Abstract
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a rapid and accurate method to identify microorganisms in clinical laboratories. This study isolates yeast-like microorganisms in the oral washes that are collected from non-bedridden nursing home residents, using CHROMagar Candida plates, and identifies them using Bruker MALDI-TOF MS. The ribosomal DNA sequences of the isolates are then examined. Three hundred and twenty yeast isolates are isolated from the oral washes. Candida species form the majority (78.1%), followed by Trichosporon/Cutaneotrichosporon species (8.8%). Bruker MALDI-TOF MS gives a high-level confidence, with a log(score) value of ≥1.8, and identifies 96.9% of the isolates. There are six inconclusive results (1.9%), and those sequences are verified as rare clinical species, including Candida ethanolica, Cutaneotrichosporon jirovecii, Exophiala dermatitidis, and Fereydounia khargensis. Almost all of the isolates have a regular color on the CHROMagar Candida plates. If the colonies are grouped by color on the plates, a specific dominant yeast species is present in each color group, except for purple or orange isolates. In conclusion, MALDI-TOF MS is verified as a fast, accurate and practical method to analyze oral yeasts in elderly subjects.
1. Introduction
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a rapid and accurate method to identify microorganisms in a clinical microbiology laboratory [1,2,3]. The process compares the proteomic fingerprint of an unknown microorganism, which mainly consists of ribosome proteins, to those in a reference spectral database. The peak profiles that are generated by Bruker MALDI-TOF MS are matched to reference libraries using the integrated patterns matching algorithm, BioTyper software (Bruker Daltonics, Bremen, Germany). This gives an arbitrary score value of 0 to 3.0 to represent the similarity between the sample and the reference spectrum. A log(score) of ≥2.0 represents successful identification of a species, and a score of 1.7 to 2.0 is acceptable for the genus level. Scores higher than 1.8 represent highly-accurate yeast identification [4,5]. Cultivation is inevitable if microorganisms are identified by MALDI-TOF MS, but the process only requires a minute number of cells, and much less culture time. Along with the development of microbial culturomics, MALDI-TOF MS plays an effective complementary role in microbiome analysis using metagenomics [6,7].
The elderly population (60 years or over) is estimated by the WHO to grow from 12.7% in 2017 to 21.3% in 2050 globally, and from 12.2% to 24.2% in Asia [8]. Therefore, the healthcare needs of elderly people are increasingly significant. A recent study on salivary microbiota in the elderly shows that general frailty is associated with composition and formulation of oral microbiota [9]. More than 100 trillion microbial cells are estimated to inhabit a healthy human body [10]. In the oral cavity, microbes live in the biofilm on oral tissues to protect against environmental stress and host immunity [11]. A dysbiotic imbalance of the indigenous oral microbial community is a cause of oral diseases, including early childhood caries, HIV-associated periodontitis, and rheumatoid arthritis [12,13,14]. A connection between distinct oral microbiota has also been observed in patients with oropharyngeal squamous cell carcinoma [15].
The oral microbiota is a complex community that is primarily composed of bacteria and fungi. Oral mycobiota are more diverse and complex than any other body site in healthy humans [16]. The intake of environmental fungi through food and breathing undoubtedly contributes to the greater fungal diversity in the oral cavity [17]. Thirteen taxa, including the prevalent Candida species, Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, and Cryptococcus, are the core mycobiota in healthy individuals [18]. However, oral mycobiota was observed to change if the host immunity changes: Candida species are particularly dominant in immunosuppressed solid organ transplant recipients [16]. In terms of the interaction dynamic between resident fungi and the innate immunity, it is worth examining the salivary mycobiota, especially the yeasts, of non-bedridden elderly subjects.
Mycobiota dysbiosis is an important factor in oral diseases, so a practical method to analyze the changes in yeast populations that are collected from saliva or oral washes is necessary. Chromogenic agar is a practical and effective tool to isolate common yeast in clinical specimens [19,20], but the reported specificity is 62% for bronchial secretion samples [21]. The color and morphology of colonies that are isolated on CHROMagar Candida is critical to the presumptive identification of clinically-important Candida species [12,21,22]. It is difficult to differentiate non-Candida yeasts using CHROMagar Candida plates, but the process allows the rapid detection of common Candida species in mycobiota [2,5].
CHROMagar Candida allows a presumptive identification of common Candida species, but the diversity of yeast in oral washes extends far beyond the genus Candida. A recent study by the authors demonstrated that Candida albicans is majorly cultured from oral washes in a high percentage (52 of 204, 25.5%) of nursing residents, but half of the residents (106/204, 52%) have at least one non-C. albicans yeast species [22]. Metagenomics with ribosomal DNA amplification is widely used to characterize of microbiota or mycobiota. MALDI-TOF MS is a rapid and accurate method to identify microorganisms in clinical laboratories [1,2,3]. This study compared different approaches to characterize all the oral yeast isolates, which are single-colony purified by CHROMagar Candida from oral washes that are collected from non-bedridden elderly subjects in nursing care institutions [22]. The results demonstrate that the Bruker MALDI-TOF MS is an effective and highly specific method for the identification of oral yeast.
2. Materials and Methods
2.1. Oral Yeast Isolates
This study was approved by the Human Experiment and Ethics Committee of the National Health Research Institute (EC1040411-E). A total of 204 residents of ≥50 years (average age: 77.1; 153 individuals >70 years old) in 10 nursing homes in central Taiwan were enrolled, after giving informed consent. The sampling method has been reported previously [22]. Figure 1 shows the 20 mL oral rinses containing saline that were collected from each person. These rinses were then centrifuged and resuspended in 1 mL of sterile saline. A total of 50 μL of suspension of each sample was streaked onto a CHROMagar Candida plate (CHROMagar Company, Paris, France) and incubated at 35 °C for 2 days. If present, colonies from each plate were selected for further analysis. Yeast colonies were identified according to the manufacturer’s instructions or the description of the morphology by Odds and Bernaerts [23].
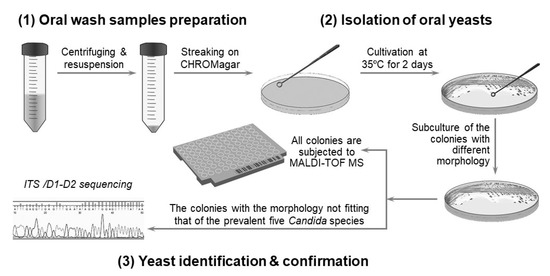
Figure 1.
Scheme for the identification of oral yeast for the study. The strategy to isolate and identify yeast involves: (1) preparation of oral wash samples, (2) yeast isolation, and (3) identification and confirmation of the yeast.
2.2. Bruker MALDI-TOF MS
A full loop of colonies was selected and suspended in 300 μL of deionized water containing 900 μL of absolute ethanol. Following centrifugation, the pellet was then dissolved in 50 μL of 70% formic acid and mixed in a vortex with 50 μL of acetonitrile. The samples were centrifuged, and 1 μL of the clear supernatant was spotted onto the MALDI target plate and air-dried at room temperature. Each spot was then overlaid with 1 μL of CHCA and completely air-dried before the Bruker MALDI-TOF MS measurement. The microorganism was identified, and data analyses were performed using the Bruker LT microflex MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) with Bruker BioTyper 3.0 system software (Bruker Daltonics) [24,25]. A MALDI Biotyper log(score) value >2.0 represents reliable identification at the genus level and a score <1.7 represents ambiguous identification [4,5].
2.3. Sequence Analysis of Ribosomal DNA
The genomic DNA of yeast was extracted and internal transcribed spacers (ITS) or D1/D2 amplification for identifications was used, as previously described [22,26]. The thermal cycling parameters were an initial denaturation for 5 min at 95 °C and 30 cycles of 1 min at 95 °C, 1 min at 55 °C, and 1 min at 72 °C. A final extension at 72 °C for 10 min was performed at the end of the amplification. The amplicons were purified and then sequenced by the Genomic Medicine Research Core Laboratory of Chang Gung Memorial Hospital Genomic Medicine Core Laboratory, Chang Gung Memorial Hospital. The nucleotide sequence results were analyzed using BLAST searches with default settings against the nonredundant (NR) database at NCBI, the ISHAM-ITS database, and the Candida Genome Database (CGD). The best BLAST hit for the query sequence of each amplicon was used to identify the isolates.
2.4. Statistic Analysis
An unpaired Student’s t-test was used to compare the value of the Biotyper score: a p value < 0.05 was considered to be significant.
3. Results
3.1. Isolation and Identification of Oral Yeast
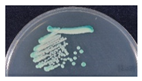
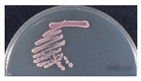
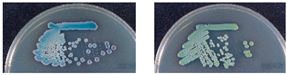
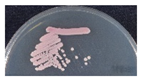
To isolate and identify the oral yeast isolates of non-bedridden elderly residents in nursing homes, the oral wash samples were collected and cultivated on CHROMagar Candida plates. Isolates with different morphologies on each plate were collected, and 320 yeast-like isolates were collected in total. All the fresh colonies of those isolates were subjected to Bruker MALDI-TOF MS (Figure 1). The genus, Candida (250 isolates, 78.1%), comprised the majority, followed by Trichosporon/Cutaneotrichosporon (28 isolates, 8.8%) (Figure 2 and Table 1). C. albicans was the most abundant (129 isolates, 40.3%) and Trichosporon asahii (18 isolates, 5.6%) was the largest species of the non-Candida isolates. In the non-Candida/non-Trichosporon group, the probiotic yeast Saccharomyces cerevisiae (7 isolates), accounted for 19.4% of the isolates in the rare species of yeasts, followed by Meyerozyma guilliermondii (6 isolates; anamorph, C. guilliermondii), and others (Figure 2 and Table 1).
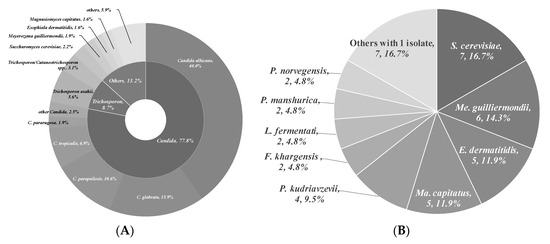
Figure 2.
Distribution of commensal oral yeasts in the residents of nursing homes. All identified yeasts were grouped together into taxa and sorted according to population size (A). The yeast isolates in the group “Others” do not belong to the genus Candida or Trichosporon/Cutaneotrichosporon. Details are shown in (B).

Table 1.
List of isolated yeasts.
3.2. Accuracy of Bruker MALDI-TOF MS Identification
Of the 320 yeast isolates, only six isolates could not be positively identified by Bruker MALDI-TOF MS with the BioTyper system. All yeast isolates were further ITS- or D1/D2-sequence verified, unless the colony color and morphology fully matched the patterns of the five major Candida species on CHROMagar Candida. A total of 302 isolates had log(score) values of more than 2.0, which represents accurate identification. Of the remaining isolates with a log(score) value of more than 1.8, six isolates showed the same results for Bruker MALDI-TOF MS and ITS-sequencing, and two were identified using Biotyper as Candida guilliermondii var. membranifaciens, which matches the corresponding anamorph Kodamaea ohmeri that was identified by ITS sequencing [27]. One isolate was identified using Biotyper as Candida fermentati, with a low log(score) value of 1.707, which is the anamorph of the ITS-sequencing result Meyerozyma caribbica. In summary, 314 of 320 isolates were accurately identified by the Bruker MALDI-TOF MS Biotyper system. The main spectrum patterns of common Candida species (C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis) and Trichosporon asahii were analyzed individually (Figures S1–S5), and the results show some highly conservative peaks that could be used as biomarker patterns for dominant yeasts in oral cavity. Only six oral wash isolates were untypable, including two each of Exophiala dermatitidis and Fereydounia khargensis, and one each of Candida ethanolica and Cutaneotrichosporon jiroveci (formerly Trichosporon jiroveci) (Table 2), which were identified by ITS or D1/D2 sequencing. Candida ethanolica and Fereydounia khargensis are not yet included in the MALDI Biotyper database version 3, which was used for this study. The results convincingly show that Bruker MALDI-TOF MS allows the effective identification of common or rare commensal, environmental yeast species of oral yeast.

Table 2.
Oral yeasts in the residents of nursing homes.
3.3. Rapid Differentiation of Yeasts by CHROMagar Candida
CHROMagar Candida plates were used to screen yeast-like colonies for this study. The color and morphology of the colonies that were isolated on CHROMagar Candida are critical for the presumptive identification of clinically-important Candida species [20,23,28]. According to the results using Bruker MALDI-TOF MS, 311 of all 320 isolates (97.5%) on CHROMagar Candida had a color that matched the reported color [20,29,30,31,32,33]. Most Candida species, especially the prevalent C. albicans (100%), C. glabrata (100%), C. parapsilosis (94.1%), and C. tropicalis (90.9%), very closely matched the color corresponding to the species on CHROMagar (Table 3), as did non-Candida yeasts, such as T. asahii (100%) (Table 4).

Table 3.
Appearance of Candida species 1 in the oral samples on CHROMagar Candida plates.

Table 4.
Appearance of non-Candida species 1 in the oral samples on CHROMagar Candida plates.
For all color-matched isolates, six color groups were identified: green, purple/pink, blue, white, gray, and orange. Almost all green colonies were C. albicans (129/130 isolates, 99.2%). C. parapsilosis accounted for more than 80% of the white group, and E. dermatitidis accounted for more than 70% of the gray colonies. In the blue color group, C. tropicalis (20/42 isolates, 47.6%) and T. asahii (18/42, 42.9%) were the main two species. In the purple/pink color group, there were 15 species, including fifty-one of C. glabrata, seven of S. cerevisiae, six of Cutaneotrichosporon mucoides (formerly Trichosporon mucoides), five of M. capitatus, four each of C. krusei (teleomorph Pichia kudriavzevii), C. pararugosa, and Me. guilliermondii (anamorph C. guilliermondii), two of P. manshurica, and one each of C. fermentati, C. intermedia, C. ethanolica, C. metapsilosis, C. utilis (teleomorph, Cyberlindnera jadinii), Clavispora lusitaniae (anamorph C. lusitaniae), and Kluyveromyces marxianus (anamorph, C. kefyr) (Table 5). A wide range of species had a purple/pink color on CHROMagar Candida, but the most abundant species was C. glabrata (51/90, 56.7%) (Table 5).

Table 5.
Proportions of color-matched species on CHROMagar Candida plates.
4. Discussion
4.1. Commensal Oral Yeasts in the Elderly
A culture-based mycobiome analysis of oral samples from elderly subjects (age above 65) in New Zealand showed that frequently isolated species included C. albicans, C. glabrata, C. parapsilosis, Clavispora lusitaniae (anamorph C. lusitaniae), Meyerozyma guilliermondii (anamorph, C. guilliermondii), Pichia fermentans, and Yarrowia lipolytica [34]. P. fermentans and Y. lipolytica were not identified by this study, but a significant population (28/55, 50.9% in non-Candida group) was of the group Trichosporon, which belongs to human skin commensals and is also frequently found in the mouths of healthy individuals [35]. The very large population of the Trichosporonaceae family (including Trichosporon and Cutaneotrichosporon species) in oral microbiota that were collected from elderly subjects in nursing homes in central Taiwan shows that the Trichosporon group is common in the environment. In a study that used samples from one-month old infants, oral mycobiome has a significantly lower alpha diversity than that of skin and anal mycobiomes, which shows that the environment of the oral cavity selects commensal fungi, including Candida, Saccharomyces, and Cladosporium, and that the mycobiome depends more on the environment than the mother’s vaginal mycobiome [36]. Candida yeasts are a type of human oral commensal that are mostly located on the tongue [37]. The carriage rate for C. albicans in the mouths of healthy individuals is estimated to be 17.7%, but this figure increases to 40.6% in hospitalized individuals [38], so health is a predisposing factor for C. albicans colonization in the oral cavity [39]. In addition to the prevalent C. albicans, C. glabrata, C. krusei (teleomorph P. kudriavzevii), C. tropicalis, and C. parapsilosis have been isolated from the oral cavity of healthy individuals [37,39,40].
Health professionals should be alerted to the presence of a biofilm that is associated with the Candida species, especially in those with reduced antifungal susceptibility. Disorders of the oral cavity are common in senior citizens who have no underlying health conditions, including dental caries, gingivitis, periodontitis, xerostomia, denture stomatitis, and candidiasis [41]. Oral disorders can contribute to poor nutritional status and health and have a negative effect on immunity in the elderly. Dental prostheses also reduce salivary flow and predispose elderly individuals to microbial colonization and biofilm formation, which results in candidiasis.
4.2. Limitation of MALDI-TOF MS, Chromogenic Agar, and Ribosomal DNA Sequencing in Microbial Pathogenesis
The results of high-throughput amplicon sequencing and shotgun metagenomics show that there are 12 million species of fungi, of which only 2.2–3.8 million species are reported to be cultivable [42]. There has been a rapid growth in sequence data, and mycobiota/microbiota dysbiosis has been identified as a critical factor in some human diseases [43,44,45]. Some bioinformatics analyses using metagenomics show the importance of some alteration of mycobiota, but pure isolates are required to determine the pathogenesis of candidate pathogens. Ribosomal DNA sequencing and MALDI-TOF MS identification all require pure cultures. Chromogenic agar plates allow better identification in a clinical laboratory because they are selective and differential. CHROMagar Candida, which is a presumptive identification method for C. albicans, C. tropicalis, C. glabrata, C. krusei (teleomorph P. kudriavzevii), and Trichosporon spp. [32], does not distinguish many environmental yeasts or some clinically-relevant yeasts. The accumulation of MS profiles for reference fungal isolates means that MALDI-TOF MS is an effective identification method for clinical examination. The Bruker database comprises 604 references for yeast and fungus and includes almost all known pathogenic fungi [46], but mix-cultures are usually misidentified in term of the microorganisms, and only monomicrobial cultures can be specifically identified [47]. In clinical, the limitation of MALDI-TOF MS is to identify different species in a sample with mix species. Recently, mix-microbial identification by MALDI-TOF MS has used specific algorithms to determine the mass spectra biomarkers for microorganisms [47]. Meanwhile, liquid chromatography–tandem MS is also used to identify mix-microbial samples [48,49]. As the mass spectral database grows significantly, yeast identification using MS, including identification of mix-culture, would be expected in the near future.
The limitation of both rDNA sequencing and MALDI-TOF MS is that they identify known species. Even though rDNA sequence is simple, it takes longer preparation time than MALDI-TOF MS. Nevertheless, the MALDI-TOF MS equipment and database are not available to all laboratories. For this study, six isolates cannot be typed using Bruker MALDI-TOF MS, including two yeast species that are not included in the MALDI Biotyper database—one C. ethanolica isolate and two F. khargensis isolates—and two yeast species that are shown in the database—one Cu. jiroveci isolate and two E. dermatitidis isolates. That Cu. jiroveci isolate was MALDI-TOF MS identified, but its log(score) value was 1.703, which is less than the identification threshold (1.8), is probably due to the limited number of isolates in the database. Five of seven E. dermatitidis isolates were identified by MALDI-TOF MS, with a wide range of scores: 2.072, 1.967, 1.951, 1.861, and 1.792, but two were MS untyped. The diversity and polymorphism of E. dermatitidis [50] might contribute to the variation in MS patterns.
4.3. Mycobiome Analysis Using Culture- and Non-Culture Methods
Advances in DNA sequencing technology mean that many microbiota and mycobiota analyses can be conducted without the need for time-consuming cultivation. However, the oral cavity is the entrance for food, drink, and air, so many environmental microbes or debris can be detected using a non-culture method. This study shows that Trichosporon is the second largest genus in the oral cavity of elderly subjects, but it is not widely distributed in samples from a previous ITS-sequencing mycobiome study that was conducted in the Cleveland area on standard Western diets [18]. Oral mycobiota is affected by environment, drink, and food styles, but the difference in yeast taxa for this study and others that use ITS-sequencing is probably due to cultivation. Malassezia yeast is a skin commensal yeast that has been reported in oral mycobiota using sequencing methods, but is not detected in oral microorganisms using culture methods, because the fatty acid synthesis pathway is incomplete [51]. Therefore, a cohort study to determine whether the culture-based MALDI-TOF MS method is compatible with analysis using DNA extraction-based sequencing analysis would be of use.
4.4. Using of CHROMagar Candida to Differentiate Yeast Species in the Oral Cavity
This study uses chromogenic agar plates to screen yeast-like microorganisms. CHROMagar Candida in isolation allows the presumptive diagnosis of clinically-important Candida species [20,28,34,37,52]. The colony color and the morphology of the isolates are categorized in Table 5. Of all colony color groups in the oral samples of elderly subjects, green precisely differentiates C. albicans from others (129/130 isolates, 99.2%) and white color represents most of the C. parapsilosis (32/38 isolates, 84.2%). C. tropicalis and T. asahii contribute to nearly half of the blue colonies, but the purple or pink colonies show no majority. The color category for this study indeed allows reliable analysis of oral mycobiome.
4.5. Use of MALDI-TOF MS to Analyze Oral Mycobiome
The presence of Candida strains with reduced susceptibility to antifungal agents, such as reduced susceptibility to azoles in C. glabrata and C. krusei (teleomorph P. kudriavzevii) and reduced susceptibility to flucytosine in C. krusei and C. rugosa, could pose a problem for immune-compromised elderly subjects who require drug treatment [34]. Therefore, fast and accurate diagnosis of yeast species is important for geriatric medicine. MALDI-TOF MS is a rapid, reliable, economical, and environmentally-friendly method for routine microbial identification. It requires a shorter culture time, because a tiny sample is sufficient, so it can be used to instigate timely and appropriate antibiotic treatment in a clinical setting [3,5]. However, oral mycobiota are highly diverse and contain some rare yeast species, such as Trichosporon jirovecii (current name: Cutaneotrichosporon jirovecii) [53] and Lachancea fermentati [54] (Table 2), which is in contrast with clinical samples. Only species with the morphology of frequently encountered, disease-associated fungi are familiar to physicians or medical technicians, so accurate diagnosis of yeast or molds is difficult. MALDI-TOF MS with a Bruker Biotyper system was used to analyze yeast taxa in the elderly subjects for this study, and a very high identification rate was achieved for oral yeasts (314/320 = 98.1%), with only six yeasts note being untyped. C. albicans and C. glabrata have a significantly higher Biotyper score (mean score = 2.34 and 2.35 respectively, Figure 3), as well as Trichosporon species (mean score = 2.23. This was probably because the Biotyper database contains a large number of MS profiles for Candida and Trichosporon species. As the number of MS profiles of fungi that can be cultured increases, MALDI-TOF MS will become an increasingly effective means of microbial identification.
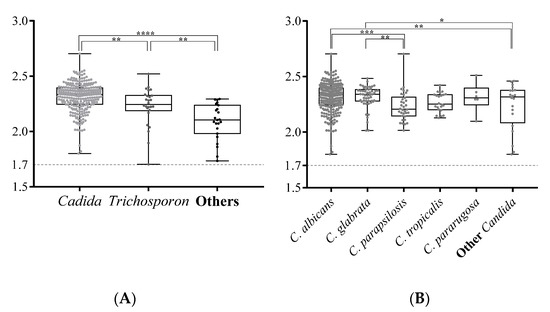
Figure 3.
The log(score) values for yeast isolates that are identified using the MALDI-TOF Bruker Biotype. The Biotyper log(score) values for Candida, Trichosporon, and other genuses are compared in (A). All scores for the isolates of the Candida species that are identified by this study are compared (B). The box plots show the diversity of the log(score) values for the yeast isolates. The upper and lower edges of the boxes correspond to the first and third quartiles. The median is represented by a horizontal line within the box. The whiskers extend from the box in order from the highest to lowest. Each score is marked by a dot. The log(score) values are compared using an unpaired Student’s t-test; a p-value <0.05 is represented as *, <0.01 as **, <0.001 as ***, and <0.0001 as ****.
5. Conclusions
This study used CHROMagar Candida to grow oral yeast colonies, which were then identified using Bruker MALDI-TOF MS. This combination better discriminates yeasts that can be cultured, and allows the study of yeasts that are associated with disease. Bruker MALDI-TOF-MS is shown to be a reliable and rapid method for the identification of yeast isolates in the oral washes of elderly residents in nursing homes, especially those of the Candida and Trichosporon species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/142/s1, Supplementary figures contains overlaid mass spectra with smoothing and baseline subtraction for Candida albicans isolates (Figure S1), Candida grablata isolates (Figure S2), Candida parapsilosis isolates (Figure S3), Candida tropicalis isolates (Figure S4) and Trichosporon asahii isolates (Figure S5).
Author Contributions
J.-J.L. and H.-J.L. made equal contributions and edited the manuscript. M.-J.C., C.-H.L., C.-C.L., and Y.-Z.C. collected samples, isolated yeast samples, and analyzed the ribosomal DNA sequence. M.-H.T. assisted with editing the manuscript. S.-H.W. designed the study, analyzed data, and wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly supported by grant no. 108-2314-B-415-002 from Ministry of Science and Technology (MOST), Taiwan to S.-H.W., by grants nos. 04A1 IVPP04-014, and 05A1 IVPP04-014 from the National Health Research Institutes to H.-J.L. and by grant nos. CMRPG3F1722, CMRPG3H0122 and CMRPG3H0123 from Chang Gung Memorial Hospital to J.-J.L. And The APC was funded by MOST 109-2314-B-415-003.
Institutional Review Board Statement
The study was approved by the Research Ethics Committee of the National Health Research Institutes (EC1040411-E).
Informed Consent Statement
204 participating nursing home residents in central Taiwan were enrolled after informed consents were obtained.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
All authors declare that they have no conflict of interest.
References
- Van Belkum, A.; Welker, M.; Pincus, D.; Charrier, J.P.; Girard, V. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: What are the current issues? Ann. Lab. Med. 2017, 37, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Wattal, C.; Oberoi, J.K.; Goel, N.; Raveendran, R.; Khanna, S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.C.; Kuo, A.J.; Liu, K.L.; Wen, Y.H.; Chia, J.H.; Chang, P.Y.; Lee, M.H.; Wu, T.L.; Chang, S.C.; Lu, J.J.; et al. Routine identification of microorganisms by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: Success rate, economic analysis, and clinical outcome. J. Microbiol. Immunol. Infect. 2017, 50, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Cassagne, C.; Normand, A.C.; L’Ollivier, C.; Ranque, S.; Piarroux, R. Performance of MALDI-TOF MS platforms for fungal identification. Mycoses 2016, 59, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Pulcrano, G.; Iula, D.V.; Vollaro, A.; Tucci, A.; Cerullo, M.; Esposito, M.; Rossano, F.; Catania, M.R. Rapid and reliable MALDI-TOF mass spectrometry identification of Candida non-albicans isolates from bloodstream infections. J. Microbiol. Methods 2013, 94, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Sung, J.Y.; Hwang, Y.; Shin, M.H.; Park, M.S.; Lee, S.H.; Yong, D.; Lee, K. Utility of conventional culture and MALDI-TOF MS for identification of microbial communities in bronchoalveolar lavage fluid in comparison with the Gs junior next generation sequencing system. Ann. Lab. Med. 2018, 38, 110–118. [Google Scholar] [CrossRef]
- United Nations. World Population Ageing 2017: Highlights; United Nations: New York, NY, USA, 2017. [Google Scholar]
- Ogawa, T.; Hirose, Y.; Honda-Ogawa, M.; Sugimoto, M.; Sasaki, S.; Kibi, M.; Kawabata, S.; Ikebe, K.; Maeda, Y. Composition of salivary microbiota in elderly subjects. Sci. Rep. 2018, 8, 414. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Kumar, P.S. Oral microbiota and systemic disease. Anaerobe 2013, 24, 90–93. [Google Scholar] [CrossRef]
- Anil, S.; Anand, P.S. Early childhood caries: Prevalence, risk factors, and prevention. Front. Pediatr. 2017, 5, 157. [Google Scholar] [CrossRef] [PubMed]
- Noguera-Julian, M.; Guillen, Y.; Peterson, J.; Reznik, D.; Harris, E.V.; Joseph, S.J.; Rivera, J.; Kannanganat, S.; Amara, R.; Nguyen, M.L.; et al. Oral microbiome in HIV-associated periodontitis. Medicine 2017, 96, 5821. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zheng, L.; Qing, P.; Zhao, H.; Li, Y.; Su, L.; Zhang, Q.; Zhao, Y.; Luo, Y.; Liu, Y.; et al. Oral microbiota perturbations are linked to high risk for rheumatoid arthritis. Front. Cell Infect. Microbiol. 2019, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dongari-Bagtzoglou, A. Shaping the oral mycobiota: Interactions of opportunistic fungi with oral bacteria and the host. Curr. Opin. Microbiol. 2015, 26, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Iliev, I.D. The mycobiota: Interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014, 14, 405–416. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, 1000713. [Google Scholar] [CrossRef]
- Rank, E.L. Chromogenic agar media in the clinical, food, and environmental testing arenas, part I. Clin. Microbiol. Newsl. 2012, 34, 43–47. [Google Scholar] [CrossRef]
- Tornai-Lehoczki, J.; Peter, G.; Dlauchy, D. CHROMagar Candida medium as a practical tool for the differentiation and presumptive identification of yeast species isolated from salads. Int. J. Food Microbiol. 2003, 86, 189–200. [Google Scholar] [CrossRef]
- Scharmann, U.; Kirchhoff, L.; Chapot, V.L.S.; Dziobaka, J.; Verhasselt, H.L.; Stauf, R.; Buer, J.; Steinmann, J.; Rath, P.M. Comparison of four commercially available chromogenic media to identify Candida albicans and other medically relevant Candida species. Mycoses 2020, 63, 823–831. [Google Scholar] [CrossRef]
- Tu, M.G.; Lin, C.C.; Chiang, Y.T.; Zhou, Z.L.; Lu, J.J.; Hsieh, L.Y.; Chen, K.T.; Chen, M.C.; Lin, H.C.; Sun, P.L.; et al. Distribution of yeast species and risk factors of oral colonization among the residents at nursing homes in Taiwan. J. Dent. Oral Health 2019, 6, 1–15. [Google Scholar]
- Odds, F.C.; Bernaerts, R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 1994, 32, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Shen, M.; Lin, H.C.; Sun, P.L.; Lo, H.J.; Lu, J.J. Molecular epidemiology of invasive Candida albicans at a tertiary hospital in northern Taiwan from 2003 to 2011. Med. Mycol. 2015, 53, 828–836. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, J.J.; Lo, H.J.; Wu, Y.M.; Chang, J.Y.; Chen, Y.Z.; Wang, S.H. DST659 genotype of Candida albicans showing positive association between biofilm formation and dominance in Taiwan. Med. Mycol. 2018, 56, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Leaw, S.N.; Chang, H.C.; Sun, H.F.; Barton, R.; Bouchara, J.P.; Chang, T.C. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 2006, 44, 693–699. [Google Scholar] [CrossRef]
- Al-Sweih, N.; Khan, Z.U.; Ahmad, S.; Devarajan, L.; Khan, S.; Joseph, L.; Chandy, R. Kodamaea ohmeri as an emerging pathogen: A case report and review of the literature. Med. Mycol. 2011, 49, 766–770. [Google Scholar] [CrossRef]
- Powell, H.L.; Sand, C.A.; Rennie, R.P. Evaluation of CHROMagar Candida for presumptive identification of clinically important Candida species. Diagn. Microbiol. Infect. Dis. 1998, 32, 201–204. [Google Scholar] [CrossRef]
- Tavanti, A.; Davidson, A.D.; Gow, N.A.; Maiden, M.C.; Odds, F.C. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 2005, 43, 284–292. [Google Scholar] [CrossRef]
- Giammanco, G.M.; Melilli, D.; Pizzo, G. Candida pararugosa isolation from the oral cavity of an Italian denture wearer. Res. Microbiol. 2004, 155, 571–574. [Google Scholar] [CrossRef]
- Eraso, E.; Sahand, I.H.; Villar-Vidal, M.; Marcos, C.; Moragues, M.D.; Madariaga, L.; Ponton, J.; Quindos, G. Usefulness of Candida ID2 agar for the presumptive identification of Candida dubliniensis. Med. Mycol. 2006, 44, 611–615. [Google Scholar] [CrossRef]
- Ghelardi, E.; Pichierri, G.; Castagna, B.; Barnini, S.; Tavanti, A.; Campa, M. Efficacy of chromogenic Candida agar for isolation and presumptive identification of pathogenic yeast species. Clin. Microbiol. Infect. 2008, 14, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Pihet, M.; Carrere, J.; Cimon, B.; Chabasse, D.; Delhaes, L.; Symoens, F.; Bouchara, J.P. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—A review. Med. Mycol. 2009, 47, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Thiyahuddin, N.M.; Lamping, E.; Rich, A.M.; Cannon, R.D. Yeast species in the oral cavities of older people: A comparison between people living in their own homes and those in rest homes. J. Fungi 2019, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Silva, R.B.; Fusco-Almeida, A.M.; Matsumoto, M.T.; Baeza, L.C.; Benaducci, T.; Mendes-Giannini, M.J. Genetic diversity and antifungal susceptibility testing of Trichosporon asahii isolated of intensive care units patients. Braz. J. Microbiol. 2008, 39, 585–592. [Google Scholar] [CrossRef]
- Ward, T.L.; Dominguez-Bello, M.G.; Heisel, T.; Al-Ghalith, G.; Knights, D.; Gale, C.A. Development of the human mycobiome over the first month of life and across body sites. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Sato, T.; Kishi, M.; Suda, M.; Sakata, K.; Shimoda, H.; Miura, H.; Ogawa, A.; Kobayashi, S. Prevalence of Candida albicans and non-albicans on the tongue dorsa of elderly people living in a post-disaster area: A cross-sectional survey. BMC Oral Health 2017, 17, 51. [Google Scholar] [CrossRef]
- Calderone, R.; Suzuki, S.; Cannon, R.; Cho, T.; Boyd, D.; Calera, J.; Chibana, H.; Herman, D.; Holmes, A.; Jeng, H.W.; et al. Candida albicans: Adherence, signaling and virulence. Med. Mycol. 2000, 38, 125–137. [Google Scholar] [CrossRef][Green Version]
- Odds, F.C. Candida and Candidosis: A Review and Bibliography; Bailliere Tindall: London, UK, 1988. [Google Scholar]
- Yang, Y.; Leaw, S.; Wang, A.; Chen, H.; Cheng, W.; Lo, H. Characterization of yeasts colonizing in healthy individuals. Med. Mycol. 2011, 49, 103–106. [Google Scholar] [CrossRef][Green Version]
- Gonsalves, W.C.; Wrightson, A.S.; Henry, R.G. Common oral conditions in older persons. Am. Fam. Phys. 2008, 78, 845–852. [Google Scholar]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Domingue, J.C.; Sears, C.L. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin. Immunol. 2017, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Normand, A.C.; Becker, P.; Gabriel, F.; Cassagne, C.; Accoceberry, I.; Gari-Toussaint, M.; Hasseine, L.; De Geyter, D.; Pierard, D.; Surmont, I.; et al. Validation of a new web application for identification of fungi by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2017, 55, 2661–2670. [Google Scholar] [CrossRef] [PubMed]
- Mortelmaier, C.; Panda, S.; Robertson, I.; Krell, M.; Christodoulou, M.; Reichardt, N.; Mulder, I. Identification performance of MALDI-ToF-MS upon mono-and bi-microbial cultures is cell number and culture proportion dependent. Anal. Bioanal. Chem. 2019, 411, 7027–7038. [Google Scholar] [CrossRef]
- Lasch, P.; Schneider, A.; Blumenscheit, C.; Doellinger, J. Identification of microorganisms by liquid chromatography-mass spectrometry (LC-MS(1)) and in silico peptide mass libraries. Mol. Cell Proteom. 2020, 19, 2125–2138. [Google Scholar] [CrossRef]
- Boulund, F.; Karlsson, R.; Gonzales-Siles, L.; Johnning, A.; Karami, N.; Al-Bayati, O.; Ahren, C.; Moore, E.R.B.; Kristiansson, E. Typing and characterization of bacteria using bottom-up tandem mass spectrometry proteomics. Mol. Cell Proteom. 2017, 16, 1052–1063. [Google Scholar] [CrossRef]
- Kirchhoff, L.; Olsowski, M.; Rath, P.M.; Steinmann, J. Exophiala dermatitidis: Key issues of an opportunistic fungal pathogen. Virulence 2019, 10, 984–998. [Google Scholar] [CrossRef]
- Diaz, P.I.; Hong, B.Y.; Dupuy, A.K.; Strausbaugh, L.D. Mining the oral mycobiome: Methods, components, and meaning. Virulence 2017, 8, 313–323. [Google Scholar] [CrossRef]
- Agwu, E.; Ihongbe, J.C.; McManus, B.A.; Moran, G.P.; Coleman, D.C.; Sullivan, D.J. Distribution of yeast species associated with oral lesions in HIV-infected patients in Southwest Uganda. Med. Mycol. 2012, 50, 276–280. [Google Scholar] [CrossRef]
- Colombo, A.L.; Padovan, A.C.; Chaves, G.M. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin. Microbiol. Rev. 2011, 24, 682–700. [Google Scholar] [CrossRef] [PubMed]
- Urubschurov, V.; Janczyk, P. Biodiversity of yeasts in the gastrointestinal ecosystem with emphasis on its importance for the host. In The Dynamical Processes of Biodiversity-Case Studies of Evolution and Spatial Distribution; IntechOpen: London, UK, 2011. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).