Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Construction of New Expression Vectors
2.3. Cloning of pECXT99A and pVWEx1-Based Expression Vectors
2.4. Fluorescence Analysis
2.5. SDS-PAGE
2.6. Enzyme Assay for Xylose Isomerase XylA
2.7. Sarcosine Quantification
3. Results
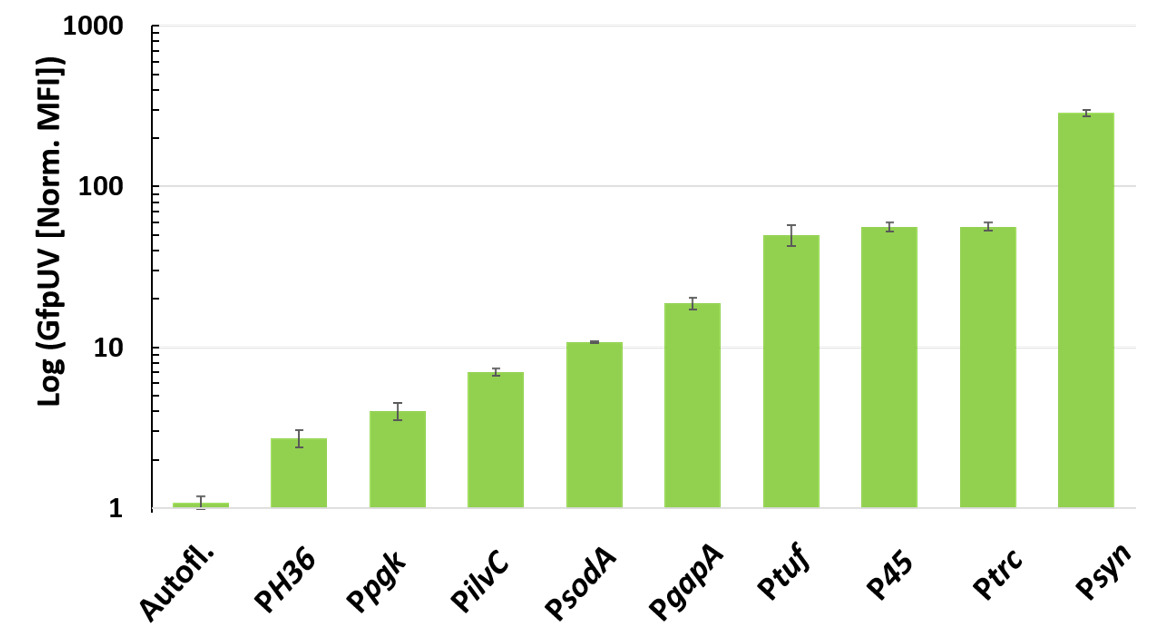
3.1. Screening of Strong Constitutive Promoters in the pECXT99A-Based Vector System
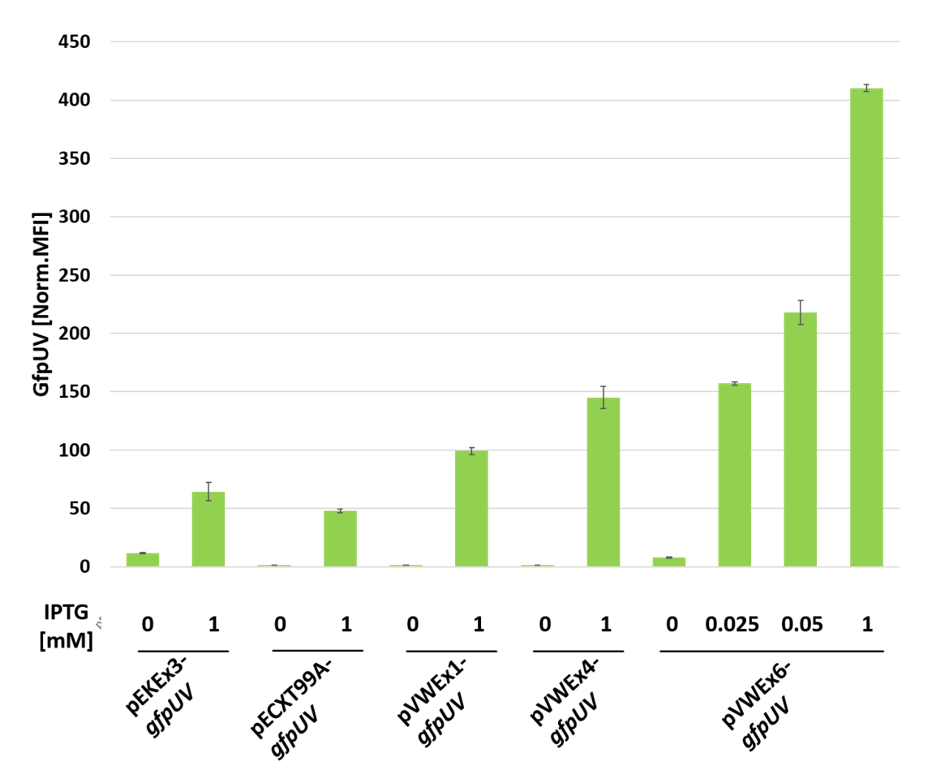
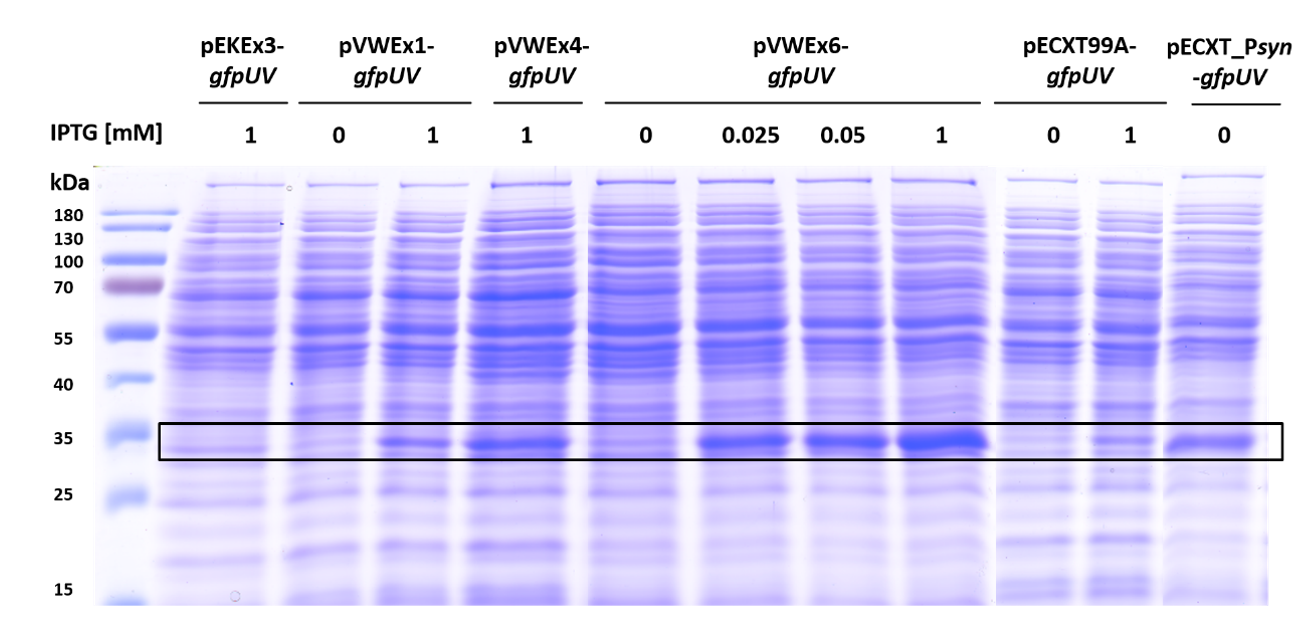
3.2. Improving Plasmid-Borne Inducible Gene Expression in C. glutamicum by Combination of a Stronger Promoter with Increased Plasmid Copy Number
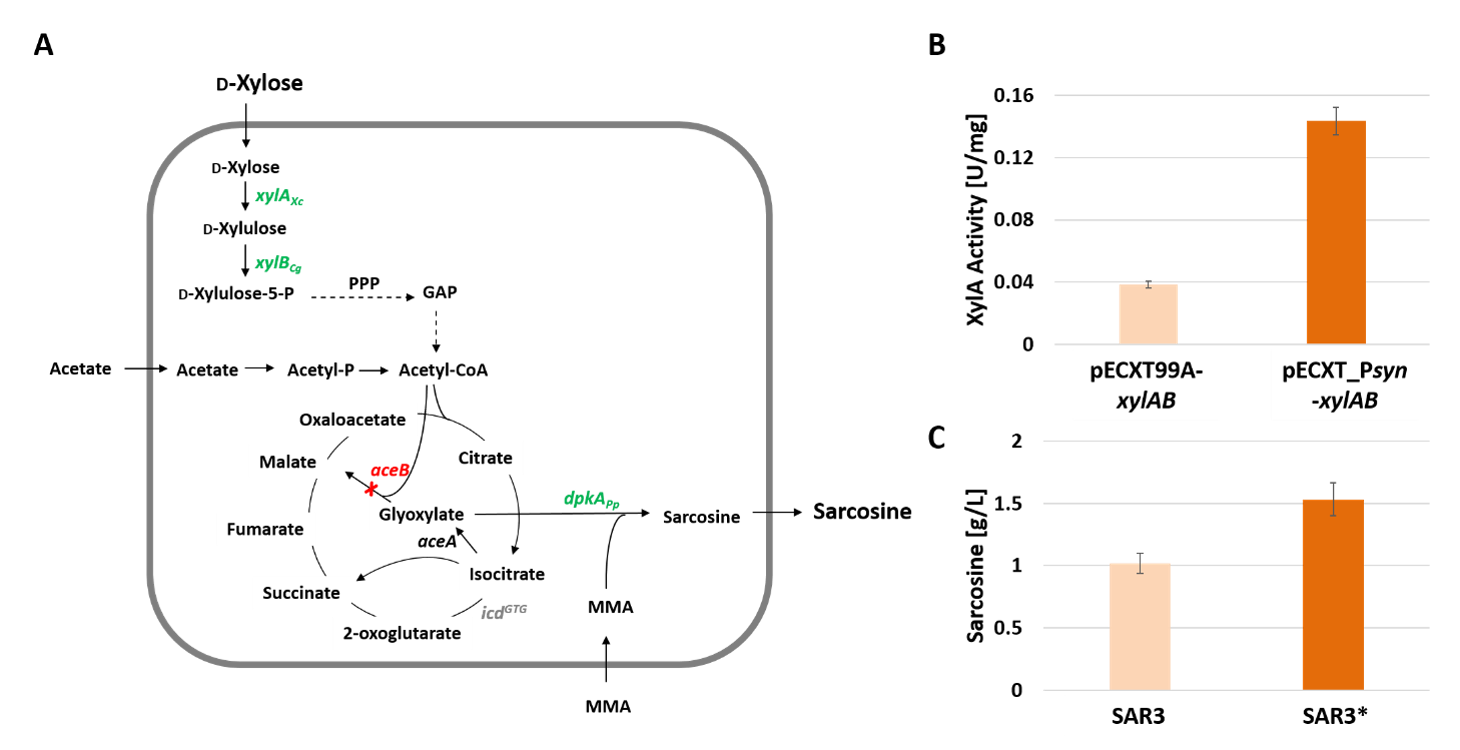
3.3. Fast Production of Sarcosine from Xylose by Application of the Newly Constructed pECXT_Psyn
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kinoshita, S.; Udaka, S.; Shimono, M. Studies on the amino acid fermentation. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 1957, 3, 193–205. [Google Scholar] [CrossRef]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Becker, J.; Rohles, C.M.; Wittmann, C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 2018, 50, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Wendisch, V.F. Engineering microbial cell factories: Metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products. Biotechnol. J. 2015, 10, 1170–1184. [Google Scholar] [CrossRef]
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, F.; Peters-Wendisch, P.; Wendisch, V.F. Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 2016, 100, 8075–8090. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, A.; Otten, A.; Wendisch, V.F. Metabolic engineering of Corynebacterium glutamicum for glycolate production. J. Biotechnol. 2014, 192, 366–375. [Google Scholar] [CrossRef]
- Wieschalka, S.; Blombach, B.; Bott, M.; Eikmanns, B.J. Bio-based production of organic acids with Corynebacterium glutamicum. Microb. Biotechnol. 2013, 6, 87–102. [Google Scholar] [CrossRef]
- Frohwitter, J.; Heider, S.A.; Peters-Wendisch, P.; Beekwilder, J.; Wendisch, V.F. Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J. Biotechnol. 2014, 191, 205–213. [Google Scholar] [CrossRef]
- Henke, N.A.; Wendisch, V.F. Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein. Mar. Drugs 2019, 17, 621. [Google Scholar] [CrossRef]
- Mindt, M.; Heuser, M.; Wendisch, V.F. Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 2019, 281, 135–142. [Google Scholar] [CrossRef]
- Wendisch, V.F.; Brito, L.F.; Gil Lopez, M.; Hennig, G.; Pfeifenschneider, J.; Sgobba, E.; Veldmann, K.H. The flexible feedstock concept in Industrial Biotechnology: Metabolic engineering of Escherichia coli, Corynebacterium glutamicum, Pseudomonas, Bacillus and yeast strains for access to alternative carbon sources. J. Biotechnol. 2016, 234, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Rittmann, D.; Lindner, S.N.; Wendisch, V.F. Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 2008, 74, 6216–6222. [Google Scholar] [CrossRef] [PubMed]
- Meiswinkel, T.M.; Rittmann, D.; Lindner, S.N.; Wendisch, V.F. Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 2013, 145, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Hennig, G.; Haupka, C.; Brito, L.F.; Rückert, C.; Cahoreau, E.; Heux, S.; Wendisch, V.F. Methanol-Essential Growth of Corynebacterium glutamicum: Adaptive Laboratory Evolution Overcomes Limitation due to Methanethiol Assimilation Pathway. Int. J. Mol. Sci. 2020, 21, 3617. [Google Scholar] [CrossRef]
- Seibold, G.; Auchter, M.; Berens, S.; Kalinowski, J.; Eikmanns, B.J. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: Growth and lysine production. J. Biotechnol. 2006, 124, 381–391. [Google Scholar] [CrossRef]
- Anusree, M.; Wendisch, V.F.; Nampoothiri, K.M. Co-expression of endoglucanase and β-glucosidase in Corynebacterium glutamicum DM1729 towards direct lysine fermentation from cellulose. Bioresour. Technol. 2016, 213, 239–244. [Google Scholar] [CrossRef]
- Gopinath, V.; Meiswinkel, T.M.; Wendisch, V.F.; Nampoothiri, K.M. Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2011, 92, 985–996. [Google Scholar] [CrossRef]
- Gopinath, V.; Murali, A.; Dhar, K.S.; Nampoothiri, K.M. Corynebacterium glutamicum as a potent biocatalyst for the bioconversion of pentose sugars to value-added products. Appl. Microbiol. Biotechnol. 2012, 93, 95–106. [Google Scholar] [CrossRef]
- Henke, N.A.; Wichmann, J.; Baier, T.; Frohwitter, J.; Lauersen, K.J.; Risse, J.M.; Peters-Wendisch, P.; Kruse, O.; Wendisch, V.F. Patchoulol Production with Metabolically Engineered Corynebacterium glutamicum. Genes (Basel) 2018, 9, 219. [Google Scholar] [CrossRef]
- Henke, N.A.; Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Production of the Marine Carotenoid Astaxanthin by Metabolically Engineered Corynebacterium glutamicum. Mar. Drugs 2016, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Gauttam, R.; Desiderato, C.; Jung, L.; Shah, A.; Eikmanns, B.J. A step forward: Compatible and dual-inducible expression vectors for gene co-expression in Corynebacterium glutamicum. Plasmid 2019, 101, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Goldbeck, O.; Seibold, G.M. Construction of pOGOduet—An inducible, bicistronic vector for synthesis of recombinant proteins in Corynebacterium glutamicum. Plasmid 2018, 95, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Knoppova, M.; Phensaijai, M.; Vesely, M.; Zemanova, M.; Nesvera, J.; Patek, M. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr. Microbiol. 2007, 55, 234–239. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Puhler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Eggeling, L.; Bott, M. (Eds.) Handbook of Corynebacterium Glutamicum; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2005. [Google Scholar]
- Tauch, A.; Kirchner, O.; Loffler, B.; Gotker, S.; Puhler, A.; Kalinowski, J. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 2002, 45, 362–367. [Google Scholar] [CrossRef]
- Peters-Wendisch, P.G.; Schiel, B.; Wendisch, V.F.; Katsoulidis, E.; Mockel, B.; Sahm, H.; Eikmanns, B.J. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 2001, 3, 295–300. [Google Scholar]
- Oehler, S.; Amouyal, M.; Kolkhof, P.; von Wilcken-Bergmann, B.; Müller-Hill, B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994, 13, 3348–3355. [Google Scholar] [CrossRef]
- Hashiro, S.; Yasueda, H. Plasmid copy number mutation in repA gene encoding RepA replication initiator of cryptic plasmid pHM1519 in Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 2018, 82, 2212–2224. [Google Scholar] [CrossRef]
- Patek, M.; Eikmanns, B.J.; Patek, J.; Sahm, H. Promoters from Corynebacterium glutamicum: Cloning, molecular analysis and search for a consensus motif. Microbiology 1996, 142, 1297–1309. [Google Scholar] [CrossRef][Green Version]
- Dostalova, H.; Holatko, J.; Busche, T.; Rucka, L.; Rapoport, A.; Halada, P.; Nesvera, J.; Kalinowski, J.; Patek, M. Assignment of sigma factors of RNA polymerase to promoters in Corynebacterium glutamicum. AMB Express 2017, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Busche, T.; Patschkowski, T.; Niehaus, K.; Patek, M.; Kalinowski, J.; Wendisch, V.F. Physiological roles of sigma factor SigD in Corynebacterium glutamicum. BMC Microbiol. 2017, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Busche, T.; Silar, R.; Pičmanová, M.; Pátek, M.; Kalinowski, J. Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum. BMC Genom. 2012, 13, 445. [Google Scholar] [CrossRef] [PubMed]
- Patek, M.; Nesvera, J. Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 2011, 154, 101–113. [Google Scholar] [CrossRef]
- Pátek, M.; Holátko, J.; Busche, T.; Kalinowski, J.; Nešvera, J. Corynebacterium glutamicum promoters: A practical approach. Microb. Biotechnol. 2013, 6, 103–117. [Google Scholar] [CrossRef]
- Yim, S.S.; An, S.J.; Kang, M.; Lee, J.; Jeong, K.J. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 2013, 110, 2959–2969. [Google Scholar] [CrossRef]
- Rytter, J.V.; Helmark, S.; Chen, J.; Lezyk, M.J.; Solem, C.; Jensen, P.R. Synthetic promoter libraries for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 2617–2623. [Google Scholar] [CrossRef]
- Abe, S.; Takayarna, K.; Kinoshita, S. Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 1967, 13, 279–301. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Stansen, C.; Uy, D.; Delaunay, S.; Eggeling, L.; Goergen, J.L.; Wendisch, V.F. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 2005, 71, 5920–5928. [Google Scholar] [CrossRef]
- Sgobba, E.; Stumpf, A.K.; Vortmann, M.; Jagmann, N.; Krehenbrink, M.; Dirks-Hofmeister, M.E.; Moerschbacher, B.; Philipp, B.; Wendisch, V.F. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour. Technol. 2018, 260, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Veldmann, K.H.; Dachwitz, S.; Risse, J.M.; Lee, J.H.; Sewald, N.; Wendisch, V.F. Bromination of L-tryptophan in a Fermentative Process With Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2019, 7, 219. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Van der Rest, M.E.; Lange, C.; Molenaar, D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biot. 1999, 52, 541–545. [Google Scholar] [CrossRef]
- Mindt, M.; Walter, T.; Risse, J.M.; Wendisch, V.F. Fermentative Production of N-Methylglutamate From Glycerol by Recombinant Pseudomonas putida. Front. Bioeng. Biotechnol. 2018, 6, 159. [Google Scholar] [CrossRef]
- Miwa, K.; Matsui, H.; Terabe, M.; Nakamori, S.; Sano, K.; Momose, H. Cryptic Plasmids in Glutamic Acid-producing Bacteria. Agric. Biol. Chem. 1984, 48, 2901–2903. [Google Scholar] [CrossRef]
- Youn, J.W.; Jolkver, E.; Kramer, R.; Marin, K.; Wendisch, V.F. Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J. Bacteriol. 2009, 191, 5480–5488. [Google Scholar] [CrossRef]
- Taniguchi, H.; Wendisch, V.F. Exploring the role of sigma factor gene expression on production by Corynebacterium glutamicum: Sigma factor H and FMN as example. Front. Microbiol. 2015, 6, 740. [Google Scholar] [CrossRef]
- Binder, D.; Frohwitter, J.; Mahr, R.; Bier, C.; Grunberger, A.; Loeschcke, A.; Peters-Wendisch, P.; Kohlheyer, D.; Pietruszka, J.; Frunzke, J.; et al. Light-Controlled Cell Factories: Employing Photocaged Isopropyl-beta-d-Thiogalactopyranoside for Light-Mediated Optimization of lac Promoter-Based Gene Expression and (+)-Valencene Biosynthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2016, 82, 6141–6149. [Google Scholar] [CrossRef]
- Wendisch, V.F.; Spies, M.; Reinscheid, D.J.; Schnicke, S.; Sahm, H.; Eikmanns, B.J. Regulation of acetate metabolism in Corynebacterium glutamicum: Transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 1997, 168, 262–269. [Google Scholar] [CrossRef]
- Cramer, A.; Gerstmeir, R.; Schaffer, S.; Bott, M.; Eikmanns, B.J. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 2006, 188, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Auchter, M.; Cramer, A.; Huser, A.; Ruckert, C.; Emer, D.; Schwarz, P.; Arndt, A.; Lange, C.; Kalinowski, J.; Wendisch, V.F.; et al. RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J. Biotechnol. 2011, 154, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.V.; Wendisch, V.F. Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb. Cell Fact. 2013, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer-Sancar, K.; Mentz, A.; Ruckert, C.; Kalinowski, J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genom. 2013, 14, 888. [Google Scholar] [CrossRef]
- Espah Borujeni, A.; Cetnar, D.; Farasat, I.; Smith, A.; Lundgren, N.; Salis, H.M. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017, 45, 5437–5448. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef]
- Kosobokova, E.N.; Skrypnik, K.A.; Kosorukov, V.S. Overview of Fusion Tags for Recombinant Proteins. Biochemistry (Mosc) 2016, 81, 187–200. [Google Scholar] [CrossRef]
- Widakowich, G.; Zhang, C.; Harris, S.; Mitri, K.; Powers, G.; Troung, K.S.; Hearn, M.T. Effects of IMAC specific peptide tags on the stability of recombinant green fluorescent protein. Biotechnol. Prog. 2011, 27, 1048–1053. [Google Scholar] [CrossRef]
- Cooper, C.D.O.; Marsden, B.D. N- and C-Terminal Truncations to Enhance Protein Solubility and Crystallization: Predicting Protein Domain Boundaries with Bioinformatics Tools. In Heterologous Gene Expression in E.coli: Methods and Protocols; Burgess-Brown, N.A., Ed.; Springer: New York, NY, USA, 2017; pp. 11–31. [Google Scholar] [CrossRef]
- Speck, J.; Hecky, J.; Tam, H.K.; Arndt, K.M.; Einsle, O.; Müller, K.M. Exploring the molecular linkage of protein stability traits for enzyme optimization by iterative truncation and evolution. Biochemistry 2012, 51, 4850–4867. [Google Scholar] [CrossRef]
- Sinha, R.; Shukla, P. Current Trends in Protein Engineering: Updates and Progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef]
- Han, S.O.; Inui, M.; Yukawa, H. Expression of Corynebacterium glutamicum glycolytic genes varies with carbon source and growth phase. Microbiology 2007, 153, 2190–2202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Liu, J.; Ni, X.; Lei, Y.; Zheng, P.; Diao, A. Screening efficient constitutive promoters in Corynebacterium glutamicum based on time-series transcriptome analysis. Sheng Wu Gong Cheng Xue Bao 2018, 34, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Development and characterization of expression vectors for Corynebacterium glutamicum. J. Microbiol. Biotechnol. 2014, 24, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Kind, S.; Jeong, W.K.; Schroder, H.; Wittmann, C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 2010. [Google Scholar] [CrossRef] [PubMed]
- Bakkes, P.J.; Ramp, P.; Bida, A.; Dohmen-Olma, D.; Bott, M.; Freudl, R. Improved pEKEx2-derived expression vectors for tightly controlled production of recombinant proteins in Corynebacterium glutamicum. Plasmid 2020, 112, 102540. [Google Scholar] [CrossRef]
- Mindt, M.; Risse, J.M.; Gruß, H.; Sewald, N.; Eikmanns, B.J.; Wendisch, V.F. One-step process for production of N-methylated amino acids from sugars and methylamine using recombinant Corynebacterium glutamicum as biocatalyst. Sci. Rep. 2018, 8, 12895. [Google Scholar] [CrossRef]
| Strain | Characteristics of strains and plasmids | Reference |
|---|---|---|
| C. glutamicum strains | ||
| WT | Wild type, ATCC 13032 | [39] |
| SAR3 | WT ΔaceB icdGTG (pVWEx1-dpkA_RBSopt) (pECXT99A-xylAB) | [11] |
| SAR3* | WT ΔaceB icdGTG (pVWEx1-dpkA_RBSopt) (pECXT-Psyn-xylAB) | This work |
| Other strains | ||
| E. coli DH5α | F-thi-1 endA1 hsdr17(r-, m-) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 | [40] |
| Plasmids | ||
| pEKEx3 | SpecR, PtrclacIq, pBL1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [41] |
| pEKEx3-gfpUV | pEKEx3 derivative for inducible expression of gfpUV from Ptac promoter | [42] |
| pECXT99A (pECXT) | TetR, PtrclacIq, pGA1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [2] |
| pECXT99A-gfpUV | pECXT99A derivative for inducible expression of gfpUV from Ptrc promoter | This work |
| pECXT99A-xylAB | pECXT99A derivative for inducible expression of xylA from Xanthomonas campestris and xylB from C. glutamicum from Ptrc promoter | [43] |
| pECXT-Ppgk | pECXT99A derivative for constitutive expression from C. glutamicum pgk promoter | This work |
| pECXT-PilvC | pECXT99A derivative for constitutive expression from C. glutamicum ilvC promoter | This work |
| pECXT-PsodA | pECXT99A derivative for constitutive expression from C. glutamicum sodA promoter | This work |
| pECXT-PgapA | pECXT99A derivative for constitutive expression from C. glutamicum gapA promoter | This work |
| pECXT-Ptuf | pECXT99A derivative for constitutive expression from C. glutamicum tuf promoter | This work |
| pECXT-PH36 | pECXT99A derivative for constitutive expression from synthetic PH36 promoter | This work |
| pECXT-P45 | pECXT99A derivative for constitutive expression from synthetic P45 promoter | This work |
| pECXT-Psyn | pECXT99A derivative for constitutive expression from synthetic Psyn promoter | This work |
| pECXT-Ppgk-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from C. glutamicum pgk promoter | This work |
| pECXT-PilvC-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from C. glutamicum ilvC promoter | This work |
| pECXT-PsodA-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from C. glutamicum sodA promoter | This work |
| pECXT-PgapA-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from C. glutamicum gapA promoter | This work |
| pECXT-Ptuf-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from C. glutamicum tuf promoter | This work |
| pECXT-PH36-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from synthetic PH36 promoter | This work |
| pECXT-P45-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from synthetic P45 promoter | This work |
| pECXT-Psyn-gfpUV | pECXT99A derivative for constitutive expression of gfpUV from synthetic Psyn promoter | This work |
| pECXT-Psyn-xylAB | pECXT99A derivative for constitutive expression of xylA from Xanthomonas campestris and xylB from C. glutamicum from synthetic Psyn promoter | This work |
| pVWEx1 | KmR, PtaclacIq, pHM1519 oriVCg, C. glutamicum/E. coli expression shuttle vector | [28] |
| pVWEx4 | pVWEx1 derivative with mutation repA | This work |
| pVWEx6 | pVWEx4 derivative with Psyn promoter and lac operator for IPTG inducible expression | This work |
| pVWEx1-gfpUV | pVWEx1 derivative for IPTG-inducible expression of gfpUV from Ptac promoter | [42] |
| pVWEx1-dpkA_RBSopt | pVWEx1 derivative for IPTG-inducible expression of dpkA from P. putida KT2440 and change of its start codon GTG to ATG and an RBS optimised for C. glutamicum | [11] |
| pVWEx4-gfpUV | pVWEx4 derivative for IPTG-inducible expression of gfpUV from Ptac promoter | This work |
| pVWEx6-gfpUV | pVWEx6 derivative for IPTG-inducible expression of gfpUV from Psyn promoter | This work |
| Oligonucleotide | Target | Sequence (5′ → 3′) |
|---|---|---|
| HN12 | Ptuf-fw | CTGTGCGGTATTTCACACCGCAGTTTTAGCGTGTCAGTAGGC |
| HN13 | Ptuf-rv | CCGGGTACCGAGCTCGAATTCCATGTTACTGAATCCTAAGGGCAACG |
| HN14 | PgapA-fw | CTGTGCGGTATTTCACACCGCAGTGTCTGTATGATTTTGCATCTG |
| HN15 | PgapA-rv | CCGGGTACCGAGCTCGAATTCCATGCACGCACCAAACCTACTCACA |
| HN16 | PilvC-fw | CTGTGCGGTATTTCACACCGCAATCCGGACAGATTGCACTCAAC |
| HN17 | PilvC-rv | CCGGGTACCGAGCTCGAATTCCATGCATTATTGTTCTACCACACACATG |
| HN30 | PsodA-fw | CTGTGCGGTATTTCACACCGCATACTTAGCTGCCAATTATTCCG |
| HN31 | PsodA-rv | CCGGGTACCGAGCTCGAATTCCATGCCGCACCGAGCATATACATCT |
| HN97 | Ppgk-fw | CTGTGCGGTATTTCACACCGCATAACGTGGGCGATCGATGC |
| HN98 | Ppgk-rv | CCGGGTACCGAGCTCGAATTCCATGGCCGTACTCCTTGGAGATTTG |
| HA02 | P45-fw | CTGTGCGGTATTTCACACCGCATTGGTCAGGGATTTTTTCCCG |
| HA03 | P45-rv | CCGGGTACCGAGCTCGAATTCCATGGAACTTCTTCGTCACTTACTTTA |
| HA04 | PH36-fw | CTGTGCGGTATTTCACACCGCACAAAAGCTGGGTACCTCTATCTG |
| HA05 | PH36-rv | CCGGGTACCGAGCTCGAATTCCATGCATGCTACTCCTACCAACCAAG |
| HA06 | Psyn-fw | GCGCCTGATGCGGTATTTTCTCCTTACGCATCTGTGCGGTATTTCACACCGCATTGACATTAATTTGAATCTGTGTTAT |
| HA07 | Psyn-rv | CTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCCATGGAACCATTATAACACAGATTCAAA |
| HA36 | repA-fw | AAAATCGCTTGACCATTGCAGGTTG |
| HA37 | repA-rv | CTTTAGCTTTCCTAGCTTGTCGTTGAC |
| HA40 | repA-seq | TGCTCGTCAGACAGAGACGCAG |
| N101 | pVWEx4-fw | ATGCATGCCGCTTCGCCTTCGATTGACATTAATTTGAATCTGTGTTATAATGGTTC |
| N102 | pVWEx4-rv | CGGCCAGTGAATTCGAGCTCGAAATTGTTATCCGCTCACAATTCCAGGAACCATTATAACACAGATTCAA |
| N103 | xylAB-fw | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGAGCAACACCGTTTTCATC |
| N104 | xylAB-rv | CTGCAGGTCGACTCTAGAGGATCTTAGTACCAACCCTGCGTTGC |
| N105 | Psyn-fw2 | ATGCATGCCGCTTCGCCTTCGTTGACATTAATTTGAATCTGTGTTATAATGGTTC |
| N106 | Psyn-rv2 | GGCCAGTGAATTCGAGCTCGCTGCAGGTCGACTCTAGAGGATC |
| HN49 | gfpUV-fw | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGAGTAAAGGAGAAGAACTTTTCA |
| HN50 | gfpUV-rv | GCATGCCTGCAGGTCGACTCTAGAGGATCTTATTTGTAGAGCTCATCCATGC |
| 582 | ATCTTCTCTCATCCTCCA |
| Plasmid | pEKEx3 | pVWEx1 | pVWEx4 | pVWEx6 | pECXT99A | pECXT_Psyn |
|---|---|---|---|---|---|---|
| Expression | Inducible | Inducible | Inducible | Inducible | Inducible | Constitutive |
| Repressor gene | lacIq | lacIq | lacIq | lacIq | lacIq | - |
| Promoter | Ptac | Ptac | Ptac | Psyn | Ptrc | Psyn |
| Induction factor | 6 | 76 | 121 | 51 | 40 | non |
| Maximal expression | 64 | 99 | 145 | 410 | 47 | 140 |
| Origin for Cg | pBL1 | pHM1519 | pHM1519 * | pHM1519 * | pGA1 mini | pGA1 mini |
| Origin for Ec | ColE1 ori | ColE1 ori | ColE1 ori | ColE1 ori | pMB1 | pMB1 |
| Resistance | Spec | Kan | Kan | Kan | Tet | Tet |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henke, N.A.; Krahn, I.; Wendisch, V.F. Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum. Microorganisms 2021, 9, 204. https://doi.org/10.3390/microorganisms9010204
Henke NA, Krahn I, Wendisch VF. Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum. Microorganisms. 2021; 9(1):204. https://doi.org/10.3390/microorganisms9010204
Chicago/Turabian StyleHenke, Nadja A., Irene Krahn, and Volker F. Wendisch. 2021. "Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum" Microorganisms 9, no. 1: 204. https://doi.org/10.3390/microorganisms9010204
APA StyleHenke, N. A., Krahn, I., & Wendisch, V. F. (2021). Improved Plasmid-Based Inducible and Constitutive Gene Expression in Corynebacterium glutamicum. Microorganisms, 9(1), 204. https://doi.org/10.3390/microorganisms9010204







