Unearthing Antibiotic Resistance Associated with Disturbance-Induced Permafrost Thaw in Interior Alaska
Abstract
1. Introduction
2. Materials and Methods
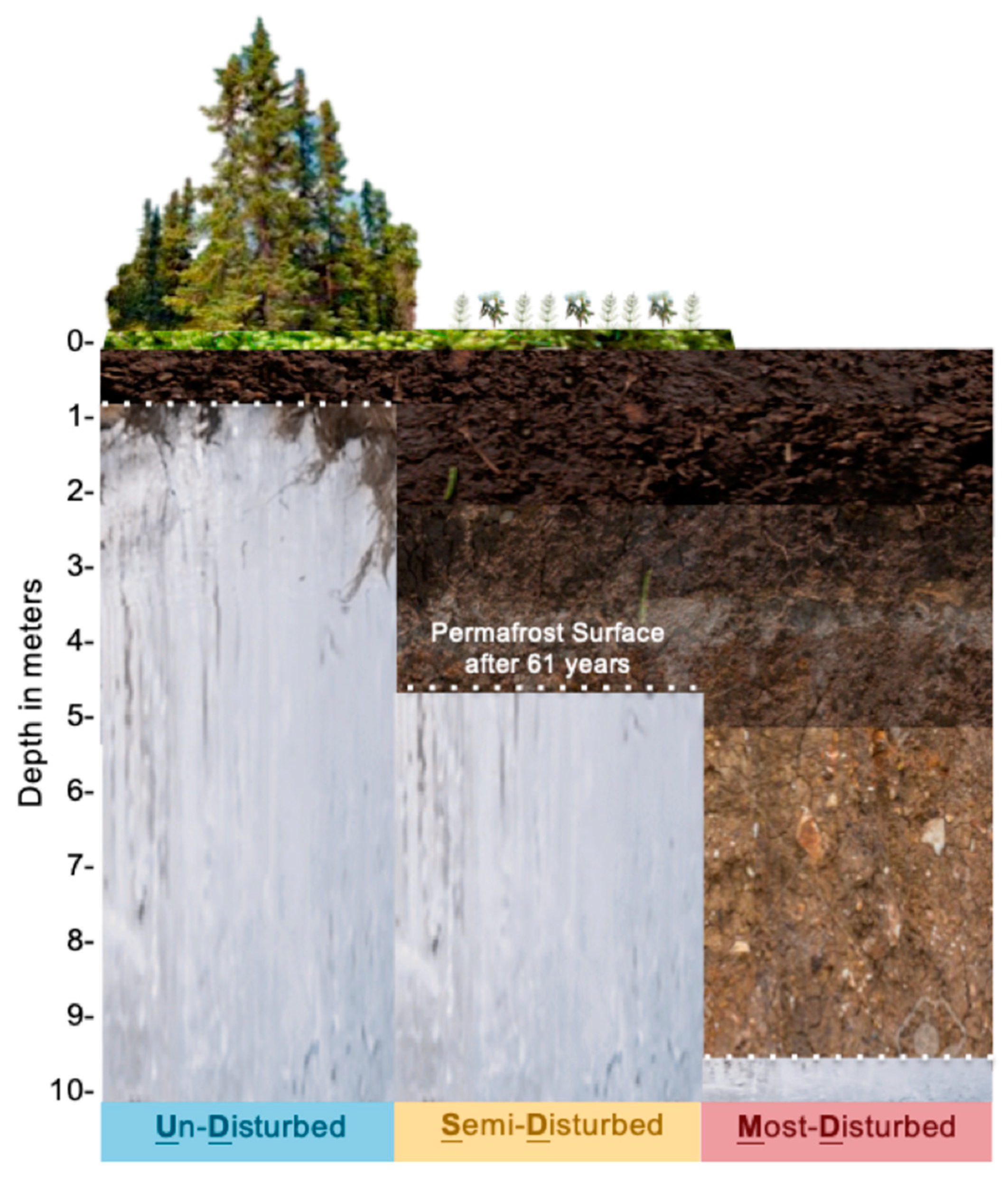
2.1. Permafrost Thaw Gradient
2.2. Bacterial Culturing
2.3. Antibiotic Susceptibility Testing
2.4. Whole Genome Sequencing, Assembly, and Taxonomic Classification
2.5. Antibiotic Resistance Gene Identification
2.6. RefSoil+ Comparison
2.7. Data Analyses and Statistics
3. Results
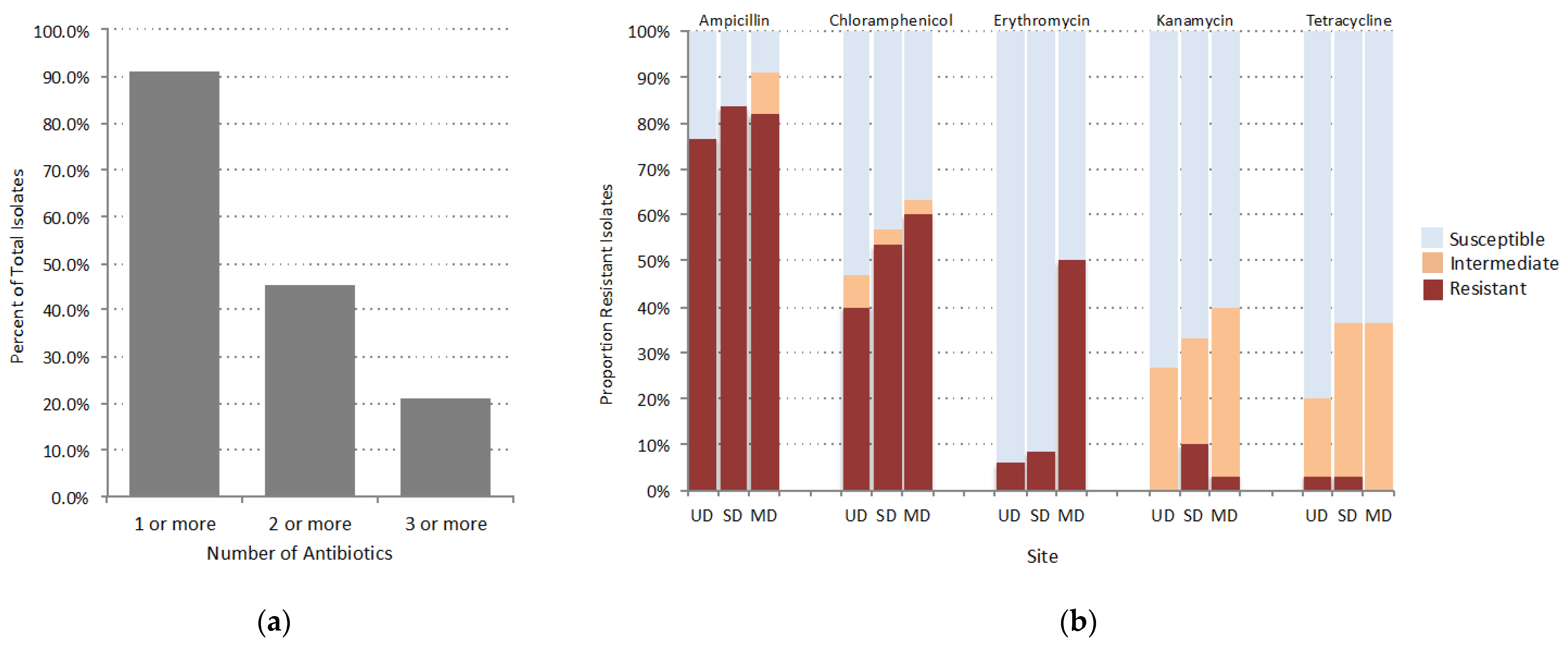
3.1. Assessment of Antibiotic Susceptibility in FPES Isolates
3.2. Genome Assembly Statistics
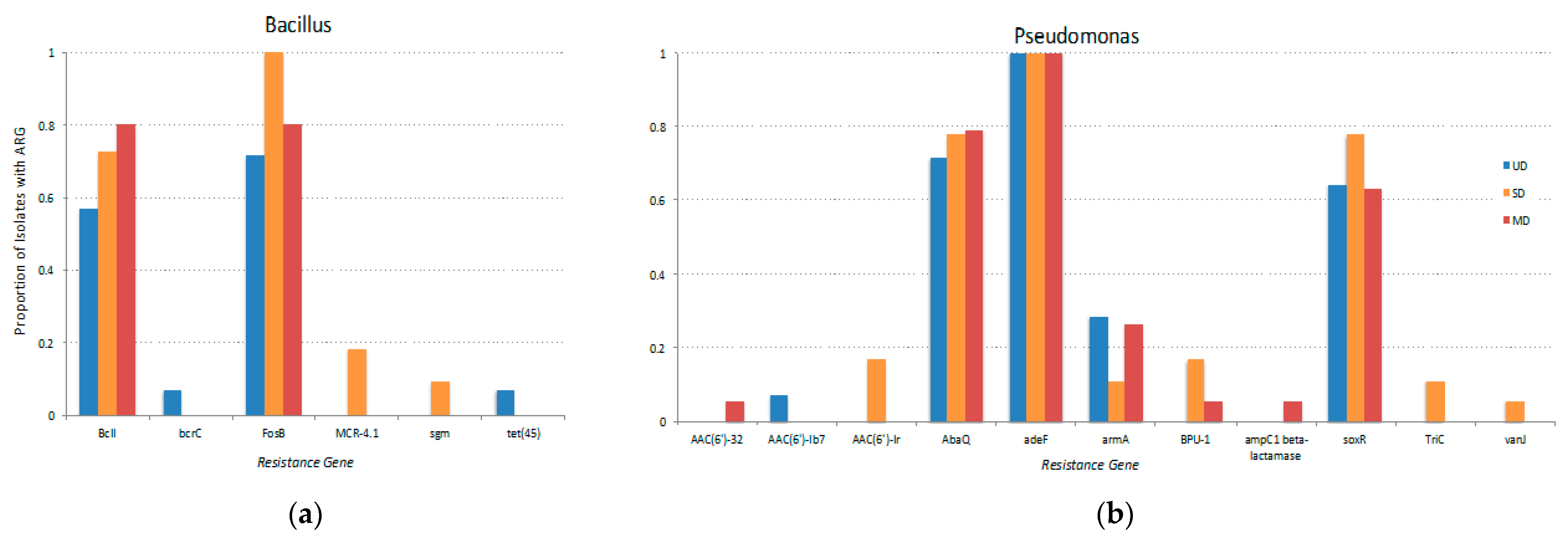
3.3. Antibiotic Resistance Genes Identified in FPES Isolates
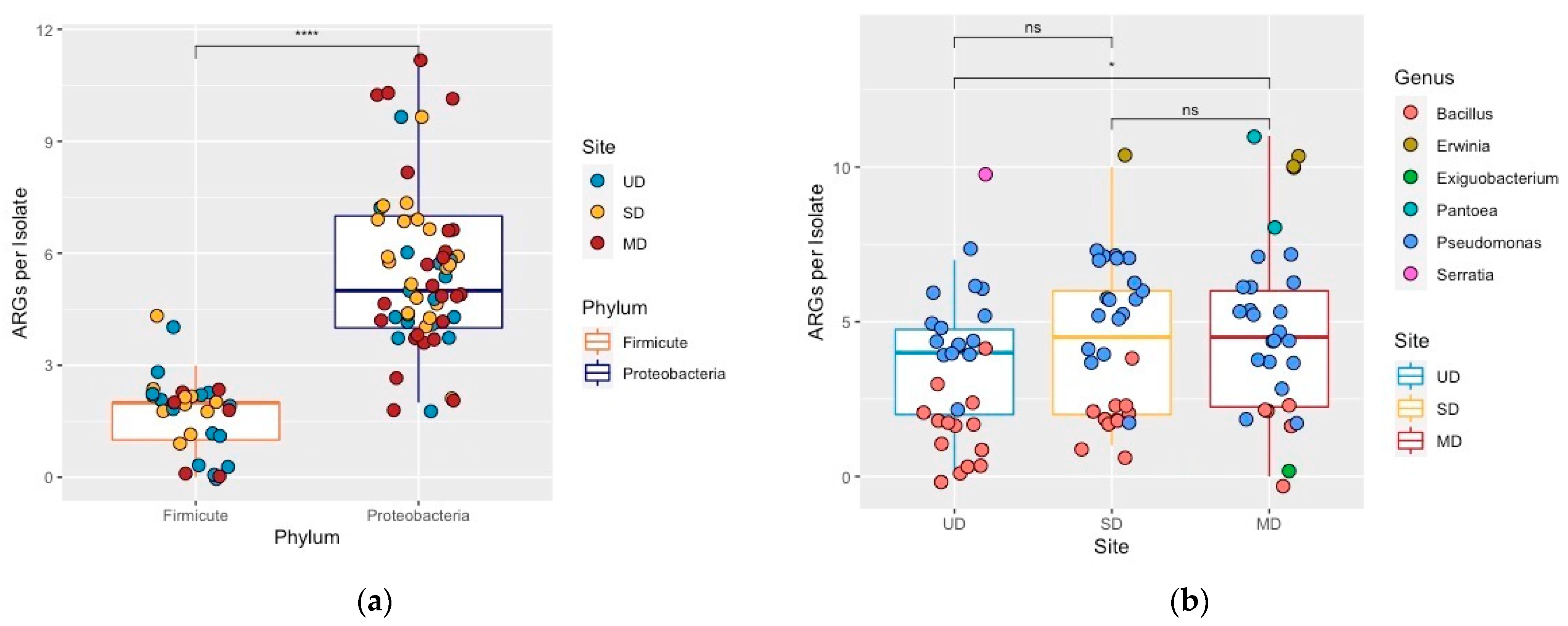
3.4. Influence of Phylogeny and Disturbance-Induced Thaw on ARGs
3.5. Comparison of ARGs in RefSoil+ and FPES Genomes from Corresponding Genera
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARG | antibiotic resistance gene |
| ARO | antibiotic resistance ontology |
| BVOC | biogenic volatile organic compounds |
| CARD | Comprehensive Antibiotic Resistance Database |
| CLSI | Clinical and Laboratory Standards Institute |
| FPES | Fairbanks Permafrost Experiment Station |
| HGT | horizontal gene transfer |
| MD | Most-disturbed |
| MGE | mobile genetic element |
| ONT | Oxford Nanopore Technologies |
| RGI | Resistance Gene Identifier |
| RND | Resistance-nodulation-cell division |
| SD | Semidisturbed |
| TSB | Tryptic Soy Broth |
| UD | Undisturbed |
Appendix A
| CLSI Breakpoint Class | FPES Isolate Genus | Ampicillin | Chloramphenicol | Erythromycin | Kanamycin | Tetracycline | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | I | S | R | I | S | R | I | S | R | I | S | R | I | S | ||
| Pseudomonas spp. | Pseudomonas | − | − | − | 12 | 13–17 | 18 | − | − | − | 12 | 13–14 | 15 | 11 | 12–14 | 15 |
| Enterobacterales | Erwinia | 13 | 14–16 | 17 | 12 | 13–17 | 18 | 12 | − | 13 | 13 | 14–17 | 18 | 11 | 12–14 | 15 |
| Enterobacterales | Pantonea | 13 | 14–16 | 17 | 12 | 13–17 | 18 | 12 | − | 13 | 13 | 14–17 | 18 | 11 | 12–14 | 15 |
| Enterobacterales | Serratia | 13 | 14–16 | 17 | 12 | 13–17 | 18 | 12 | − | 13 | 13 | 14–17 | 18 | 11 | 12–14 | 15 |
| Enterococcus spp. | Bacillus | 16 | − | 17 | 12 | 13–17 | 18 | 13 | 14–22 | 23 | − | − | − | 14 | 15–18 | 19 |
| Enterococcus spp. | Exiguobacterium | 16 | − | 17 | 12 | 13–17 | 18 | 13 | 14–22 | 23 | − | − | − | 14 | 15–18 | 19 |
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review on Antimicrobial Resistance: London, UK, 2016; pp. 1–84. [Google Scholar]
- Ghosh, S.; LaPara, T.M. The Effects of Subtherapeutic Antibiotic Use in Farm Animals on the Proliferation and Persistence of Antibiotic Resistance among Soil Bacteria. ISME J. 2007, 1, 191–203. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. NPJ Clean Water 2020, 3, 1–11. [Google Scholar] [CrossRef]
- Berg, J.; Tom-Petersen, A.; Nybroe, O. Copper Amendment of Agricultural Soil Selects for Bacterial Antibiotic Resistance in the Field. Lett. Appl. Microbiol. 2005, 40, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, S.P.; Stokes, H.W.; Chowdhury, P.R. Mobile Elements, Zoonotic Pathogens and Commensal Bacteria: Conduits for the Delivery of Resistance Genes into Humans, Production Animals and Soil Microbiota. Front. Microbiol. 2013, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. The Role of Antibiotics and Antibiotic Resistance in Nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef]
- Caruso, G.; Giammanco, A.; Cardamone, C.; Oliveri, G.; Mascarella, C.; Capra, G.; Fasciana, T. Extra-Intestinal Fluoroquinolone-Resistant Escherichia Coli Strains Isolated from Meat. BioMed. Res. Int. 2018, 2018, e8714975. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in Antibacterial Discovery from Actinomycetes. Curr. Opin. Pharm. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Barlow, M. Evolution of the Serine Beta-Lactamases: Past, Present and Future. Drug Resist. Updat. 2004, 7, 111–123. [Google Scholar] [CrossRef] [PubMed]
- de Lima Procópio, R.E.; da Silva, I.R.; Martins, M.K.; de Azevedo, J.L.; de Araújo, J.M. Antibiotics Produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How Many Antibiotics Are Produced by the Genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.G.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The Scourge of Antibiotic Resistance: The Important Role of the Environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial Phylogeny Structures Soil Resistomes across Habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Mackie, R.I. Evolution and Ecology of Antibiotic Resistance Genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Schuur, E.A.G.; Mack, M.C. Ecological Response to Permafrost Thaw and Consequences for Local and Global Ecosystem Services. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 279–301. [Google Scholar] [CrossRef]
- Box, J.E.; Colgan, W.T.; Christensen, T.R.; Schmidt, N.M.; Lund, M.; Parmentier, F.-J.W.; Brown, R.; Bhatt, U.S.; Euskirchen, E.S.; Romanovsky, V.E.; et al. Key Indicators of Arctic Climate Change: 1971–2017. Environ. Res. Lett. 2019, 14, 045010. [Google Scholar] [CrossRef]
- McGuire, A.D.; Lawrence, D.M.; Koven, C.; Clein, J.S.; Burke, E.; Chen, G.; Jafarov, E.; MacDougall, A.H.; Marchenko, S.; Nicolsky, D.; et al. Dependence of the Evolution of Carbon Dynamics in the Northern Permafrost Region on the Trajectory of Climate Change. Proc. Natl. Acad. Sci. USA 2018, 115, 3882–3887. [Google Scholar] [CrossRef]
- Thoman, R.; Walsh, J. Alaska’s Changing Environment—International Arctic Research Center. Available online: https://uaf-iarc.org/our-work/alaskas-changing-environment/ (accessed on 16 December 2020).
- Douglas, T.; Kanevskiy, M.; Romanovsky, V.; Shur, Y.; Yoshikawa, K. Permafrost Dynamics at the Fairbanks Permafrost Experimental Station near Fairbanks, Alaska. Inst. North. Eng. Univ. Alsk. Fairbanks 2008, 1, 373–378. [Google Scholar]
- Schuur, E.A.G.; Crummer, K.G.; Vogel, J.G.; Mack, M.C. Plant Species Composition and Productivity Following Permafrost Thaw and Thermokarst in Alaskan Tundra. Ecosystems 2007, 10, 280–292. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.C.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef] [PubMed]
- Seitz, T.J.; Schütte, U.M.E.; Drown, D.M. Soil Disturbance Affects Plant Growth via Soil Microbial Community Shifts. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dunivin, T.K.; Choi, J.; Howe, A.; Shade, A. RefSoil+: A Reference Database for Genes and Traits of Soil Plasmids. mSystems 2019, 4, e00349-18. [Google Scholar] [CrossRef]
- Linell, K.A. Long-term effects of vegetation cover on permafrost stability in an area of discontinuous permafrost. In Proceedings of the Permafrost: North American Contribution to the Second Internal Conference, Yakutsk, Russia, 13–28 July 1973; National Academy Press: Washington, DC, USA; pp. 688–693. [Google Scholar]
- Johnstone, J.F.; Hollingsworth, T.N.; Chapin, F.S. A Key for Predicting Postfire Successional Trajectories in Black Spruce Stands of Interior Alaska. Gen. Tech. Rep. PNW-GTR-767 Portland OR US Dep. Agric. For. Serv. Pac. Northwest. Res. Stn. 2008, 767, 4–8. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Available online: https://www.asmscience.org/content/education/protocol/protocol.3189 (accessed on 16 December 2020).
- Krueger, F. TrimGalore. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 16 December 2020).
- Wick, R. Filtlong. Available online: https://github.com/rrwick/filtlong (accessed on 16 December 2020).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Haan, T.; McDougall, S.; Drown, D.M. Complete Genome Sequence of Bacillus Mycoides TH26, Isolated from a Permafrost Thaw Gradient. Microbiol. Resour. Announc. 2019, 8, e00507-19. [Google Scholar] [CrossRef]
- Humphrey, J.; Seitz, T.; Haan, T.; Ducluzeau, A.-L.; Drown, D.M. Complete Genome Sequence of Pantoea Agglomerans TH81, Isolated from a Permafrost Thaw Gradient. Microbiol. Resour. Announc. 2019, 8, e01486-18. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.; Lago, B.; Dave, B.; Pereira, S.; Sharma, A.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Shade, A. RefSoil_plasmids. Available online: https://github.com/ShadeLab/RefSoil_plasmids (accessed on 16 December 2020).
- RStudio Team. RStudio: Integrated Development for R; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Use R! Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2018. Available online: http://CRAN.R-project.org/package=pheatmap (accessed on 16 December 2020).
- Wright, G.D.; Poinar, H. Antibiotic Resistance Is Ancient: Implications for Drug Discovery. Trends Microbiol. 2012, 20, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Plasmid-Mediated Horizontal Gene Transfer Is a Coevolutionary Process. Trends Microbiol. 2012, 20, 262–267. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Touchon, M.; Cury, J.; Rocha, E.P.C. The Chromosomal Organization of Horizontal Gene Transfer in Bacteria. Nat. Commun. 2017, 8, 841. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. The Role of Natural Environments in the Evolution of Resistance Traits in Pathogenic Bacteria. Proc. Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.A.; Lee, S.A.; Manoil, C. Importance of Core Genome Functions for an Extreme Antibiotic Resistance Trait. mBio 2017, 8. [Google Scholar] [CrossRef]
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea Agglomerans, a Plant Pathogen Causing Human Disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar] [CrossRef]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and Pathogenesis of Bacillus Cereus Infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef]
- Lax, S.; Gilbert, J.A. Hospital-Associated Microbiota and Implications for Nosocomial Infections. Trends Mol. Med. 2015, 21, 427–432. [Google Scholar] [CrossRef]
- Ramos, J.L.; Gallegos, M.T.; Marqués, S.; Ramos-González, M.I.; Espinosa-Urgel, M.; Segura, A. Responses of Gram-Negative Bacteria to Certain Environmental Stressors. Curr. Opin. Microbiol. 2001, 4, 166–171. [Google Scholar] [CrossRef]
- Kramshøj, M.; Albers, C.N.; Holst, T.; Holzinger, R.; Elberling, B.; Rinnan, R. Biogenic Volatile Release from Permafrost Thaw Is Determined by the Soil Microbial Sink. Nat. Commun. 2018, 9, 3412. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.A.; Tomczak, A.; Khatri, P. Gene Annotation Bias Impedes Biomedical Research. Sci. Rep. 2018, 8, 1362. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Moe, L.A.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional Metagenomics Reveals Diverse β-Lactamases in a Remote Alaskan Soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Das, A.J.; Cochrane, G.; De Ganck, S.; Kumar-Singh, S.; Aarestrup, F.M.; Goossens, H.; Malhotra-Kumar, S. Consolidating and Exploring Antibiotic Resistance Gene Data Resources. J. Clin. Microbiol. 2016, 54, 851–859. [Google Scholar] [CrossRef]
- Fasciana, T.; Gentile, B.; Aquilina, M.; Ciammaruconi, A.; Mascarella, C.; Anselmo, A.; Fortunato, A.; Fillo, S.; Petralito, G.; Lista, F.; et al. Co-Existence of Virulence Factors and Antibiotic Resistance in New Klebsiella Pneumoniae Clones Emerging in South of Italy. BMC Infect. Dis. 2019, 19, 928. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.-C.; Feller, G.; Gerday, C. Psychrophilic Microorganisms: Challenges for Life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
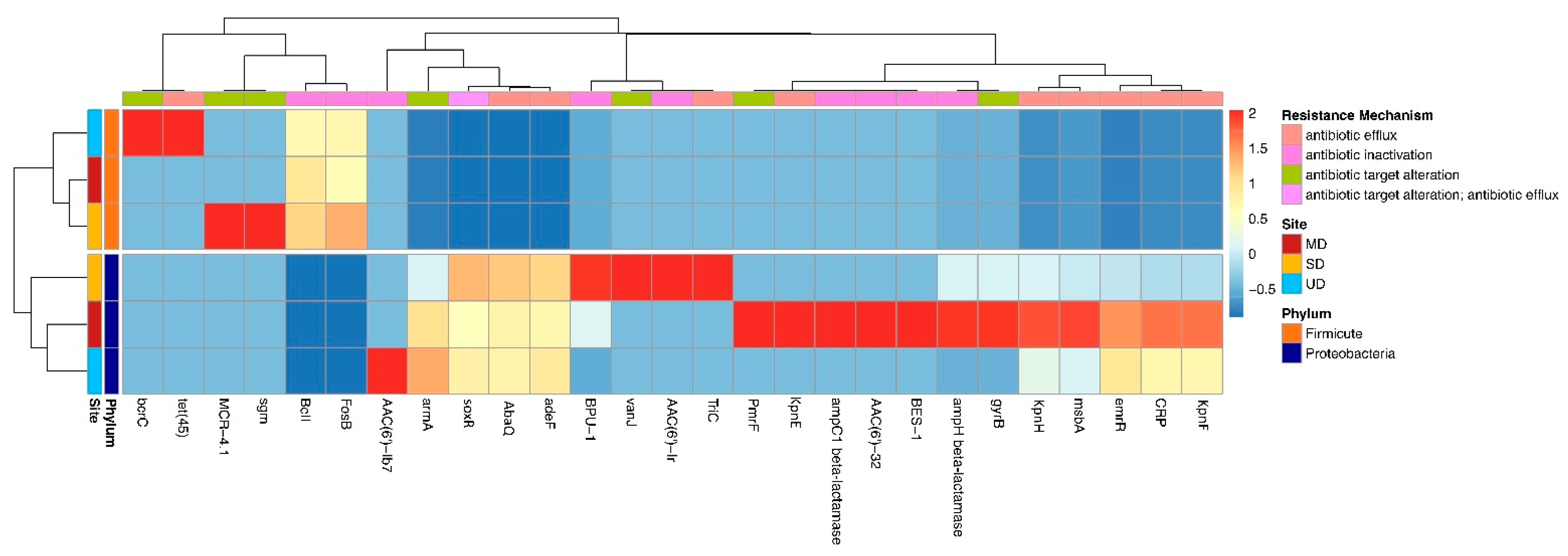
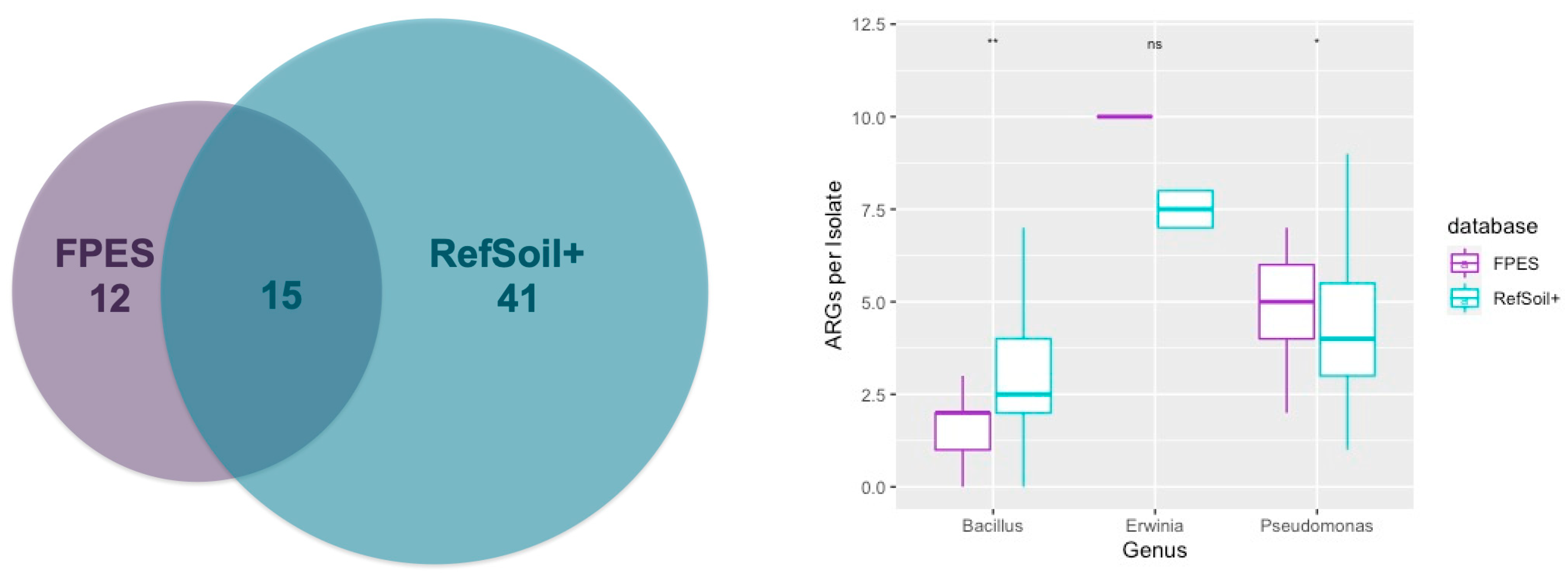
| Taxonomy | FPES Site | ||||
|---|---|---|---|---|---|
| Phylum | Genus | UD | SD | MD | Total |
| Firmicute | Bacillus | 14 | 11 | 5 | 30 |
| Firmicute | Exiguobacterium | 0 | 0 | 1 | 1 |
| Proteobacteria | Erwinia | 1 | 1 | 3 | 5 |
| Proteobacteria | Pantoea | 0 | 0 | 2 | 2 |
| Proteobacteria | Pseudomonas | 14 | 18 | 19 | 51 |
| Proteobacteria | Serratia | 1 | 0 | 0 | 1 |
| Total | 30 | 30 | 30 | 90 | |
| PLASMID BORNE ARGs IN FPES ISOLATES | |||||
|---|---|---|---|---|---|
| Best Hit ARO | Resistance Mechanism | Drug Class | AMR Gene Family | Genus Origin | Count |
| BES-1 | antibiotic inactivation | carbapenem; cephalosporin; penam | SIM beta-lactamase | Pantoea | 1 |
| bcrC | antibiotic target alteration | peptide antibiotic | udecaprenyl pyrophosphate related proteins | Bacillus | 2 |
| TriC | antibiotic efflux | triclosan | RND antibiotic efflux pump | Pseudomonas | 2 |
| KpnF | antibiotic efflux | Broad Spectrum | MFS antibiotic efflux pump | Erwinia | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haan, T.J.; Drown, D.M. Unearthing Antibiotic Resistance Associated with Disturbance-Induced Permafrost Thaw in Interior Alaska. Microorganisms 2021, 9, 116. https://doi.org/10.3390/microorganisms9010116
Haan TJ, Drown DM. Unearthing Antibiotic Resistance Associated with Disturbance-Induced Permafrost Thaw in Interior Alaska. Microorganisms. 2021; 9(1):116. https://doi.org/10.3390/microorganisms9010116
Chicago/Turabian StyleHaan, Tracie J., and Devin M. Drown. 2021. "Unearthing Antibiotic Resistance Associated with Disturbance-Induced Permafrost Thaw in Interior Alaska" Microorganisms 9, no. 1: 116. https://doi.org/10.3390/microorganisms9010116
APA StyleHaan, T. J., & Drown, D. M. (2021). Unearthing Antibiotic Resistance Associated with Disturbance-Induced Permafrost Thaw in Interior Alaska. Microorganisms, 9(1), 116. https://doi.org/10.3390/microorganisms9010116





