Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms
Abstract
1. Introduction
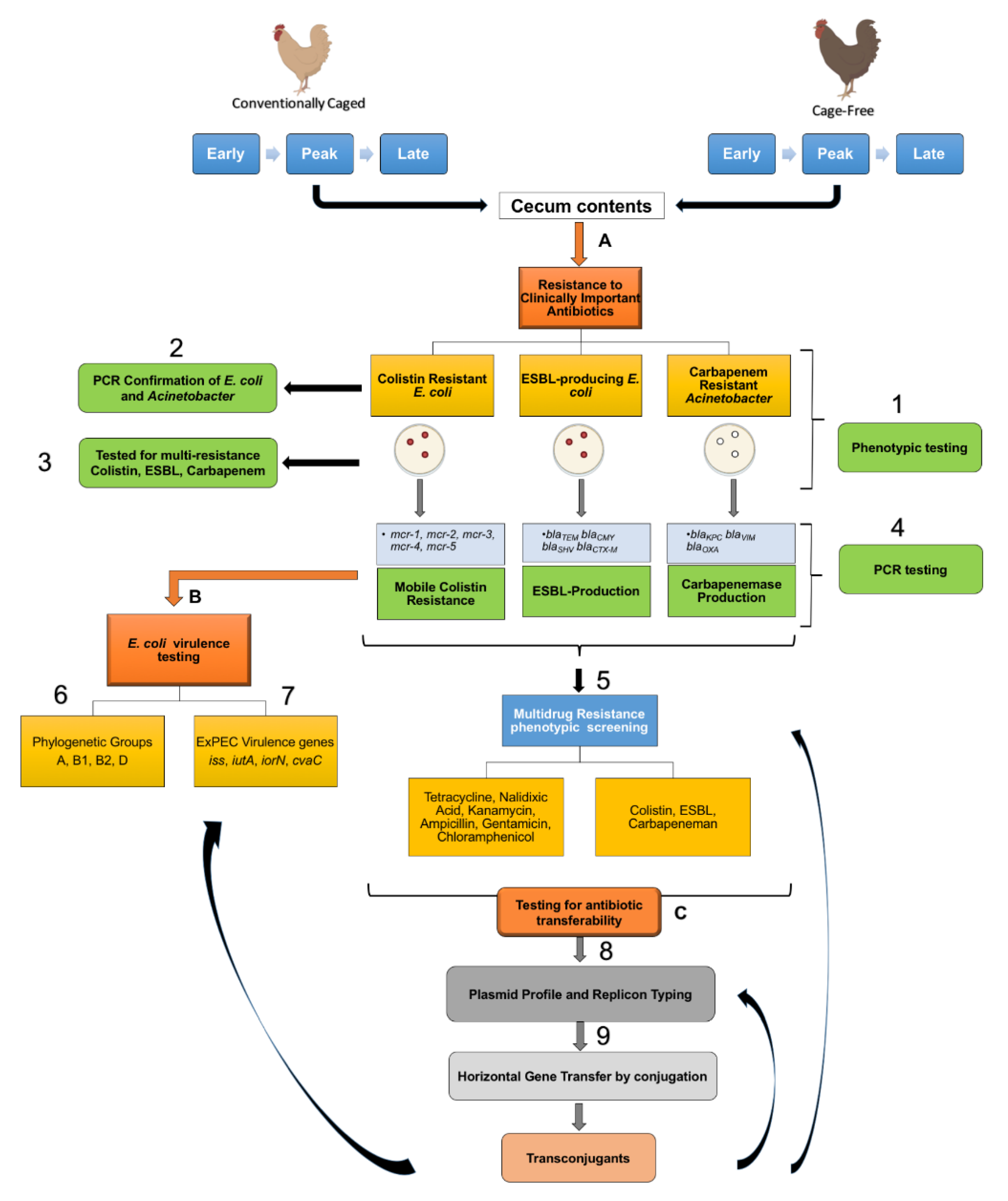
2. Materials and Methods
2.1. Source Material
2.2. Identification of Last Resort Antibiotic-Resistant Isolates
2.3. Plasmid Profiling and Typing
2.4. PCR Screenings for ExPEC
2.5. AMR Transfer Assays
2.6. Statistics
3. Results
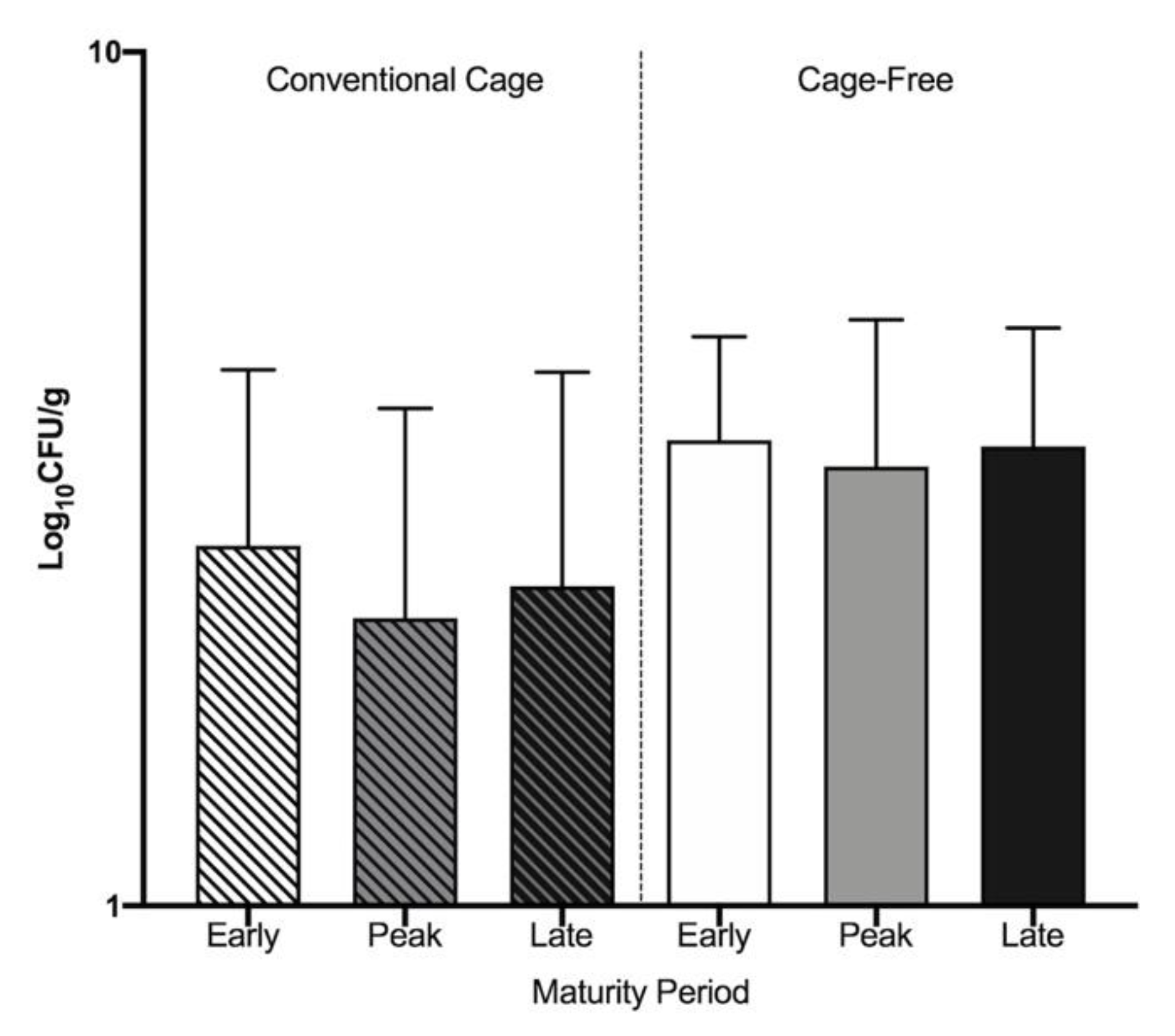
3.1. Widespread Antibiotic Resistance Detected in Poultry Fecal Isolates
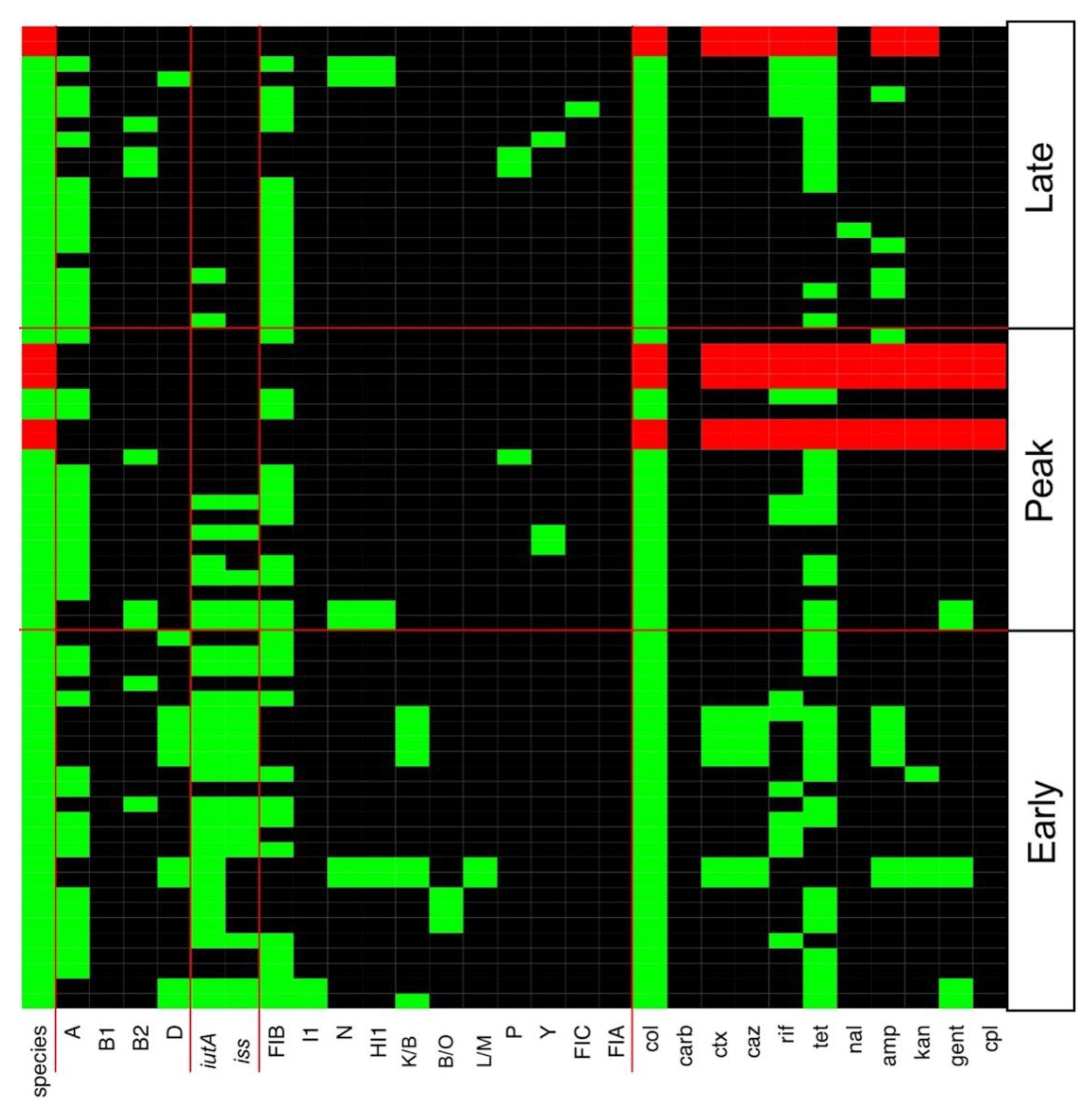
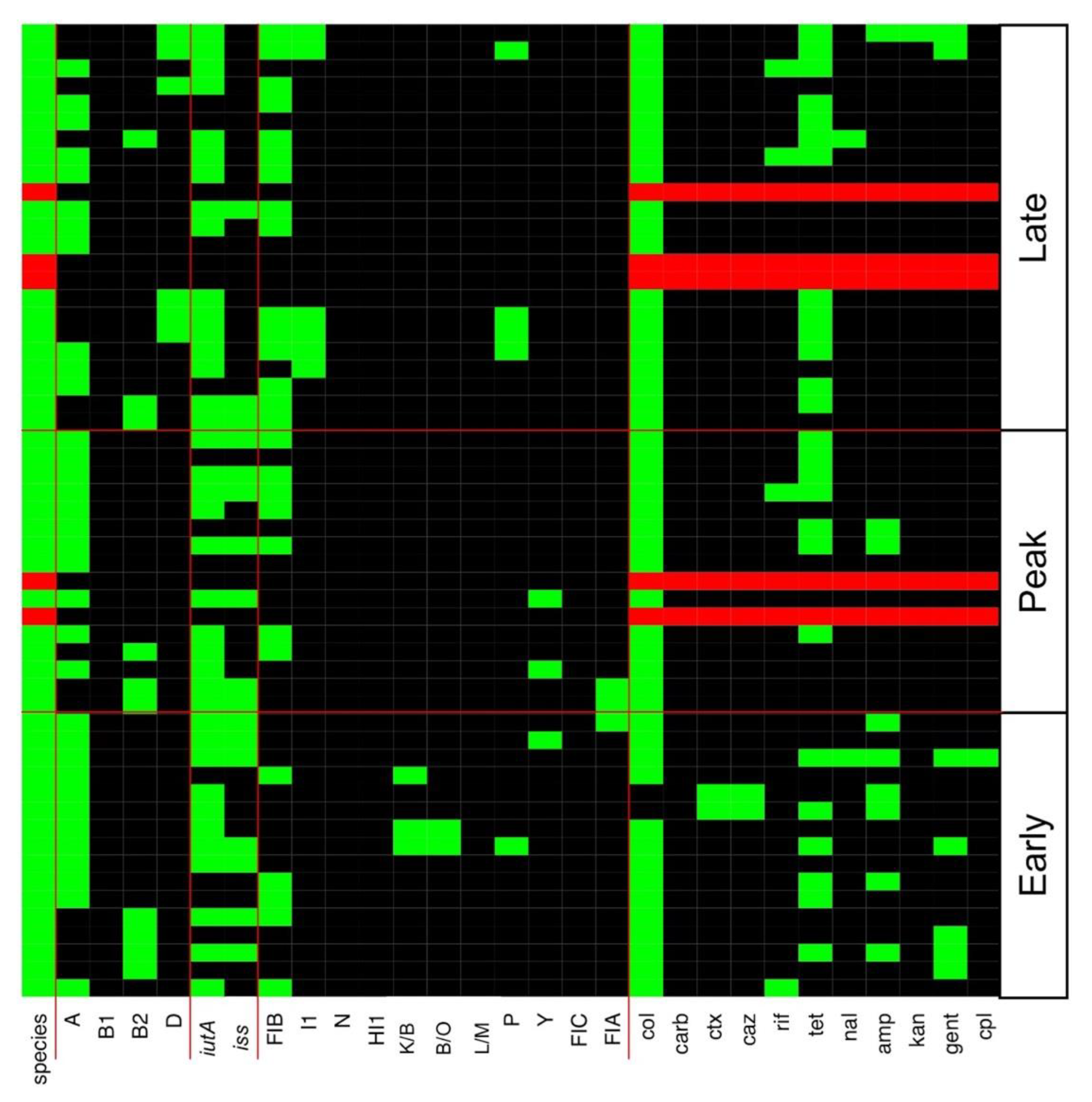
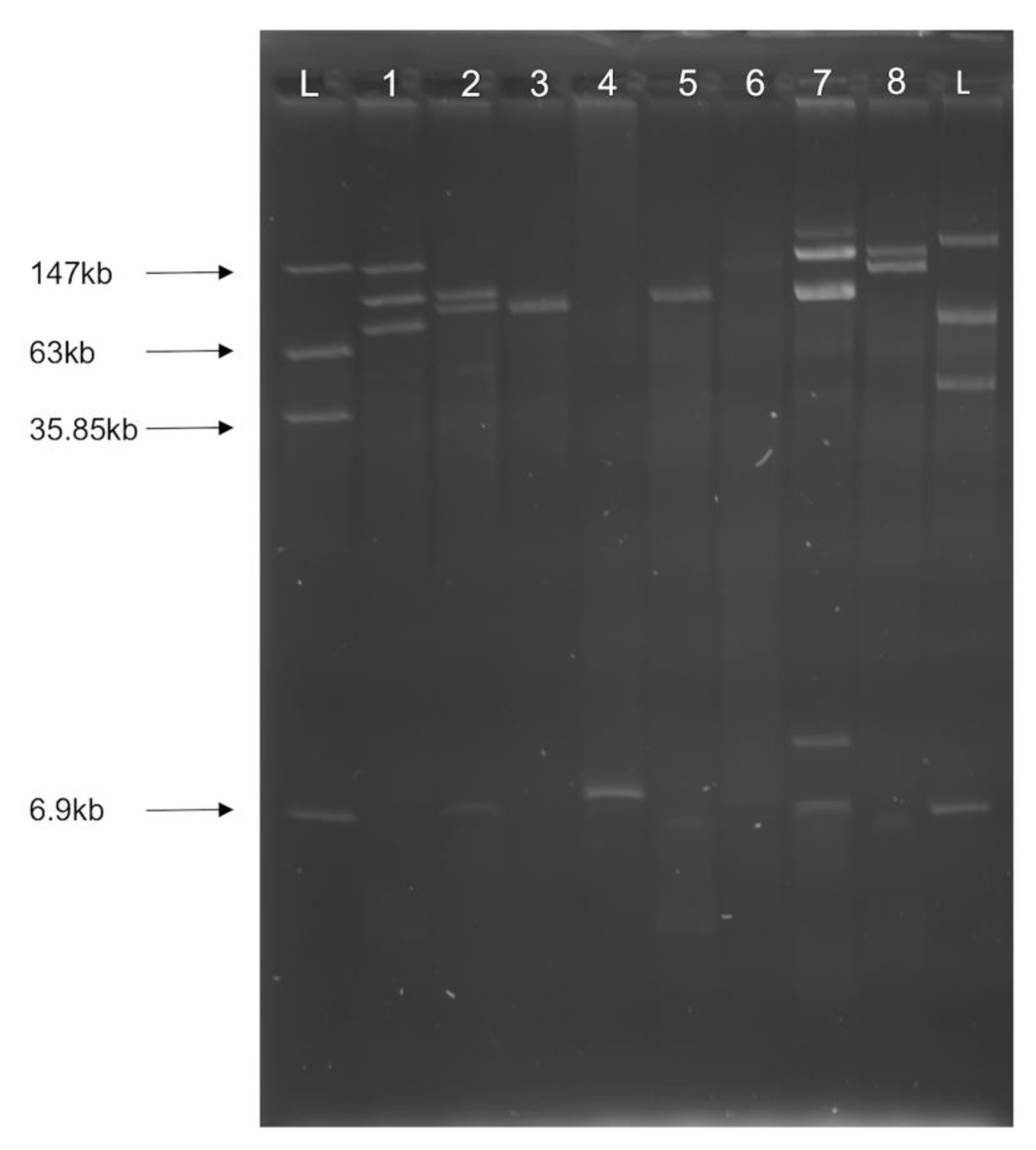
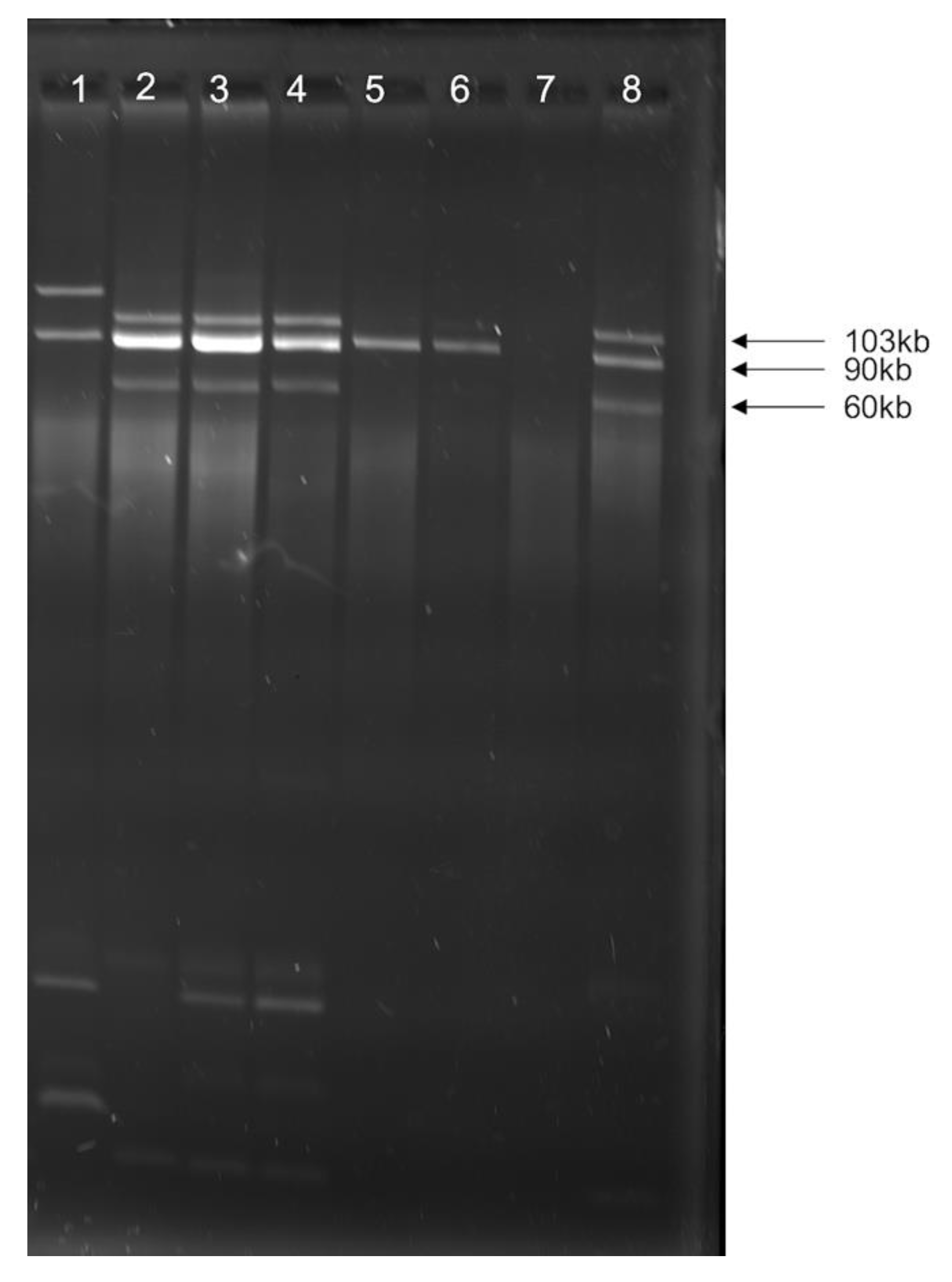
3.2. Multiple Plasmid Types Found in AMR E. coli Isolates
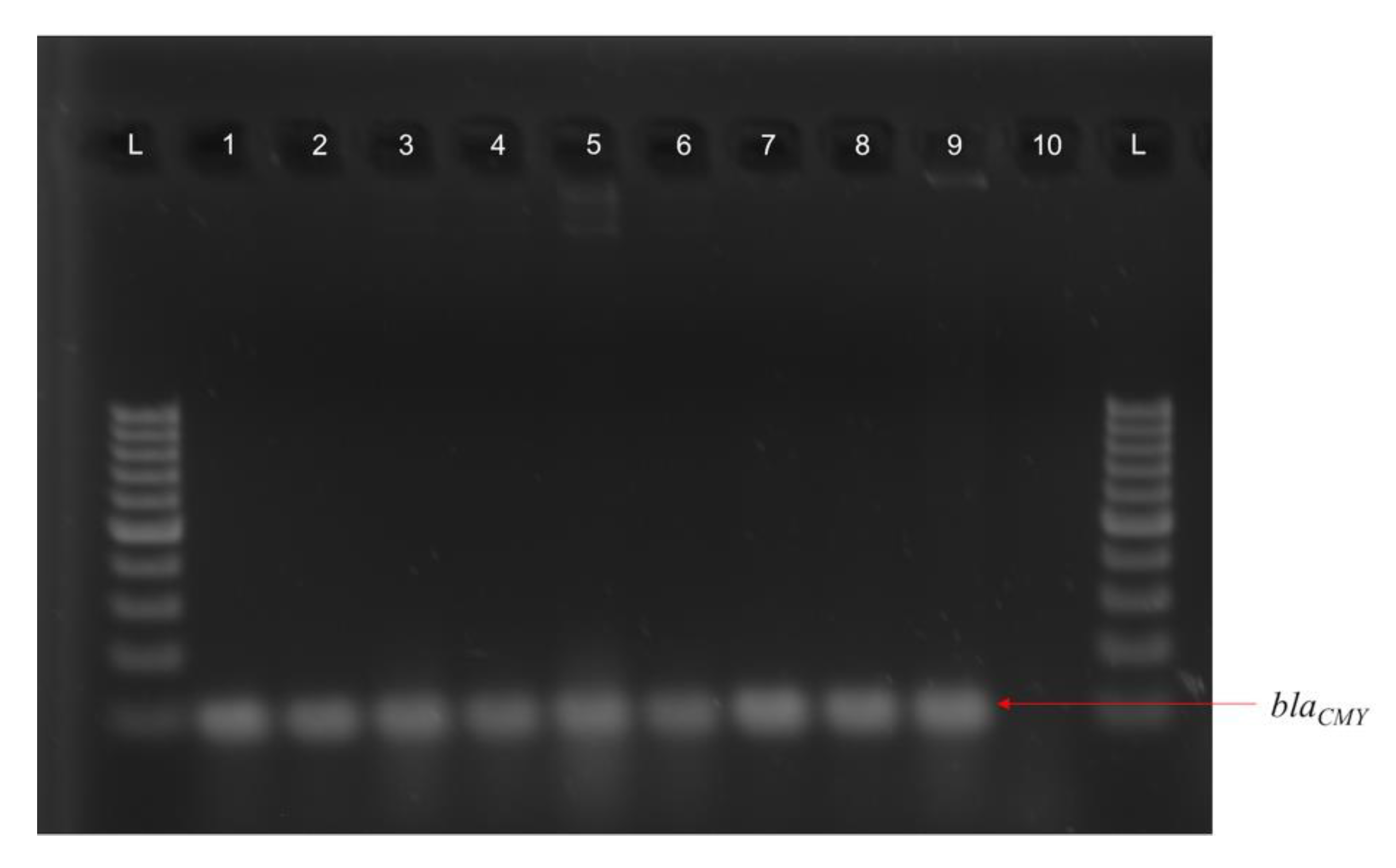
3.3. Various ExPEC Phylogenetic Groups and Virulence Markers Genes Identified in AMR E. coli Isolates
3.4. blaCMY Can Be Exchanged between Virulent and Non-Virulent E. coli through an IncK/B Plasmid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Naylor, N.R.; Pouwels, K.B.; Hope, R.; Green, N.; Henderson, K.L.; Knight, G.M.; Atun, R.; Robotham, J.V.; Deeny, S.R. The Health and Cost Burden of Antibiotic Resistant and Susceptible Escherichia coli Bacteraemia in the English Hospital Setting: A National Retrospective Cohort Study. PLoS ONE 2019, 14, e0221944. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Gousia, P. Agriculture and Food Animals as a Source of Antimicrobial-Resistant Bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Hegde, N.V.; Kariyawasam, S.; DebRoy, C. Comparison of Antimicrobial Resistant Genes in Chicken Gut Microbiome Grown on Organic and Conventional Diet. Vet. Anim. Sci. 2016, 1–2, 9–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Wang, J.; Wang, J.; Zhu, L.; Yang, L.; Yang, R. Distribution Characteristics of Antibiotic Resistant Bacteria and Genes in Fresh and Composted Manures of Livestock Farms. Sci. Total Environ. 2019, 695, 133781. [Google Scholar] [CrossRef]
- Ghaffoori Kanaan, M.H.; Al-Shadeedi, S.M.J.; Al-Massody, A.J.; Ghasemian, A. Drug Resistance and Virulence Traits of Acinetobacter baumannii from Turkey and Chicken Raw Meat. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101451. [Google Scholar] [CrossRef]
- Monte, D.F.; Mem, A.; Fernandes, M.R.; Cerdeira, L.; Esposito, F.; Galvão, J.A.; Lincopan, N.; Landgraf, M. Chicken Meat as a Reservoir of Colistin-Resistant Escherichia coli Strains Carrying Mcr-1 Genes in South America. Antimicrob. Agents Chemother. 2017, 61, 4. [Google Scholar] [CrossRef]
- Vitas, A.I.; Naik, D.; Pérez-Etayo, L.; González, D. Increased Exposure to Extended-Spectrum β-Lactamase-Producing Multidrug-Resistant Enterobacteriaceae through the Consumption of Chicken and Sushi Products. Int. J. Food Microbiol. 2018, 269, 80–86. [Google Scholar] [CrossRef]
- CDC. The Biggest Antibiotic-Resistant Threats in the U.S. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 22 October 2019).
- Mendelson, M.; Brink, A.; Gouws, J.; Mbelle, N.; Naidoo, V.; Pople, T.; Schellack, N.; van Vuuren, M.; Rees, H.; Banoo, S.; et al. The One Health Stewardship of Colistin as an Antibiotic of Last Resort for Human Health in South Africa. Lancet Infect. Dis. 2018, 18, e288–e294. [Google Scholar] [CrossRef]
- FAO’s Animal Production and Health Division: Meat & Meat Products. Available online: http://www.fao.org/ag/againfo/themes/en/meat/backgr_sources.html (accessed on 11 November 2020).
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W.; et al. Gut Inflammation Can Boost Horizontal Gene Transfer between Pathogenic and Commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the Virulence Plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef]
- Norman, A.; Hansen, L.H.; She, Q.; Sørensen, S.J. Nucleotide Sequence of POLA52: A Conjugative IncX1 Plasmid from Escherichia coli Which Enables Biofilm Formation and Multidrug Efflux. Plasmid 2008, 60, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High Prevalence of Mcr-1 Encoding Colistin Resistance and First Identification of BlaCTX-M-55 in ESBL/CMY-2-Producing Escherichia coli Isolated from Chicken Faeces and Retail Meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid Rise of the ESBL and Mcr-1 Genes in Escherichia coli of Chicken Origin in China, 2008–2014. Emerg. Microbes Infect. 2018, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ballou, A.L.; Ali, R.A.; Mendoza, M.A.; Ellis, J.C.; Hassan, H.M.; Croom, W.J.; Koci, M.D. Development of the Chick Microbiome: How Early Exposure Influences Future Microbial Diversity. Front. Vet. Sci. 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Card, R.M.; Cawthraw, S.A.; Nunez-Garcia, J.; Ellis, R.J.; Kay, G.; Pallen, M.J.; Woodward, M.J.; Anjum, M.F. An In Vitro Chicken Gut Model Demonstrates Transfer of a Multidrug Resistance Plasmid from Salmonella to Commensal Escherichia coli. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Haberecht, S.; Bajagai, Y.S.; Moore, R.J.; Van, T.T.H.; Stanley, D. Poultry Feeds Carry Diverse Microbial Communities That Influence Chicken Intestinal Microbiota Colonisation and Maturation. AMB Express 2020, 10. [Google Scholar] [CrossRef]
- Lupo, A.; Vogt, D.; Seiffert, S.N.; Endimiani, A.; Perreten, V. Antibiotic Resistance and Phylogenetic Characterization of Acinetobacter baumannii Strains Isolated from Commercial Raw Meat in Switzerland. J. Food Prot. 2014, 77, 1976–1981. [Google Scholar] [CrossRef] [PubMed]
- Mellata, M. Human and Avian Extraintestinal Pathogenic Escherichia coli: Infections, Zoonotic Risks, and Antibiotic Resistance Trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Bardon, J.; Htoutou Sedlakova, M.; Prochazkova, P.; Kolar, M. Identification of Novel OXA-134-like β-Lactamases in Acinetobacter Lwoffii and Acinetobacter Schindleri Isolated from Chicken Litter. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 2019, 163, 141–146. [Google Scholar] [CrossRef]
- Zilberberg, M.D.; Nathanson, B.H.; Sulham, K.; Fan, W.; Shorr, A.F. Multidrug Resistance, Inappropriate Empiric Therapy, and Hospital Mortality in Acinetobacter baumannii Pneumonia and Sepsis. Crit. Care 2016, 20. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18,. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, Q.; Liu, S.; Sun, R.; Zhou, Y.; Li, Y. Age-Related Variations in Intestinal Microflora of Free-Range and Caged Hens. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Gole, V.C.; Caraguel, C.G.B.; Sexton, M.; Fowler, C.; Chousalkar, K.K. Shedding of Salmonella in Single Age Caged Commercial Layer Flock at an Early Stage of Lay. Int. J. Food Microbiol. 2014, 189, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Van Goor, A.; Redweik, G.A.J.; Stromberg, Z.R.; Treadwell, C.G.; Xin, H.; Mellata, M. Microbiome and Biological Blood Marker Changes in Hens at Different Laying Stages in Conventional and Cage Free Housings. Poult. Sci. 2020, 99, 2362–2374. [Google Scholar] [CrossRef] [PubMed]
- Yang, X. Moraxellaceae. In Encyclopedia of Food Microbiology; Elsevier: Lacombe, AB, Canada, 2014; pp. 826–833. [Google Scholar]
- Gomes-Neto, J.C.; Mantz, S.; Held, K.; Sinha, R.; Segura Munoz, R.R.; Schmaltz, R.; Benson, A.K.; Walter, J.; Ramer-Tait, A.E. A Real-Time PCR Assay for Accurate Quantification of the Individual Members of the Altered Schaedler Flora Microbiota in Gnotobiotic Mice. J. Microbiol. Methods 2017, 135, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Fernando, D.M.; Khan, I.U.H.; Patidar, R.; Lapen, D.R.; Talbot, G.; Topp, E.; Kumar, A. Isolation and Characterization of Acinetobacter baumannii Recovered from Campylobacter Selective Medium. Front Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- CLSI Performance Standards for Antimicrobial Susceptibility Testing 2020; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020.
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, Mcr-1, Mcr-2, Mcr-3, Mcr-4 and Mcr-5 for Surveillance Purposes. Eurosurveillance 2018, 23, 17–00672. [Google Scholar] [CrossRef]
- Singh, P.; Pfeifer, Y.; Mustapha, A. Multiplex Real-Time PCR Assay for the Detection of Extended-Spectrum β-Lactamase and Carbapenemase Genes Using Melting Curve Analysis. J. Microbiol. Methods 2016, 124, 72–78. [Google Scholar] [CrossRef]
- Mellata, M.; Ameiss, K.; Mo, H.; Curtiss, R. Characterization of the Contribution to Virulence of Three Large Plasmids of Avian Pathogenic Escherichia coli Χ7122 (O78:K80:H9). IAI 2010, 78, 1528–1541. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.M.; Johnson, S.J.; Logue, C.M.; White, D.G.; Doetkott, C.; Nolan, L.K. Plasmid Replicon Typing of Commensal and Pathogenic Escherichia coli Isolates. Appl. Environ. Microbiol. 2007, 73, 1976–1983. [Google Scholar] [CrossRef]
- Brown, P.K.; Curtiss, R. Unique Chromosomal Regions Associated with Virulence of an Avian Pathogenic Escherichia coli Strain. Proc. Natl. Acad. Sci. USA 1996, 93, 11149–11154. [Google Scholar] [CrossRef] [PubMed]
- Mellata, M.; Touchman, J.W.; Curtiss, R. Full Sequence and Comparative Analysis of the Plasmid PAPEC-1 of Avian Pathogenic E. coli Χ7122 (O78:K80:H9). PLoS ONE 2009, 4, e4232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stromberg, Z.R.; Van Goor, A.; Redweik, G.A.J.; Wymore Brand, M.J.; Wannemuehler, M.J.; Mellata, M. Pathogenic and Non-Pathogenic Escherichia coli Colonization and Host Inflammatory Response in a Defined Microbiota Mouse Model. Dis. Models Mech. 2018, 11, dmm035063. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.C.; Stromberg, Z.R.; Redweik, G.A.J.; Wannemuehler, M.J.; Mellata, M. Mouse Genetic Background Affects Transfer of an Antibiotic Resistance Plasmid in the Gastrointestinal Tract. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Nowrouzian, F.L.; Wold, A.E.; Adlerberth, I. Escherichia coli Strains Belonging to Phylogenetic Group B2 Have Superior Capacity to Persist in the Intestinal Microflora of Infants. J. Infect. Dis. 2005, 191, 1078–1083. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Leekitcharoenphon, P.; Hendriksen, R.S.; Aarestrup, F.M.; Wasyl, D. A Metagenomic Glimpse into the Gut of Wild and Domestic Animals: Quantification of Antimicrobial Resistance and More. PLoS ONE 2020, 15, e0242987. [Google Scholar] [CrossRef]
- Al Azad, M.A.R.; Rahman, M.M.; Amin, R.; Begum, M.I.A.; Fries, R.; Husna, A.; Khairalla, A.S.; Badruzzaman, A.T.M.; El Zowalaty, M.E.; Lampang, K.N.; et al. Susceptibility and Multidrug Resistance Patterns of Escherichia coli Isolated from Cloacal Swabs of Live Broiler Chickens in Bangladesh. Pathogens 2019, 8, 118. [Google Scholar] [CrossRef]
- Ievy, S.; Islam, M.S.; Sobur, M.A.; Talukder, M.; Rahman, M.B.; Khan, M.F.R.; Rahman, M.T. Molecular Detection of Avian Pathogenic Escherichia coli (APEC) for the First Time in Layer Farms in Bangladesh and Their Antibiotic Resistance Patterns. Microorganisms 2020, 8, 1021. [Google Scholar] [CrossRef]
- Grami, R.; Mansour, W.; Mehri, W.; Bouallègue, O.; Boujaâfar, N.; Madec, J.-Y.; Haenni, M. Impact of Food Animal Trade on the Spread of mcr-1-Mediated Colistin Resistance, Tunisia, July 2015. Eurosurveillance 2016, 21. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive Resistome Analysis Reveals the Prevalence of NDM and MCR-1 in Chinese Poultry Production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, Y.; Sun, Y.; Ma, L.; Zeng, Q.; Jiang, X.; Li, A.; Zeng, Z.; Zhang, T. Antibiotic-Mediated Changes in the Fecal Microbiome of Broiler Chickens Define the Incidence of Antibiotic Resistance Genes. Microbiome 2018, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- The European Committee of Antimicrobial Susceptibilitiy Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 10.0 2020; The European Committee of Antimicrobial Susceptibility Testing: Växjö, Sweden, 2020.
- Thiry, D.; Berrah, A.; Evrard, J.; Duprez, J.-N.; Mainil, J.G.; Saulmont, M. Assessment of Two Selective Agar Media to Isolate Colistin-Resistant Bovine Escherichia coli: Correlation with Minimal Inhibitory Concentration and Presence of Mcr Genes. J. Microbiol. Methods 2019, 159, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, L.; Chen, X.; Zhi, C.; Yao, X.; Liu, Y.; Wu, S.; Guo, Z.; Yi, L.; Zeng, Z.; et al. High Prevalence of Colistin Resistance and Mcr-1 Gene in Escherichia coli Isolated from Food Animals in China. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a Novel Mobile Colistin Resistance Gene, Mcr-8, in NDM-Producing Klebsiella Pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Nesporova, K.; Jamborova, I.; Valcek, A.; Medvecky, M.; Literak, I.; Dolejska, M. Various Conjugative Plasmids Carrying the Mcr-5 Gene in Escherichia coli Isolates from Healthy Chickens in Paraguay. J. Antimicrob. Chemother. 2019, 74, 3394–3397. [Google Scholar] [CrossRef]
- Nation, R.L.; Li, J. Colistin in the 21st Century. Curr. Opin. Infect. Dis. 2009, 22, 535–543. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Skurnik, M. Temperature-Regulated Efflux Pump/Potassium Antiporter System Mediates Resistance to Cationic Antimicrobial Peptides in Yersinia. Mol. Microbiol. 2000, 37, 67–80. [Google Scholar] [CrossRef]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Albertí, S.; Bengoechea, J.A. Capsule Polysaccharide Mediates Bacterial Resistance to Antimicrobial Peptides. IAI 2004, 72, 7107–7114. [Google Scholar] [CrossRef]
- Makris, D.; Petinaki, E.; Tsolaki, V.; Manoulakas, E.; Mantzarlis, K.; Apostolopoulou, O.; Sfyras, D.; Zakynthinos, E. Colistin versus Colistin Combined with Ampicillin-Sulbactam for Multiresistant Acinetobacter baumannii Ventilator-Associated Pneumonia Treatment: An Open-Label Prospective Study. Indian J. Crit. Care Med. 2018, 22, 67–77. [Google Scholar] [CrossRef]
- Ur Rahman, S.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. Available online: https://www.hindawi.com/journals/bmri/2018/9519718/ (accessed on 10 December 2020).
- Aruhomukama, D.; Najjuka, C.F.; Kajumbula, H.; Okee, M.; Mboowa, G.; Sserwadda, I.; Mayanja, R.; Joloba, M.L.; Kateete, D.P. BlaVIM- and BlaOXA-Mediated Carbapenem Resistance among Acinetobacter baumannii and Pseudomonas Aeruginosa Isolates from the Mulago Hospital Intensive Care Unit in Kampala, Uganda. BMC Infect Dis 2019, 19, 853. [Google Scholar] [CrossRef]
- Caneiras, C.; Calisto, F.; Jorge da Silva, G.; Lito, L.; Melo-Cristino, J.; Duarte, A. First Description of Colistin and Tigecycline-Resistant Acinetobacter baumannii Producing KPC-3 Carbapenemase in Portugal. Antibiotics 2018, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The Role of ISAba1 in Expression of OXA Carbapenemase Genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, Efflux Pumps and Multidrug Resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P. Carbapenem Resistance in Acinetobacter baumannii: Mechanisms and Epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.S.; Wester, A.L.; Ahrenfeldt, J.; Mo, S.S.; Slettemeås, J.S.; Steinbakk, M.; Samuelsen, Ø.; Grude, N.; Simonsen, G.S.; Løhr, I.H.; et al. Norwegian Patients and Retail Chicken Meat Share Cephalosporin-Resistant Escherichia coli and IncK/BlaCMY-2 Resistance Plasmids. Clin. Microbiol. Infect. 2017, 23, 407.e9–407.e15. [Google Scholar] [CrossRef]
- Song, H.-J.; Moon, D.C.; Mechesso, A.F.; Kang, H.Y.; Kim, M.H.; Choi, J.-H.; Kim, S.-J.; Yoon, S.-S.; Lim, S.-K. Resistance Profiling and Molecular Characterization of Extended-Spectrum/Plasmid-Mediated AmpC β-Lactamase-Producing Escherichia coli Isolated from Healthy Broiler Chickens in South Korea. Microorganisms 2020, 8, 1434. [Google Scholar] [CrossRef]
- Robberts, F.J.L.; Kohner, P.C.; Patel, R. Unreliable Extended-Spectrum β-Lactamase Detection in the Presence of Plasmid-Mediated AmpC in Escherichia coli Clinical Isolates. J. Clin. Microbiol. 2009, 47, 358–361. [Google Scholar] [CrossRef]
- Stewart, A.G.; Harris, P.N.A.; Henderson, A.; Schembri, M.A.; Paterson, D.L. Oral Cephalosporin and β-Lactamase Inhibitor Combinations for ESBL-Producing Enterobacteriaceae Urinary Tract Infections. J. Antimicrob. Chemother. 2020, 75, 2384–2393. [Google Scholar] [CrossRef]
- Gordon, D.M.; Clermont, O.; Tolley, H.; Denamur, E. Assigning Escherichia coli Strains to Phylogenetic Groups: Multi-Locus Sequence Typing versus the PCR Triplex Method. Environ. Microbiol. 2008, 10, 2484–2496. [Google Scholar] [CrossRef]
- Jafari, R.A.; Motamedi, H.; Maleki, E.; Ghanbarpour, R.; Mayahi, M. Phylogenetic Typing and Detection of Extended-Spectrum β-Lactamases in Escherichia coli Isolates from Broiler Chickens in Ahvaz, Iran. Vet. Res. Forum. 2016, 7, 227–233. [Google Scholar]
- Logue, C.M.; Wannemuehler, Y.; Nicholson, B.A.; Doetkott, C.; Barbieri, N.L.; Nolan, L.K. Comparative Analysis of Phylogenetic Assignment of Human and Avian ExPEC and Fecal Commensal Escherichia coli Using the (Previous and Revised) Clermont Phylogenetic Typing Methods and Its Impact on Avian Pathogenic Escherichia coli (APEC) Classification. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Van Overbeek, L.S.; Wichers, J.H.; van Amerongen, A.; van Roermund, H.J.W.; van der Zouwen, P.; Willemsen, P.T.J. Circulation of Shiga Toxin-Producing Escherichia coli Phylogenetic Group B1 Strains Between Calve Stable Manure and Pasture Land With Grazing Heifers. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Páramo, P.; Menac’h, A.L.; Gall, T.L.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of Forces Shaping the Commensal Escherichia coli Genetic Structure by Comparing Animal and Human Isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, S.A.; Cruz-Córdova, A.; Luna-Pineda, V.M.; Reyes-Grajeda, J.P.; Cázares-Domínguez, V.; Escalona, G.; Sepúlveda-González, M.E.; López-Montiel, F.; Arellano-Galindo, J.; López-Martínez, B.; et al. Multidrug- and Extensively Drug-Resistant Uropathogenic Escherichia coli Clinical Strains: Phylogenetic Groups Widely Associated with Integrons Maintain High Genetic Diversity. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Hashemizadeh, Z.; Kalantar-Neyestanaki, D.; Mansouri, S. Association between Virulence Profile, Biofilm Formation and Phylogenetic Groups of Escherichia coli Causing Urinary Tract Infection and the Commensal Gut Microbiota: A Comparative Analysis. Microb. Pathog. 2017, 110, 540–545. [Google Scholar] [CrossRef]
- Landgraf, T.N.; Berlese, A.; Fernandes, F.F.; Milanezi, M.L.; Martinez, R.; Panunto-Castelo, A. The Ferric Aerobactin Receptor IutA, a Protein Isolated on Agarose Column, Is Not Essential for Uropathogenic Escherichia coli Infection. Rev. Lat. Am. Enferm. 2012, 20, 340–345. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.M.; Nolan, L.K. Evolution of the Iss Gene in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2360–2369. [Google Scholar] [CrossRef]
- Benz, F.; Huisman, J.S.; Bakkeren, E.; Herter, J.A.; Stadler, T.; Ackermann, M.; Diard, M.; Egli, A.; Hall, A.R.; Hardt, W.-D.; et al. Plasmid- and Strain-Specific Factors Drive Variation in ESBL-Plasmid Spread In Vitro and In Vivo. ISME J. 2020. [Google Scholar] [CrossRef]
- Jy, B.; Be, F. Plasmid Localization and Partition in Enterobacteriaceae. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Kubasova, T.; Cejkova, D.; Matiasovicova, J.; Sekelova, Z.; Polansky, O.; Medvecky, M.; Rychlik, I.; Juricova, H. Antibiotic Resistance, Core-Genome and Protein Expression in IncHI1 Plasmids in Salmonella Typhimurium. Genome Biol. Evol. 2016, 8, 1661–1671. [Google Scholar] [CrossRef]
- Robertson, J.; Bessonov, K.; Schonfeld, J.; Nash, J.H.E. Universal Whole-Sequence-Based Plasmid Typing and Its Utility to Prediction of Host Range and Epidemiological Surveillance. Microb. Genom. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Botelho, L.A.B.; Kraychete, G.B.; Rocha, P.B.; de Souza da-Silva, A.P.; Picão, R.C.; Moreira, B.M.; Bonelli, R.R. CTX-M- and PAmpC-Encoding Genes Are Associated with Similar Mobile Genetic Elements in Escherichia coli Isolated from Different Brands of Brazilian Chicken Meat. Microb. Drug Resist. 2019, 26, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in Prevalence and Characteristics of ESBL/PAmpC Producing E. Coli in Food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef]
- Maamar, E.; Hammami, S.; Alonso, C.A.; Dakhli, N.; Abbassi, M.S.; Ferjani, S.; Hamzaoui, Z.; Saidani, M.; Torres, C.; Boutiba-Ben Boubaker, I. High Prevalence of Extended-Spectrum and Plasmidic AmpC Beta-Lactamase-Producing Escherichia coli from Poultry in Tunisia. Int. J. Food Microbiol. 2016, 231, 69–75. [Google Scholar] [CrossRef]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; et al. Whole Genome Analyses of CMY-2-Producing Escherichia coli Isolates from Humans, Animals and Food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Moran, R.A.; Hall, R.M. Evolution of Regions Containing Antibiotic Resistance Genes in FII-2-FIB-1 ColV-Colla Virulence Plasmids. Microb. Drug Resist. 2017, 24, 411–421. [Google Scholar] [CrossRef]
- Turton, J.; Davies, F.; Turton, J.; Perry, C.; Payne, Z.; Pike, R. Hybrid Resistance and Virulence Plasmids in “High-Risk” Clones of Klebsiella Pneumoniae, Including Those Carrying BlaNDM-5. Microorganisms 2019, 7, 326. [Google Scholar] [CrossRef]
| E. coli Strain | Notable Characteristics | Selected Plasmidic Virulence Factors | Source |
|---|---|---|---|
| Recipients | |||
| 7122 | APEC O78:K80:H9, nalR, strR, lac+ | cvaC, iss, iutA, iroN | [35] |
| 7368 | Plasmids-cured 7122: pChi7122-1 pChi7122-2 pChi7122-3, nalR, lac+ | − | [33] |
| 6092 | E. coli K-12, tetR, lac− | − | [36] |
| MG1655 | E. coli K-12, nalR, lac+ | − | [37] |
| HS-4 | Human commensal E. coli, rifR, lac− | − | [38] |
| Donor | |||
| IA-EC-0010 (CC) | Lac+, ctxR, cazR, colR | iutA | This Study |
| IA-EC-0018 (CC) | Lac+, ctxR, caz, colR | iss, iutA | This Study |
| IA-EC-0075 (CF) | Lac+, ctxR, cazR | iutA | This Study |
| IA-EC-0076 (CF) | Lac+, ctxR | iutA | This Study |
| Transconjugants | |||
| 7122 (pIA-EC-0010-2) | nalR, strR, ctxR, cazR lac+, pIA-EC-0010-2 (Donor IA-EC-0010) | cvaC, iss, iutA, iroN | This Study |
| 7368 (pIA-EC-0010-2) | nalR, ctxR, cazR, lac+, pIA-EC-0010-2 (Donor IA-EC-0010) | − | This Study |
| 7122 (pIA-EC-0018-2) | nalR, strR, ctxR, cazR lac+, pIA-EC-0018-2 (Donor IA-EC-0018) | cvaC, iss, iutA, iroN | This Study |
| 7368 (pIA-EC-0018-2) | nalR, ctxR, cazR, lac+, pIA-EC-0018-2 (Donor IA-EC-0018) | − | This Study |
| 6092 (pIA-EC-0018-2) | tetR, ctxR, cazR,lac−, pIA-EC-0018-2 (Donor 7122 (pIA-EC-0018-2) | − | This Study |
| Environment | Lay Period | Positively Identified Antibiotic Resistant Isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| Pre-Enrichment | Post-Enrichment | |||||||
| Total ColistinR (CFU/g) | CRE CFU/g (% Total) | Total-LactamR (CFU/g) | -LactamR E. coli CFU/g (% Total) | -LactamR Acinetobacter CFU/g(% Total) | Total CarbaR (CFU/g) | CRA CFU/g (% Total) | ||
| CC | Early | 3.7 × 103 | 2 × 102 (5.57) | 41.7 | 41.7 (100) | 0 | 0 | 0 (0) |
| Peak | 1.9 × 103 | 1 × 102 (6.21) | 55.6 | 0 (0) | 41.5 (76.4) | 0 | 0 (0) | |
| Late | 7 × 103 | 1 × 102 (1.67) | 5.6 102 | 0 (0) | 13.3 (36.2) | 0 | 0 (0) | |
| CF | Early | 2 × 104 | 1 × 102 (0.52) | 1.7 102 | 17.9 (10.5) | 0 (0) | 50.5 | 0 (0) |
| Peak | 2 × 104 | 1 × 102 (0.57) | 2 102 | 0 (0) | 0 (0) | 1.3 × 102 | 22.5 (16.7) | |
| Late | 1 × 104 | 1.5 × 102 (1.19) | 1.5 102 | 0 (0) | 0 (0) | 4 × 102 | 25.9 (6.3) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jochum, J.M.; Redweik, G.A.J.; Ott, L.C.; Mellata, M. Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms. Microorganisms 2021, 9, 141. https://doi.org/10.3390/microorganisms9010141
Jochum JM, Redweik GAJ, Ott LC, Mellata M. Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms. Microorganisms. 2021; 9(1):141. https://doi.org/10.3390/microorganisms9010141
Chicago/Turabian StyleJochum, Jared M., Graham A. J. Redweik, Logan C. Ott, and Melha Mellata. 2021. "Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms" Microorganisms 9, no. 1: 141. https://doi.org/10.3390/microorganisms9010141
APA StyleJochum, J. M., Redweik, G. A. J., Ott, L. C., & Mellata, M. (2021). Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms. Microorganisms, 9(1), 141. https://doi.org/10.3390/microorganisms9010141





