
| MJA2.1 | 394 ± 2 | 343 ± 2 | 16.0 ± 0.0 (75′) | 15.8 ± 1.1 (75′) |
| ME5FP10 | 314 ± 3 | 428 ± 1 | 13.2 ± 1.8 (60′) | 19.3 ± 0.4 (60′) |
| ME7FP6 | 373 ± 5 | 442 ± 1 | 13.3 ± 1.7 (60′) | 20.4 ± 0.4 (60′) |
| Ag2 | 25 ± 0 | 73 ± 5 | 1.3 ± 0.4 (90′) | 3.84 ± 0.2 (90′) |
| H.S.1.1 | 153 ± 19 | 174 ± 18 | 9.3 ± 0.7 (90′) | 13.4 ± 0.2 (90′) |
| YMAS2 | 292 ± 3 | 452 ± 2 | 12.6 ± 1.6 (60′) | 21.0 ± 2.0 (60′) |
| YMAS5 | 211 ± 3 | 327 ± 1 | 10.3 ± 1.3 (75′) | 16.3 ± 0.7 (75′) |
| YMAS12 | 337 ± 6 | 443 ± 3 | 14.8 ± 0.2 (60′) | 20.6 ± 2.2 (60′) |
| YMAS23 | 294 ± 5 | 431 ± 2 | 13.6 ± 2.0 (60′) | 19.8 ± 1.4 (60′) |
| YMAS36 | 329 ± 14 | 488 ± 6 | 11.8 ± 1.4 (60′) | 19.3 ± 0.7 (60′) |
| YMAS44 | 314 ± 1 | 381 ± 2 | 14.5 ± 0.7 (75′) | 18.1 ± 0.4 (60′) |
| YMAS60 | 403 ± 30 | 623 ± 54 | 15.0 ± 0.7 (45′) | 26.1 ± 3.4 (60′) |
| CS | 343 ± 1 | 426 ± 2 | 14.2 ± 2.0 (60′) | 20.0 ± 0.8 (60′) |
| CO2 Production Rate(cm3/h) | Volume of CO2 (cm3) | |||
|---|---|---|---|---|
| Strain | 10 h | 12 h | 18 h | |
| YMAS2 | 44 ± 3.4 | 329 ± 21 | 335 ± 35 | 343 ± 40 |
| YMAS5 | 36 ± 2.9 | 405 ± 16 | 423 ± 14 | 565 ± 25 |
| YMAS12 | 43 ± 3.4 | 388 ± 50 | 402 ± 30 | 466 ± 94 |
| YMAS23 | 37 ± 2.4 | 322 ± 15 | 339 ± 9 | 376 ± 19 |
| YMAS36 | 29 ± 0.5 | 309 ± 9 | 329 ± 38 | 400 ± 13 |
| YMAS44 | 37 ± 2.2 | 374 ± 52 | 390 ± 28 | 471 ± 12 |
| YMAS60 | 31 ± 0.3 | 327 ± 14 | 337 ± 1 | 411 ± 90 |
| Ay2 | 42.± 0.4 | 354 ± 20 | 363 ± 6 | 379 ± 16 |
| Ent1 | 42 ± 3.0 | 348 ± 16 | 358 ± 1 | 369 ± 1 |
| SFG1 | 44 ± 1.5 | 353 ± 35 | 367 ± 15 | 385 ± 19 |
| SFG3 | 42 ± 2.3 | 331 ± 49 | 341 ± 35 | 361 ± 51 |
| MJA2.1 | 43 ± 1.0 | 340 ± 16 | 354 ± 4 | 375 ± 0 |
| ME5FP10 | 39 ± 1.3 | 302 ± 66 | 323 ± 37 | 380 ± 1 |
| ME7FP6 | 42 ± 0.5 | 319 ± 61 | 342 ± 29 | 397 ± 6.2 |
| Ag2 | 12 ± 0.8 | 93 ± 17 | 100 ± 23 | 118 ± 49 |
| H.S.1.1 | 15 ± 0.2 | 101 ± 31 | 119 ± 49 | 173 ± 11 |
| CS | 39 ± 3.1 | 305± 16 | 345 ± 6 | 403 ± 12 |
| (U) nM pNP/min/mg Protein | ||
|---|---|---|
| Strain | YPD | YPM |
| YMAS2 | 108 ± 4.5 | 4897 ± 444 |
| YMAS5 | 50 ± 2.0 | 4127 ± 134 |
| YMAS12 | 98 ± 3.1 | 3475 ± 344 |
| YMAS23 | 51 ± 7.1 | 5636 ± 319 |
| YMAS44 | 77 ± 2.7 | 4082 ± 250 |
| Ay2 | 88 ± 2.0 | 5092 ± 173 |
| Ent1 | 252 ± 2.9 | 3773 ± 330 |
| SFG1 | 80 ± 0.8 | 5475 ± 200 |
| SFG3 | 71 ± 10 | 4613 ± 314 |
| MJA2.1 | 11 ± 6.7 | 3513 ± 143 |
| ME5FP10 | 61 ± 6.8 | 4149 ± 203 |
| ME7FP6 | 97 ± 0.2 | 3099 ± 101 |
| CS | 103 ± 6.5 | 4540 ± 265 |
| (U) mM Glucose /min/mg WCE | ||
|---|---|---|
| Strain | YPD | YPS |
| YMAS1 | 1412 | 5618 |
| YMAS2 | 5109 | 9383 |
| YMAS3 | 3061 | 7738 |
| YMAS 5 | 5908 | 9300 |
| YMAS5 | 3110 | 7259 |
| YMAS7 | 1504 | 4512 |
| YMAS8 | 2802 | 11,567 |
| YMAS12 | 3711 | 7671 |
| YMAS23 | 4211 | 14,184 |
| YMAS 35 | 357 | 3602 |
| YMAS 36 | 2404 | 6832 |
| YMAS 44 | 2758 | 6147 |
| YMAS 47 | 2512 | 3464 |
| YMAS 52 | 348 | 3997 |
| YMAS 59 | 3862 | 9392 |
| YMAS 60 | 3546 | 9111 |
| Ay2 | 4374 | 8595 |
| Ag4 | 1864 | 2088 |
| Ag7 | 2232 | 4808 |
| Ent1 | 5129 | 13,453 |
| Ent3 | 3663 | 7222 |
| Ent5 | 2198 | 4519 |
| Br1 | 2854 | 4235 |
| Br7 | 4295 | 8404 |
| Gal1 | 1283 | 6667 |
| SFG1 | 4327 | 6104 |
| SFG2 | 3506 | 5058 |
| SFG3 | 5164 | 7153 |
| ME7FP6 | 521 | 4660 |
| Pc1 | 4277 | 7873 |
| Pc2 | 12,886 | 13,646 |
| MFa6 | 4448 | 8079 |
| MFa7 | 4018 | 6131 |
| MFb3 | 2721 | 5447 |
| MFb5 | 4000 | 6029 |
| Bc4 | 3950 | 12,541 |
| Bc5 | 4719 | 8231 |
| Ag2 | 1027 | 1524 |
| H.S.1.1 | 786 | 817 |
| CD | 1029 | 4236 |
| Yeast Species | Number of Isolates | Thiamine (B1) | Nicotinic Acid (B3) | Biotin (B7) | |
|---|---|---|---|---|---|
| Kazachstania bulderi | 33 (MD) | − | − | + | |
| K. bulderi | 2 (MD) | + | − | − | |
| K. bulderi | 1 (MD) | − | − | −/+ | |
| Kazachstania humilis | 7 (MD) | − | − | + | |
| Kazachstania servazzii | 22 (21 MD, 1F) | + | + | + | |
| Meyerozyma carpophila | 2 (BD) | − | − | + | |
| M. carpophila | 2 (F) | + | − | − | |
| Meyerozyma guilliermondii | 8 (MD) | − | − | −/+ | |
| M. guilliermondii | 2 (G) | − | − | + | |
| Pichia fermentans | 47 (MD) | + | − | − | |
| P. fermentans | 3 (MD) | − | − | + | |
| P. fermentans | 3 (MD) | + | + | + | |
| P. fermentans | 9 (F) | + | − | − | |
| Saccharomyces cerevisiae | 36 (MD) | − | − | + | |
| S. cerevisiae | 121 (BD) | − | − | + | |
| S. cerevisiae | 1 (G) | + | − | − | |
| Torulaspora delbrueckii | 73 (71 MD, 2 F) | − | − | + | |
| T. delbrueckii | 37 (35 MD, 2 F) | − | − | −/+ | |
| T. delbrueckii | 4 (3 MD, 1 F) | − | − | − | |
| Wickerhamomyces anomalus | 19 (8 MD, 7 H, 4 G) | − | − | − |
| Species and Strain Codes(Matrix) * | Riboflavin Concentration ** (µg/mL Medium) | ||
|---|---|---|---|
| Iron Concentration (Fe+++) | |||
| 1.23 µM | 12.3 µM | 61.5 µM | |
| Meyerozyma carpophila H6.1 (F) | 4.15 ± 0.13 | 4.30 ± 0.19 | 4.40 ± 0.17 |
| M. carpophila YMAS59 (BD) | 4.20 ± 0.10 | 4.40 ± 0.30 | 4.30 ± 0.20 |
| M. carpophila Ag2 (BD) | 4.10 ± 0.11 | 4.00 ± 0.12 | 4.10 ± 0.30 |
| Meyerozyma guilliermondii GTi7 (F) | 14.82 ± 0.10 | 3.80 ± 0.21 | 0.40 ± 0.01 |
| M. guilliermondii P4A6 (MD) | 16.10 ± 0.31 | 4.20 ± 0.51 | 0.50 ± 0.00 |
| M. guilliermondii P4A9 (MD) | 16.08 ± 0.42 | 4.20 ± 0.34 | 0.50 ± 0.01 |
| Species and Strain Codes (Source) * | Bioassays (halo in mm **) | Activities by Growth *** | ||||
|---|---|---|---|---|---|---|
| Esterase | Protease | Glyadinase | Pectinase | Phytase | Cellobiase | |
| Kazachstania bulderi P1FP9 (MD) | 4 | 0 | 0 | 0 | − | − |
| K. bulderi P3FP5 (MD) | 4 | 0 | 0 | 0 | − | − |
| Kazachstania humilis MTB−1 (MD) | 6 | 0 | 0 | 0 | + | − |
| K. humilis MBE−4 (MD) | 6 | 0 | 0 | 0 | + | − |
| Kazachstania servazzii H4.1 (F) | 5 | 6 | 0 | 0 | − | − |
| K. servazzii P2A5 (MD) | 5 | 6 | 0 | 0 | − | − |
| Meyerozyma carpophila H6.3 (F) | 2 | 5 | 0 | 0 | − | + |
| M. carpophila Ag2 (BD) | 2 | 5 | 0 | 0 | − | + |
| Meyerozyma guilliermondii GTi7 (F) | 2 | 4 | 0 | 0 | − | + |
| M. guilliermondii P4A6 (MD) | 2 | 4 | 0 | 0 | − | + |
| M. guilliermondii P4A9 (MD) | 2 | 4 | 0 | 0 | ||
| Pichia fermentans EE1A (F) | 0 | 15 | 4 | 0 | − | + |
| P. fermentans HRTi7 (F) | 0 | 15 | 4 | 0 | − | + |
| P. fermentans ME2FP5 (MD) | 0 | 15 | 4 | 0 | − | + |
| P. fermentans P6FP4 (MD) | 0 | 15 | 4 | 0 | − | + |
| Torulaspora delbrueckii H1.2 (F) | 5 | 0 | 0 | 0 | − | + |
| T. delbrueckii ME2FP7 (MD) | 5 | 0 | 0 | 0 | − | + |
| T. delbrueckii ME6FP10 (MD) | 5 | 0 | 0 | 0 | − | + |
| T. delbrueckii P6A10 (MD) | 5 | 0 | 0 | 0 | − | + |
| Wickerhamomyces anomalus T9 (G) | 12 | 21 | 4 | 4 | + | + |
| W. anomalus HRTi1 (F) | 12 | 21 | 4 | 4 | + | + |
| W. anomalus ME1FP9 (MD) | 12 | 21 | 4 | 4 | + | + |
| W. anomalus YMAT1 (MD) | 12 | 21 | 4 | 4 | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiva, R.; Celador-Lera, L.; Uña, J.A.; Jiménez-López, A.; Espinosa-Alcantud, M.; Mateos-Horganero, E.; Vega, S.; Santos, M.Á.; Velázquez, E.; Tamame, M. Yeast Biodiversity in Fermented Doughs and Raw Cereal Matrices and the Study of Technological Traits of Selected Strains Isolated in Spain. Microorganisms 2021, 9, 47. https://doi.org/10.3390/microorganisms9010047
Chiva R, Celador-Lera L, Uña JA, Jiménez-López A, Espinosa-Alcantud M, Mateos-Horganero E, Vega S, Santos MÁ, Velázquez E, Tamame M. Yeast Biodiversity in Fermented Doughs and Raw Cereal Matrices and the Study of Technological Traits of Selected Strains Isolated in Spain. Microorganisms. 2021; 9(1):47. https://doi.org/10.3390/microorganisms9010047
Chicago/Turabian StyleChiva, Rosana, Lorena Celador-Lera, José Antonio Uña, Ana Jiménez-López, María Espinosa-Alcantud, Enrique Mateos-Horganero, Soledad Vega, María Ángeles Santos, Encarna Velázquez, and Mercedes Tamame. 2021. "Yeast Biodiversity in Fermented Doughs and Raw Cereal Matrices and the Study of Technological Traits of Selected Strains Isolated in Spain" Microorganisms 9, no. 1: 47. https://doi.org/10.3390/microorganisms9010047
APA StyleChiva, R., Celador-Lera, L., Uña, J. A., Jiménez-López, A., Espinosa-Alcantud, M., Mateos-Horganero, E., Vega, S., Santos, M. Á., Velázquez, E., & Tamame, M. (2021). Yeast Biodiversity in Fermented Doughs and Raw Cereal Matrices and the Study of Technological Traits of Selected Strains Isolated in Spain. Microorganisms, 9(1), 47. https://doi.org/10.3390/microorganisms9010047







