The Effect of Antimicrobial Resistance Plasmids Carrying blaCMY-2 on Biofilm Formation by Escherichia coli from the Broiler Production Chain
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Media
2.2. Crystal Violet Biofilm Assay
2.3. Congo Red Biofilm Assay
2.4. Determination of Phylotypes
2.5. Conjugation of QREC Strains
2.6. Motility Assay and Microtiter Plate Growth Assay
2.7. Statistics
3. Results
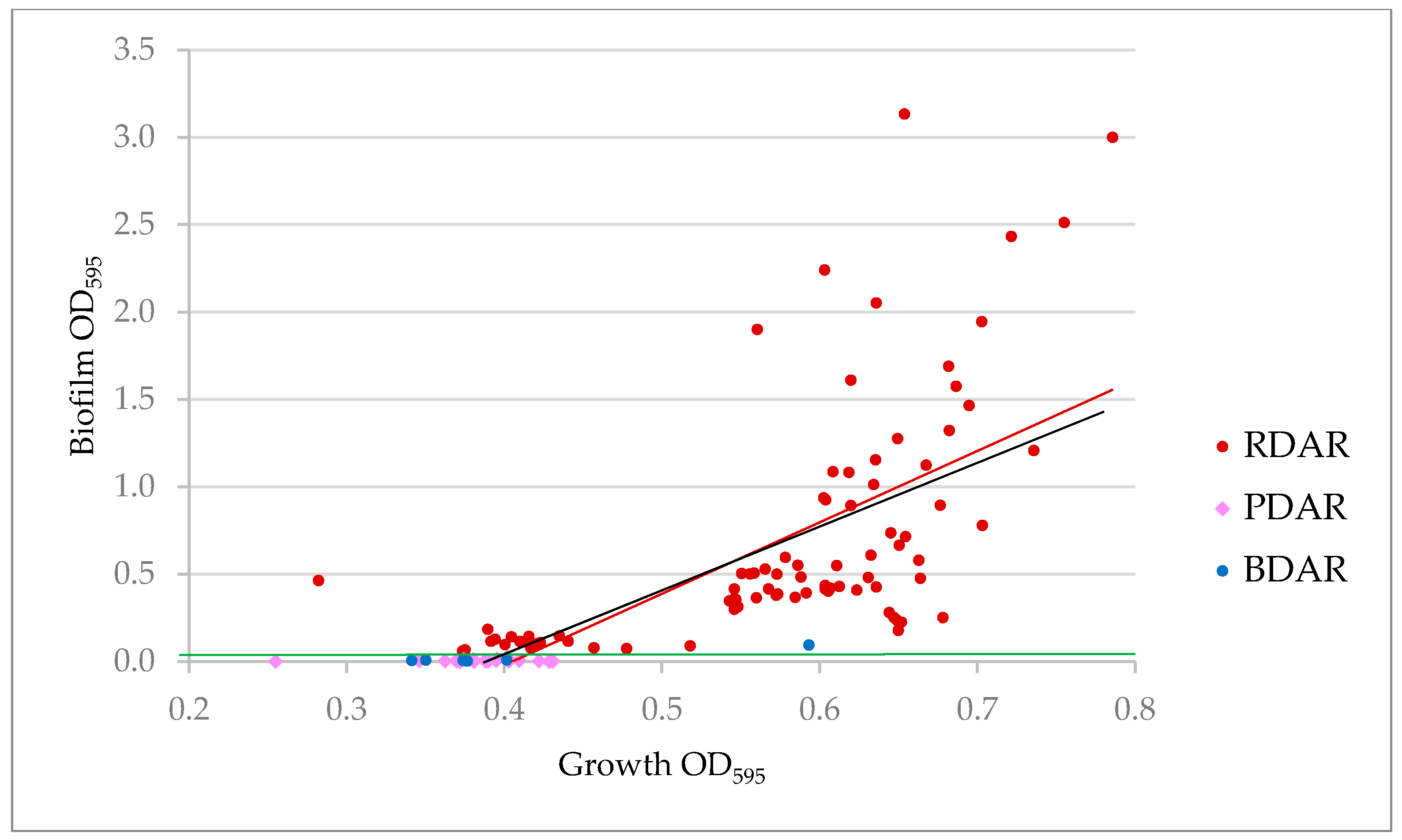
3.1. Biofilm Production by ESCR E. coli Related to Biofilm Assay and Morphotype
3.2. Biofilm Production by ESCR E. coli Related to Phylotype
3.3. Biofilm Production by ESCR E. coli Related to Source
3.4. Conjugation Experiments with QREC Recipients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinas, L.; Moreno, M.A.; Zarazaga, M.; Porrero, C.; Saenz, Y.; Garcia, M.; Dominguez, L.; Torres, C. Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 2003, 47, 2056–2058. [Google Scholar] [CrossRef]
- Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 117–123. [Google Scholar] [CrossRef]
- Sunde, M.; Tharaldsen, H.; Slettemeas, J.S.; Norstrom, M.; Carattoli, A.; Bjorland, J. Escherichia coli of animal origin in Norway contains a blaTEM-20-carrying plasmid closely related to blaTEM-20 and blaTEM-52 plasmids from other European countries. J. Antimicrob. Chemother. 2009, 63, 215–216. [Google Scholar] [CrossRef]
- Simonsen, G.S.; Urdahl, A.M. NORM/NORM-VET 2013 Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway; 1502-2307 (print)/1890-9965 (electronic); Veterinærinstituttet: Tromsø, Norway; Folkehelseinstituttet: Oslo, Norway, 2014.
- Mo, S.S.; Urdahl, A.M.; Nesse, L.L.; Slettemeas, J.S.; Ramstad, S.N.; Torp, M.; Norstrom, M. Occurrence of and risk factors for extended-spectrum cephalosporin-resistant Enterobacteriaceae determined by sampling of all Norwegian broiler flocks during a six month period. PLoS ONE 2019, 14, e0223074. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.S.; Kristoffersen, A.B.; Sunde, M.; Nodtvedt, A.; Norstrom, M. Risk factors for occurrence of cephalosporin-resistant Escherichia coli in Norwegian broiler flocks. Prev. Vet. Med. 2016, 130, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- LeChevallier, M.W.; Cawthon, C.D.; Lee, R.G. Inactivation of biofilm bacteria. Appl. Environ. Microbiol. 1988, 54, 2492–2499. [Google Scholar] [CrossRef]
- Moretro, T.; Vestby, L.K.; Nesse, L.L.; Storheim, S.E.; Kotlarz, K.; Langsrud, S. Evaluation of efficacy of disinfectants against Salmonella from the feed industry. J. Appl. Microbiol. 2009, 106, 1005–1012. [Google Scholar] [CrossRef]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.E. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Gander, S. Bacterial biofilms: Resistance to antimicrobial agents. J. Antimicrob. Chemother. 1996, 37, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Nesse, L.L.; Sekse, C.; Berg, K.; Johannesen, K.C.; Solheim, H.; Vestby, L.K.; Urdahl, A.M. Potentially pathogenic Escherichia coli can form a biofilm under conditions relevant to the food production chain. Appl. Environ. Microbiol. 2014, 80, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Schiebel, J.; Bohm, A.; Nitschke, J.; Burdukiewicz, M.; Weinreich, J.; Ali, A.; Roggenbuck, D.; Rodiger, S.; Schierack, P. Genotypic and Phenotypic Characteristics Associated with Biofilm Formation by Human Clinical Escherichia coli Isolates of Different Pathotypes. Appl. Environ. Microbiol. 2017, 83, e01660-17. [Google Scholar] [CrossRef]
- Nesse, L.L.; Osland, A.M.; Mo, S.S.; Sekse, C.; Slettemeas, J.S.; Bruvoll, A.E.E.; Urdahl, A.M.; Vestby, L.K. Biofilm forming properties of quinolone resistant Escherichia coli from the broiler production chain and their dynamics in mixed biofilms. BMC Microbiol. 2020, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef]
- Gualdi, L.; Tagliabue, L.; Bertagnoli, S.; Ierano, T.; De Castro, C.; Landini, P. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 2008, 154, 2017–2024. [Google Scholar] [CrossRef]
- Solano, C.; Garcia, B.; Valle, J.; Berasain, C.; Ghigo, J.M.; Gamazo, C.; Lasa, I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002, 43, 793–808. [Google Scholar] [CrossRef]
- Cimdins, A.; Simm, R. Semiquantitative Analysis of the Red, Dry, and Rough Colony Morphology of Salmonella enterica Serovar Typhimurium and Escherichia coli Using Congo Red. Methods Mol. Biol. 2017, 1657, 225–241. [Google Scholar] [CrossRef]
- Romling, U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. CMLS 2005, 62, 1234–1246. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Moretro, T.; Langsrud, S.; Heir, E.; Nesse, L.L. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 2009, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Romling, U. Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Sanz-Garcia, F.; Blanco, P.; Martinez, J.L. Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem. 2017, 61, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.S.; Slettemeas, J.S.; Berg, E.S.; Norstrom, M.; Sunde, M. Plasmid and Host Strain Characteristics of Escherichia coli Resistant to Extended-Spectrum Cephalosporins in the Norwegian Broiler Production. PLoS ONE 2016, 11, e0154019. [Google Scholar] [CrossRef]
- Mo, S.S.; Sunde, M.; Ilag, H.K.; Langsrud, S.; Heir, E. Transfer Potential of Plasmids Conferring Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Poultry. Appl. Environ. Microbiol. 2017, 83, e00654-17. [Google Scholar] [CrossRef] [PubMed]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Vestby, L.K.; Moretro, T.; Ballance, S.; Langsrud, S.; Nesse, L.L. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 2009, 5, 43. [Google Scholar] [CrossRef]
- Doumith, M.; Day, M.J.; Hope, R.; Wain, J.; Woodford, N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J. Clin. Microbiol. 2012, 50, 3108–3110. [Google Scholar] [CrossRef]
- Sunde, M.; Simonsen, G.S.; Slettemeas, J.S.; Bockerman, I.; Norstrom, M. Integron, Plasmid and Host Strain Characteristics of Escherichia coli from Humans and Food Included in the Norwegian Antimicrobial Resistance Monitoring Programs. PLoS ONE 2015, 10, e0128797. [Google Scholar] [CrossRef][Green Version]
- Schmidt, G.V.; Mellerup, A.; Christiansen, L.E.; Stahl, M.; Olsen, J.E.; Angen, O. Sampling and Pooling Methods for Capturing Herd Level Antibiotic Resistance in Swine Feces using qPCR and CFU Approaches. PLoS ONE 2015, 10, e0131672. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Janssens, J.C.; Steenackers, H.; Robijns, S.; Gellens, E.; Levin, J.; Zhao, H.; Hermans, K.; De Coster, D.; Verhoeven, T.L.; Marchal, K.; et al. Brominated furanones inhibit biofilm formation by Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2008, 74, 6639–6648. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Ko, K.S. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int. J. Antimicrob. Agents 2015, 46, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Schaufler, K.; Semmler, T.; Pickard, D.J.; de Toro, M.; de la Cruz, F.; Wieler, L.H.; Ewers, C.; Guenther, S. Carriage of Extended-Spectrum Beta-Lactamase-Plasmids Does Not Reduce Fitness but Enhances Virulence in Some Strains of Pandemic E. coli Lineages. Front. Microbiol. 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K.; Gonzalez Barrios, A.F.; Herzberg, M.; Lee, J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, R.S.; Vogel, N.; Aizenberg, J. Role of Flagella in Adhesion of Escherichia coli to Abiotic Surfaces. Langmuir 2015, 31, 6137–6144. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, R.S.; Vlamakis, H.; Kim, P.; Khan, M.; Kolter, R.; Aizenberg, J. Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc. Natl. Acad. Sci. USA 2013, 110, 5624–5629. [Google Scholar] [CrossRef]
- Vestby, L.K.; Lonn-Stensrud, J.; Moretro, T.; Langsrud, S.; Aamdal-Scheie, A.; Benneche, T.; Nesse, L.L. A synthetic furanone potentiates the effect of disinfectants on Salmonella in biofilm. J. Appl. Microbiol. 2010, 108, 771–778. [Google Scholar] [CrossRef][Green Version]
- Padilla-Vaca, F.; Vargas-Maya, N.I.; Elizarraras-Vargas, N.U.; Rangel-Serrano, A.; Cardoso-Reyes, L.R.; Razo-Soria, T.; Membrillo-Hernandez, J.; Franco, B. Flotillin homologue is involved in the swimming behavior of Escherichia coli. Arch. Microbiol. 2019, 201, 999–1008. [Google Scholar] [CrossRef]
- Wu, M.C.; Lin, T.L.; Hsieh, P.F.; Yang, H.C.; Wang, J.T. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS ONE 2011, 6, e23500. [Google Scholar] [CrossRef] [PubMed]
- Galie, S.; Garcia-Gutierrez, C.; Miguelez, E.M.; Villar, C.J.; Lombo, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Saldana, Z.; Xicohtencatl-Cortes, J.; Avelino, F.; Phillips, A.D.; Kaper, J.B.; Puente, J.L.; Giron, J.A. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 2009, 11, 992–1006. [Google Scholar] [CrossRef] [PubMed]

| Strain No | Source | Phylo-Type | Morpho-Type | Plasmids | Plasmid Size | Ref. |
|---|---|---|---|---|---|---|
| Donor strains | ||||||
| 2012-01-1292 | Retail meat | D | RDAR | pNVI1292/IncK (accession no KU312044) | 79.3 kB | [7] |
| 2012-01-2798 | Retail meat | A | RDAR | pNVI2798/IncI1 | 100–110 kB | [28] |
| Recipient strains | ||||||
| 2014-01-5835 | Retail meat | A | RDAR | [18] | ||
| 2014-01-11 | Retail meat | B1 | RDAR | [18] | ||
| 2014-01-1268 | Caecum | B1 | RDAR | [18] | ||
| 2014-01-1499 | Retail meat | B1 | RDAR | [18] | ||
| 2014-01-5572 | Retail meat | B1 | RDAR | [18] | ||
| 2014-01-6035 | Retail meat | B1 | RDAR | [18] | ||
| 2014-01-7132 | Retail meat | B1 | RDAR | [18] | ||
| 2014-01-4271 | Caecum | D | RDAR | [18] | ||
| 2014-01-5565 | Caecum | D | RDAR | [18] | ||
| 2014-01-5792 | Caecum | D | RDAR | [18] | ||
| 2014-01-6041 | Caecum | D | RDAR | [18] | ||
| 2014-01-6123 | Caecum | D | RDAR | [18] | ||
| Primer | Sequence | Size |
|---|---|---|
| gadA.F | 5′-GATGAAATGGCGTTGGCGCAAG-3′ | 373 bp |
| gadA.R | 5′-GGCGGAAGTCCCAGACGATATCC-3′ | |
| chuA.F | 5′-ATGATCATCGCGGCGTGCTG-3′ | 281 bp |
| chuA.R | 5′-AAACGCGCTCGCGCCTAAT-3′ | |
| yjaA.F | 5′-TGTTCGCGATCTTGAAAGCAAACGT-3′ | 216 bp |
| yjaA.R | 5′-ACCTGTGACAAACCGCCCTCA-3′ | |
| TSPE4.C2 F | 5′-GCGGGTGAGACAGAAACGCG-3′ | 152 bp |
| TSPE4.C2 R | 5′-TTGTCGTGAGTTGCGAACCCG-3′ |
| Biofilm | Growth | |||||||
|---|---|---|---|---|---|---|---|---|
| Morpho-Type | No | % | Mean OD595 ± SD | Min. OD595 | Max. OD595 | Mean OD595 ± SD | Min. OD595 | Max. OD595 |
| RDAR | 86 | 68.8 | 0.693 A ± 0.687 | 0.062 | 3.134 | 0.575 A ± 0.104 | 0.282 | 0.786 |
| BDAR | 8 | 6.4 | 0.018 B ± 0.032 | 0.000 | 0.096 | 0.388 B ± 0.087 | 0.316 | 0.593 |
| PDAR | 27 | 21.6 | 0.005 B ± 0.006 | −0.006 | 0.026 | 0.386 B ± 0.033 | 0.255 | 0.431 |
| SAW | 4 | 3.2 | 0.006 B ± 0.005 | 0.003 | 0.013 | 0.500 A ± 0.082 | 0.389 | 0.584 |
| TOTAL | 125 | 100.0 | 0.479 ± 0.652 | −0.006 | 3.134 | 0.520 ± 0.124 | 0.255 | 0.786 |
| Biofilm | % of Each Morphotype | |||||||
|---|---|---|---|---|---|---|---|---|
| Phylo-Type | No | Mean OD595 ± SD | Min. OD595 | Max OD595 | RDAR | BDAR | PDAR | SAW |
| A | 18 | 0.924 A ± 1.066 | 0.003 | 3.134 | 88.9 | 0.0 | 0.0 | 11.1 |
| B1 | 3 | 0.982 * ± 1.327 | 0.179 | 2.513 | 100.0 | 0.0 | 0.0 | 0.0 |
| B2 | 1 | 0.096 * | - | - | 100.0 | 0.0 | 0.0 | 0.0 |
| D | 103 | 0.391 B ± 0.491 | −0.006 | 2.433 | 65.0 | 6.8 | 26.2 | 1.9 |
| Biofilm | % of Each Morphotype | % of Each Phylotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | No | Mean OD595 ± SD | RDAR | BDAR | PDAR | SAW | A | B1 | B2 | D |
| Caecum | 73 | 0.540 ± 0.650 | 76.7 | 4.1 | 17.8 | 1.4 | 15.1 | 1.4 | 1.4 | 82.2 |
| Meat | 52 | 0.393 ± 0.651 | 57.7 | 9.6 | 26.9 | 5.8 | 13.5 | 3.8 | 0.0 | 82.7 |
| Chicken Caecal Samples | Chicken Retail Meat Samples | |||||
|---|---|---|---|---|---|---|
| Phylo-Type | No | Mean Biofilm OD595 ± SD | No | Mean Biofilm OD595 ± SD | p-Value * | |
| All strains | A | 11 | 1.076 ± 1.066 | 7 | 0.686 ± 1.104 | 0.47 |
| D | 60 | 0.417 ± 0.430 | 43 | 0.354 ± 0.569 | 0.53 | |
| RDAR strains | A | 11 | 1.076 ± 1.066 | 5 | 0.959 ± 1.225 | 0.85 |
| D | 45 | 0.566 ± 0.410 | 23 | 0.657 ± 0.641 | 0.48 | |
| Recipient Quinolone-Resistant E. coli (QREC) Strains | Transconjugant Strains | |
|---|---|---|
| With Plasmid pNVI1292/IncK | With Plasmid pNVI2798/IncI1 | |
| 2014-01-5835 | - | - |
| 2014-01-11 | 11-IncK | 11-IncI1 |
| 2014-01-1268 | 1268-IncK | - |
| 2014-01-1499 | - | - |
| 2014-01-5572 | 5572-IncK | 5572-IncI1 |
| 2014-01-6035 | 6035-IncK | 6035-IncI1 |
| 2014-01-7132 | - | 7132-IncI1 |
| 2014-01-4271 | - | - |
| 2014-01-5565 | - | 5565-IncI1 |
| 2014-01-5792 | - | 5792-IncI1 |
| 2014-01-6041 | - | 6041-IncI1 |
| 2014-01-6123 | - | 6123-IncI1 |
| Parameter | Biofilm | Motility | Growth | Growth Rate | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Assay | CVBA | CVBA | CR-BA | CR-BA | MA | CVBA | CVBA | MGA | MGA | MGA |
| Temperature | 20 °C | 37 °C | 20 °C | 37 °C | 37 °C | 20 °C | 37 °C | 20 °C | 37 °C | 37 °C |
| TC-IncK | ||||||||||
| 11-IncK | 77.7 | 75.3 | 85.7 | 100.0 | 81.0 | 94.5 | 97.8 | 98.0 | 98.0 | 98.7 |
| 1268-IncK | 68.6 | 41.6 | 83.3 | 100.0 | 82.4 | 81.0 | 136.1 | 101.4 | 105.9 | 102.8 |
| 5572-IncK | 76.6 | 75.4 | 150.0 | 120.0 | 66.7 | 91.4 | 98.3 | 100.2 | 99.4 | 99.3 |
| 6035-IncK | 92.8 | 54.1 | 100.0 | 133.3 | 110.0 | 96.6 | 98.5 | 101.4 | 99.4 | 102.2 |
| Mean | 78.9 | 61.6 | 104.8 | 113.3 | 85.0 | 90.9 | 107.7 | 100.2 | 100.7 | 100.7 |
| p-value * | 0.02 | 0.03 | 0.89 | 0.22 | 0.15 | 0.11 | 0.45 | 0.78 | 0.71 | 0.53 |
| TC-IncI1 | ||||||||||
| 11-IncI1 | 71.3 | 83.5 | 85.7 | 90.0 | 69.0 | 96.1 | 97.6 | 100.4 | 94.0 | 96.8 |
| 5565-IncI1 | 60.9 | 65.2 | 93.8 | 81.3 | 82.2 | 102.2 | 100.0 | 99.2 | 103.1 | 99.5 |
| 5572-IncI1 | 65.4 | 93.3 | 120.0 | 120.0 | 63.0 | 91.7 | 101.1 | 100.2 | 98.7 | 96.7 |
| 5792-IncI1 | 91.6 | 113.6 | 86.7 | 100.0 | 83.6 | 111.1 | 109.4 | 97.4 | 98.5 | 97.5 |
| 6035-IncI1 | 67.5 | 72.8 | 100.0 | 108.3 | 75.9 | 92.5 | 89.0 | 100.8 | 98.2 | 97.7 |
| 6041-IncI1 | 55.1 | 29.2 | 82.4 | 92.3 | 96.4 | 96.1 | 89.9 | 100.0 | 96.8 | 102.3 |
| 6123-IncI1 | 96.6 | 89.7 | 100.0 | 93.8 | 86.0 | 117.7 | 105.9 | 97.4 | 102.4 | 99.4 |
| 7132-IncI1 | 81.0 | 52.6 | 100.0 | 73.3 | 83.3 | 109.6 | 105.5 | 100.6 | 97.1 | 96.9 |
| Mean | 73.7 | 75.0 | 96.1 | 94.9 | 79.9 | 102.1 | 99.8 | 99.5 | 98.6 | 98.4 |
| p-value * | 0.03 | 0.02 | 0.22 | 0.25 | <0.01 | 0.48 | 0.24 | 0.34 | 0.21 | 0.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nesse, L.L.; Mo, S.S.; Ramstad, S.N.; Witsø, I.L.; Sekse, C.; Bruvoll, A.E.E.; Urdahl, A.M.; Vestby, L.K. The Effect of Antimicrobial Resistance Plasmids Carrying blaCMY-2 on Biofilm Formation by Escherichia coli from the Broiler Production Chain. Microorganisms 2021, 9, 104. https://doi.org/10.3390/microorganisms9010104
Nesse LL, Mo SS, Ramstad SN, Witsø IL, Sekse C, Bruvoll AEE, Urdahl AM, Vestby LK. The Effect of Antimicrobial Resistance Plasmids Carrying blaCMY-2 on Biofilm Formation by Escherichia coli from the Broiler Production Chain. Microorganisms. 2021; 9(1):104. https://doi.org/10.3390/microorganisms9010104
Chicago/Turabian StyleNesse, Live L., Solveig S. Mo, Silje N. Ramstad, Ingun L. Witsø, Camilla Sekse, Anna Eline E. Bruvoll, Anne Margrete Urdahl, and Lene K. Vestby. 2021. "The Effect of Antimicrobial Resistance Plasmids Carrying blaCMY-2 on Biofilm Formation by Escherichia coli from the Broiler Production Chain" Microorganisms 9, no. 1: 104. https://doi.org/10.3390/microorganisms9010104
APA StyleNesse, L. L., Mo, S. S., Ramstad, S. N., Witsø, I. L., Sekse, C., Bruvoll, A. E. E., Urdahl, A. M., & Vestby, L. K. (2021). The Effect of Antimicrobial Resistance Plasmids Carrying blaCMY-2 on Biofilm Formation by Escherichia coli from the Broiler Production Chain. Microorganisms, 9(1), 104. https://doi.org/10.3390/microorganisms9010104






