Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Bacteriophage Propagation
2.3. Quantitative Real-Time PCR (qPCR)
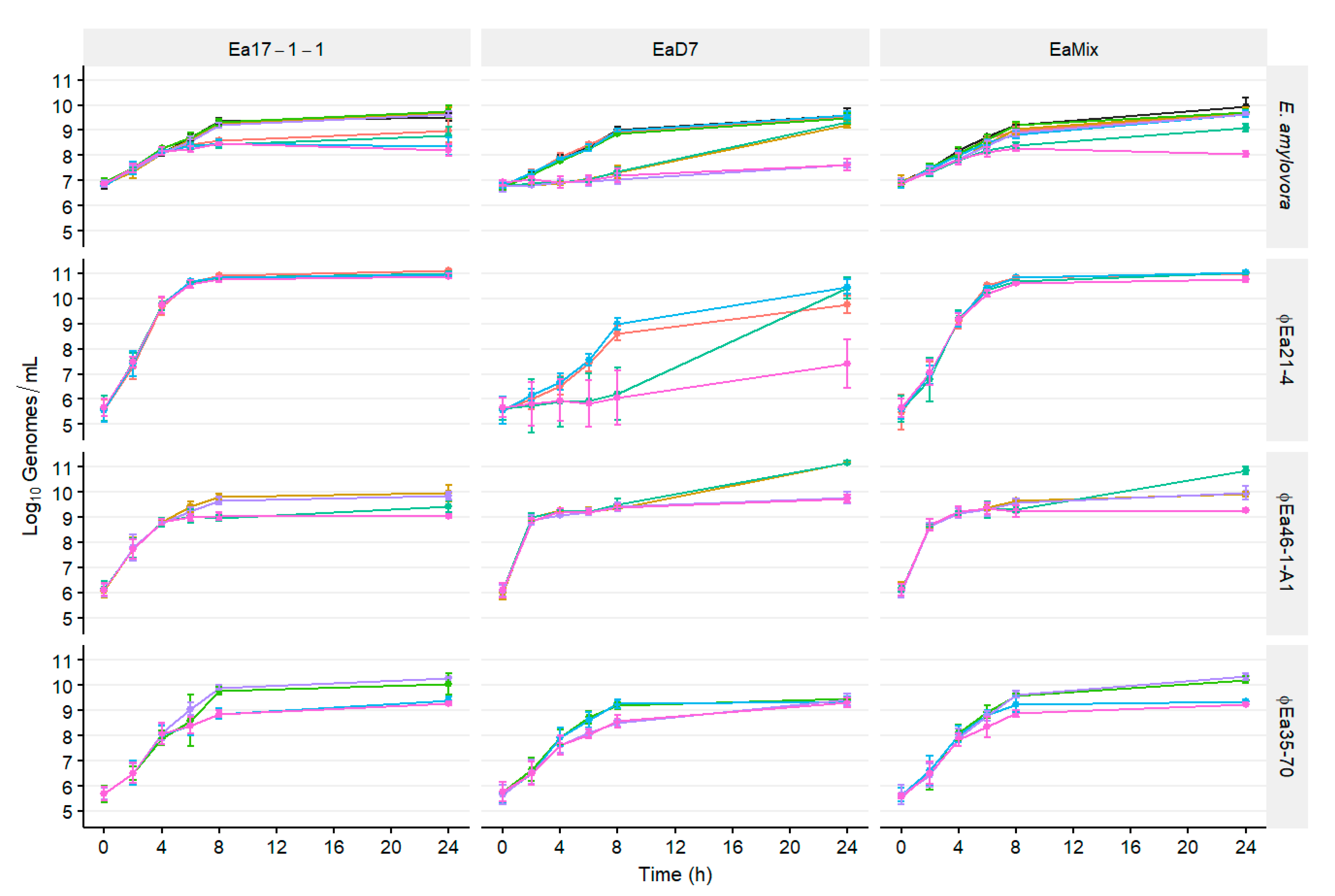
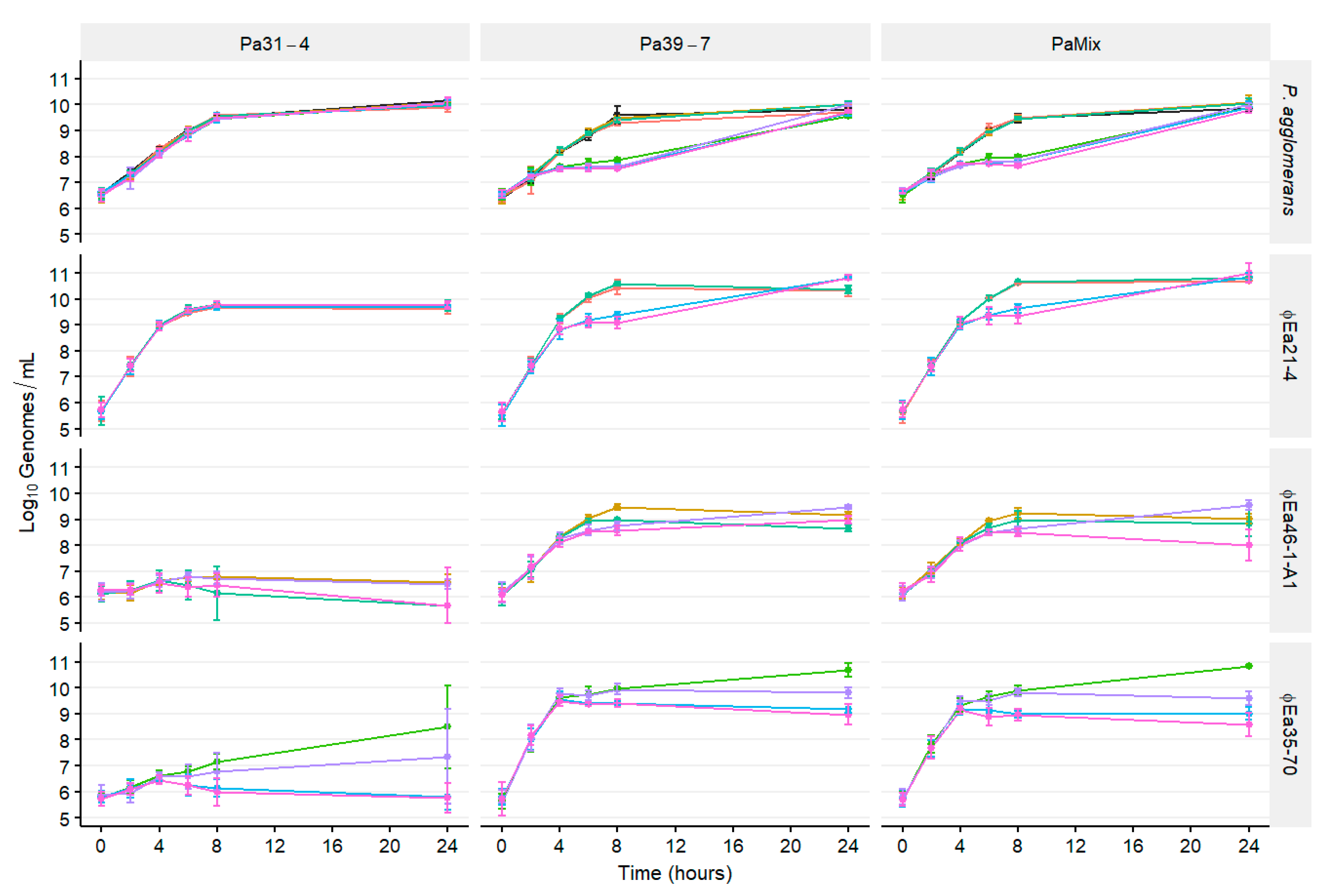
2.4. Population Dynamics of E. amylovora, P. agglomerans, and Different Phage Cocktails
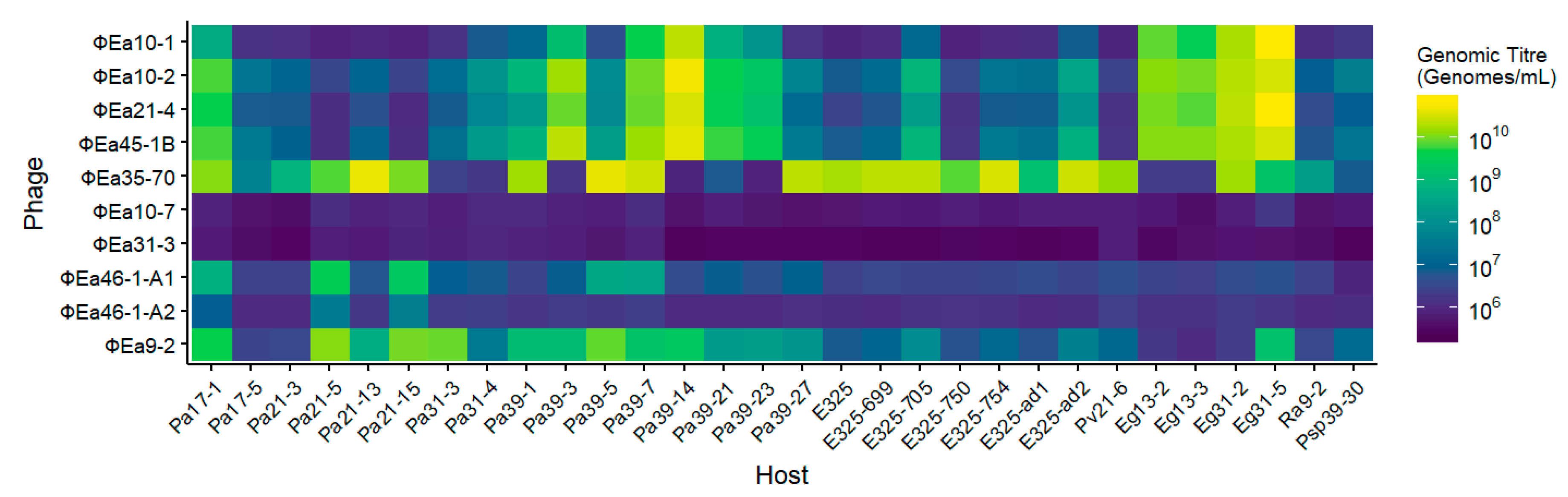
2.5. Phage Host Range Assay
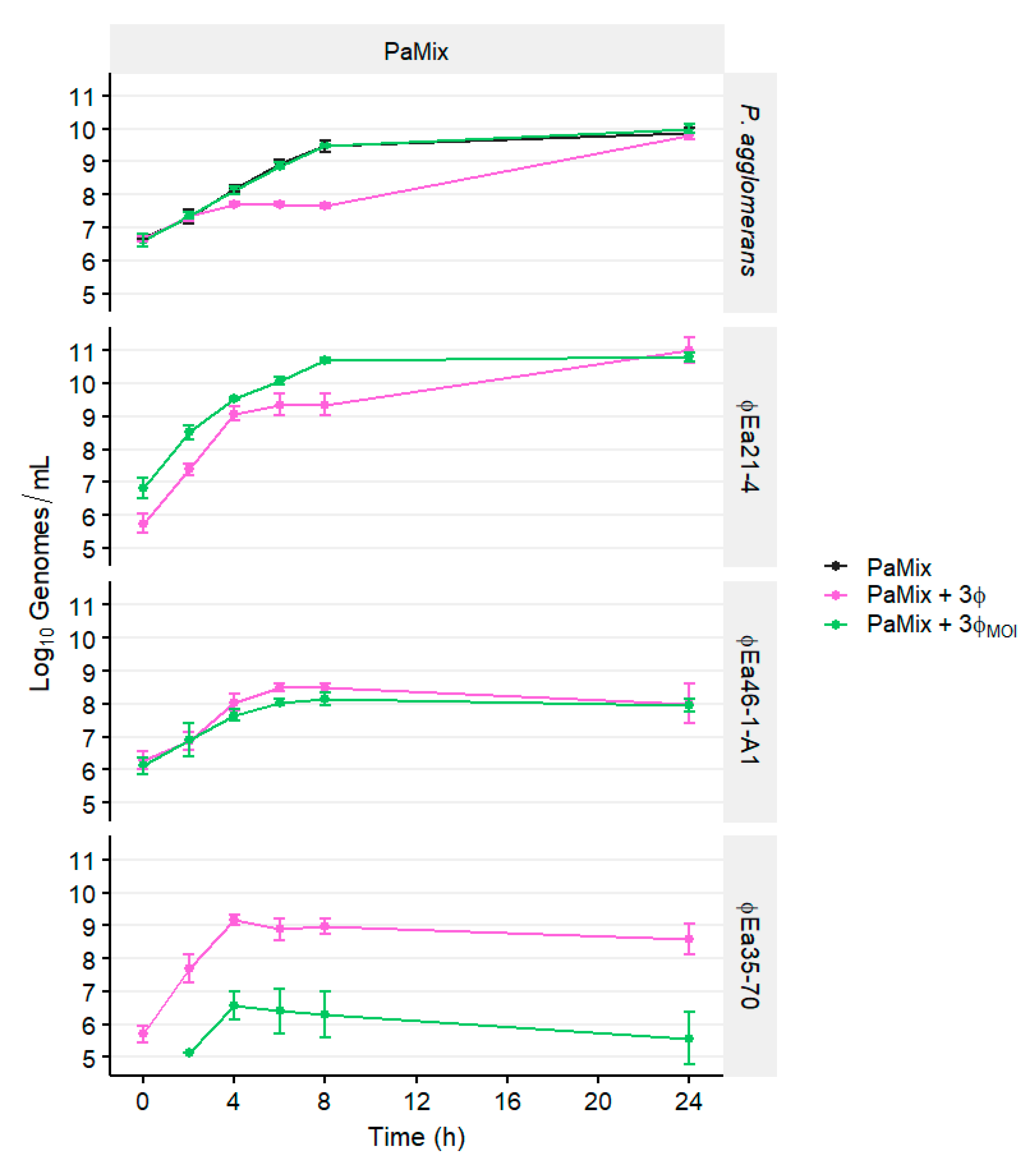
2.6. Phage and Carrier Combinations for E. amylovora Reduction
2.7. Genomic Sequencing of P. agglomerans and other Epiphytes
2.8. Data Analysis
3. Results
3.1. Population Dynamics of E. amylovora and Different Phage Cocktails
3.2. Host Range of Phages on Potential Carriers
3.3. Population Dynamics of P. agglomerans and Different Phage Cocktails
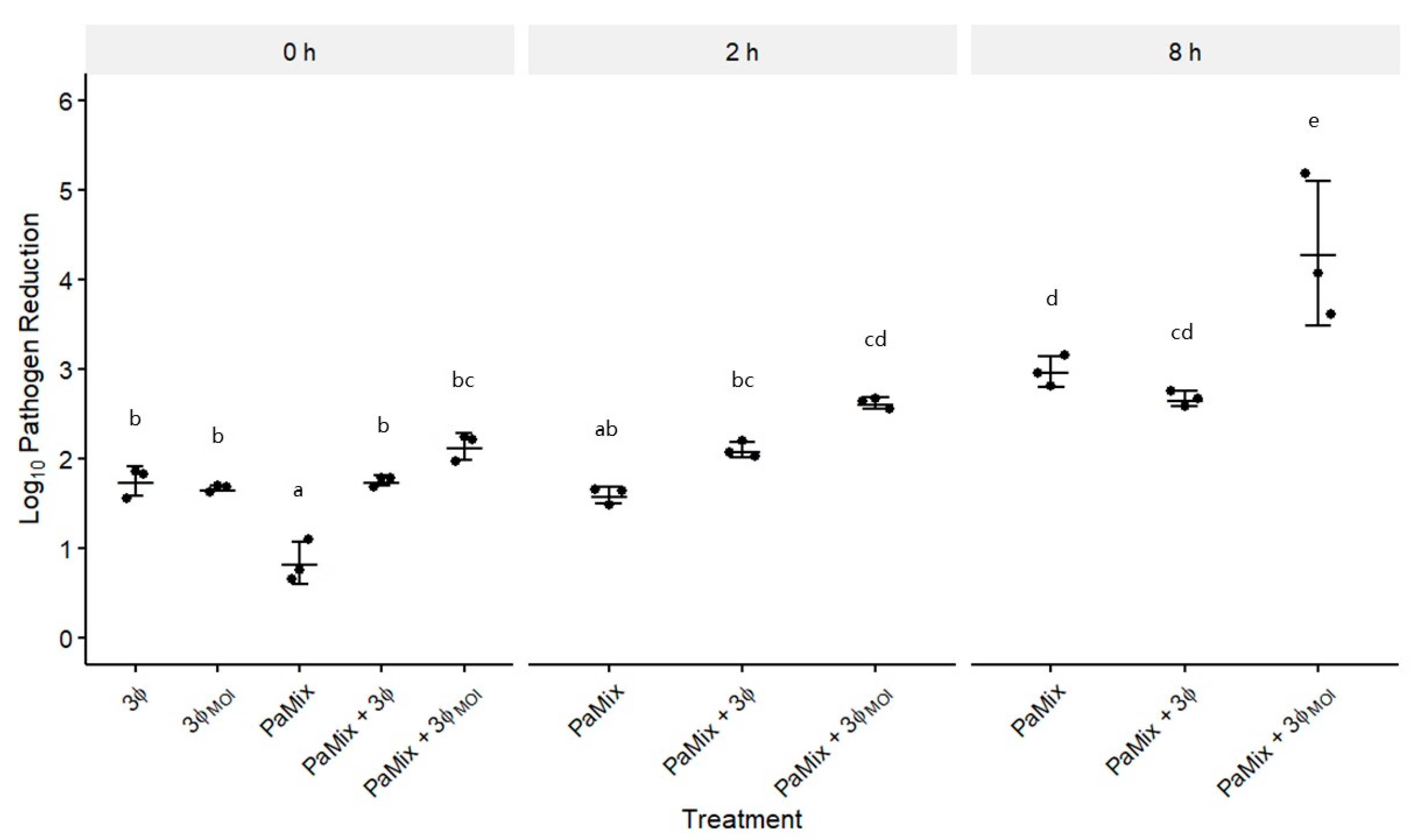
3.4. Phage and Carrier Combinations for E. amylovora Reduction
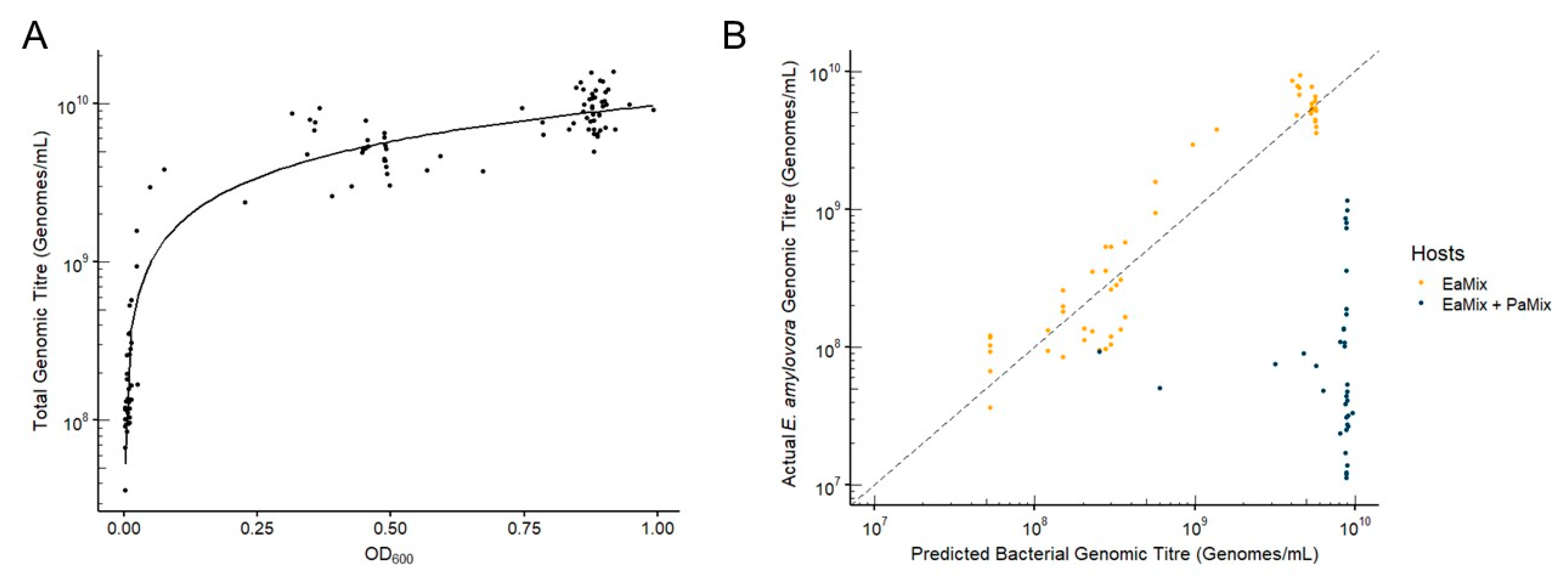
3.5. Comparison of qPCR and Spectrophotometry for Quantification of Mixed Bacterial Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the soil environment-degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Gill, J.J. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 2014, 5, 618. [Google Scholar] [CrossRef] [PubMed]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Krysiak-Baltyn, K.; Martin, G.J.; Stickland, A.D.; Scales, P.J.; Gras, S.L. Computational models of populations of bacteria and lytic phage. Crit. Rev. Microbiol. 2016, 42, 942–968. [Google Scholar] [CrossRef]
- van der Zwet, T.; Orolaza-Halbrendt, N.; Zeller, W. Spread and current distribution of fire blight. In Fire Blight: History, Biology, and Management; The American Phytopathological Society: Saint Paul, MN, USA, 2016; pp. 15–36. [Google Scholar]
- Pusey, P.L. The role of water in epiphytic colonization and infection of pomaceous flowers by Erwinia amylovora. Phytopathology 2000, 90, 1352–1357. [Google Scholar] [CrossRef]
- van der Zwet, T.; Orolaza-Halbrendt, N.; Zeller, W. The disease cycle of fire blight. In Fire Blight: History, Biology, and Management; The American Phytopathological Society: Saint Paul, MN, USA, 2016; pp. 155–182. [Google Scholar]
- Cui, Z.; Huntley, R.B.; Zeng, Q.; Steven, B. Temporal and spatial dynamics in the apple flower microbiome in the presence of the phytopathogen Erwinia amylovora. bioRxiv 2020. [Google Scholar] [CrossRef]
- Steven, B.; Huntley, R.B.; Zeng, Q. The influence of flower anatomy and apple cultivar on the apple flower phytobiome. Phytobiomes. J. 2018, 2, 171–179. [Google Scholar] [CrossRef]
- Stockwell, V.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [CrossRef]
- Pusey, P.L.; Stockwell, V.O.; Mazzola, M. Epiphytic bacteria and yeasts on apple blossoms and their potential as antagonists of Erwinia amylovora. Phytopathology 2009, 99, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Pusey, P.L.; Stockwell, V.O.; Reardon, C.L.; Smits, T.H.; Duffy, B. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 2011, 101, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M. Development of a Bacteriophage-Based Biopesticide for Fire Blight. Ph.D. Thesis, Biological Sciences, Brock University, St. Catharines, ON, Canada, 2007. [Google Scholar]
- Gayder, S.; Parcey, M.; Castle, A.J.; Svircev, A.M. Host range of bacteriophages against a world-wide collection of Erwinia amylovora determined using a quantitative PCR assay. Viruses 2019, 11, 910. [Google Scholar] [CrossRef]
- Roach, D.R.; Sjaarda, D.R.; Castle, A.J.; Svircev, A.M. Host exopolysaccharide quantity and composition impact Erwinia amylovora bacteriophage pathogenesis. Appl. Environ. Microbiol. 2013, 79, 3249–3256. [Google Scholar] [CrossRef]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 2016, 9, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Yan, T.; Willias, S.P.; Mia, M.Z.; Bei, W.; Connerton, I.F.; Fischetti, V.A.; et al. Application of a broad range lytic phage LPST94 for biological control of Salmonella in foods. Microorganisms 2020, 8, 247. [Google Scholar] [CrossRef]
- Xie, Y.; Wahab, L.; Gill, J.J. Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 2018, 10, 189. [Google Scholar] [CrossRef]
- Zaczek-Moczydlowska, M.A.; Young, G.K.; Trudgett, J.; Plahe, C.; Fleming, C.C.; Campbell, K.; O’Hanlon, R. Phage cocktail containing Podoviridae and Myoviridae bacteriophages inhibits the growth of Pectobacterium spp. under in vitro and in vivo conditions. PLoS ONE 2020, 15, e0230842. [Google Scholar] [CrossRef]
- Kifelew, L.G.; Warner, M.S.; Morales, S.; Thomas, N.; Gordon, D.L.; Mitchell, J.G.; Speck, P.G. Efficacy of lytic phage cocktails on Staphylococcus aureus and Pseudomonas aeruginosa in mixed-species planktonic cultures and biofilms. Viruses 2020, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Duyvejonck, H.; Merabishvili, M.; Pirnay, J.P.; De Vos, D.; Verbeken, G.; Van Belleghem, J.; Gryp, T.; De Leenheer, J.; Van der Borght, K.; Van Simaey, L.; et al. Development of a qPCR platform for quantification of the five bacteriophages within bacteriophage cocktail 2 (BFC2). Sci. Rep. 2019, 9, 13893. [Google Scholar] [CrossRef] [PubMed]
- Parcey, M.; Gayder, S.; Castle, A.J.; Svircev, A.M. Molecular profile of phage infection: A novel approach for the characterization of Erwinia phages through qPCR. Int. J. Mol. Sci. 2020, 21, 553. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.K. Survey results of Erwinia amylovora in Utah for resistance to streptomycin and investigations comparing kasugamycin (kasumin) to streptomycin and oxytetracycline for control of fire blight. Acta Hortic. 2008, 793, 433–437. [Google Scholar] [CrossRef]
- Roach, D.R.; Sjaarda, D.R.; Sjaarda, C.P.; Ayala, C.J.; Howcroft, B.; Castle, A.J.; Svircev, A.M. Absence of lysogeny in wild populations of Erwinia amylovora and Pantoea agglomerans. Microb. Biotechnol. 2015, 8, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Svircev, A.M.; Smith, R.; Castle, A.J. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 2003, 69, 2133–21388. [Google Scholar] [CrossRef]
- Lehman, S.M.; Kropinski, A.M.; Castle, A.J.; Svircev, A.M. Complete genome of the broad-host-range Erwinia amylovora phage ϕEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 2009, 75, 2139–2147. [Google Scholar] [CrossRef]
- Yagubi, A.I.; Castle, A.J.; Kropinski, A.M.; Banks, T.W.; Svircev, A.M. Complete genome sequence of Erwinia amylovora bacteriophage vB_EamM_Ea35-70. Genome Announc. 2014, 2, e00413–e00414. [Google Scholar] [CrossRef]
- Del Fabbro, C.; Scalabrin, S.; Morgante, M.; Giorgi, F.M. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS ONE 2013, 8, e85024. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files, Version 1.33; 2011. Available online: https://github.com/najoshi/sickle. (accessed on 4 March 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; Version 3.5.1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation, R Package Version 0.7.6; 2018. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 13 September 2018).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; R Package Version 3.0.0; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H. Scales: Scale Functions for Visualization, R Package Version 1.0.0; 2018. Available online: https://CRAN.R-project.org/package=scales (accessed on 6 October 2018).
- Arnold, J.B. ggthemes: Extra Themes, Scales and Geoms for ‘ggplot2’, R Package Version 4.0.1; 2018. Available online: https://CRAN.R-project.org/package=ggthemes (accessed on 6 October 2018).
- Danelishvili, L.; Young, L.S.; Bermudez, L.E. In vivo efficacy of phage therapy for Mycobacterium avium infection as delivered by a nonvirulent mycobacterium. Microb. Drug Resist. 2006, 12, 1–6. [Google Scholar] [CrossRef]
- Tanaka, H.; Negishi, H.; Maeda, H. Control of tobacco bacterial wilt by an avirulent strain of Pseudomonas solanacearum M4S and its bacteriophage. Jpn. J. Phytopathol. 1990, 56, 243–246. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 2019, 179, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell 2017, 66, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef]
- Rousset, F.; Cui, L.; Siouve, E.; Becavin, C.; Depardieu, F.; Bikard, D. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet. 2018, 14, e1007749. [Google Scholar] [CrossRef]
- Zeng, Z.; Salmond, G.P.C. Bacteriophage host range evolution through engineered enrichment bias, exploiting heterologous surface receptor expression. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Jensen, D.F.; Karlsson, M.; Sarrocco, S.; Vannacci, G. Biological control using microorganisms as an alternative to disease resistance. In Plant Pathogen Resistance Biotechnology; Collinge, D.B., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 341–363. [Google Scholar]
- Pratama, A.A.; Terpstra, J.; de Oliveria, A.L.M.; Salles, J.F. The role of rhizosphere bacteriophages in plant health. Trends Microbiol. 2020, 28, 709–718. [Google Scholar] [CrossRef]
- Barr, J.J. A bacteriophages journey through the human body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef]
- Papaianni, M.; Paris, D.; Woo, S.L.; Fulgione, A.; Rigano, M.M.; Parrilli, E.; Tutino, M.L.; Marra, R.; Manganiello, G.; Casillo, A.; et al. Plant dynamic metabolic response to bacteriophage treatment after Xanthomonas campestris pv. campestris infection. Front. Microbiol. 2020, 11, 732. [Google Scholar] [CrossRef]

| Strain | Accession Number | Reference |
|---|---|---|
| Erwinia amylovora | ||
| Ea17-1-1 | JAAEVW000000000 | [16] |
| EaD7 | JAAEUT000000000 | [16] |
| Ea6-4 | JAAEVD000000000 | [16] |
| Ea29-7 | JAAEVM000000000 | [16] |
| 20060013 | JAAEXH000000000 | [16,27] |
| 20070126 | JAAEXF000000000 | [16,27] |
| Pantoea agglomerans | ||
| Pa17-1 | JACSXN000000000 | [28] |
| Pa17-5 | JACSXM000000000 | [28] |
| Pa21-3 | JACSXL000000000 | [28] |
| Pa21-5 | JACSXK000000000 | [28] |
| Pa21-13 | JACSXI000000000 | [28] |
| Pa21-15 | JACSXH000000000 | [28] |
| Pa31-3 | JACSXF000000000 | [28] |
| Pa31-4 | JACSXE000000000 | [28] |
| Pa39-1 | JACSXC000000000 | [28] |
| Pa39-3 | JACSXB000000000 | [28] |
| Pa39-5 | JACSXA000000000 | [28] |
| Pa39-7 | JACSWZ000000000 | [28] |
| Pa39-14 | JACSWY000000000 | [28] |
| Pa39-21 | JACSWX000000000 | [28] |
| Pa39-23 | JACSWW000000000 | [28] |
| Pa39-27 | JACSWV000000000 | [28] |
| E325 | JACSWT000000000 | [14] |
| E325-699 | JACSWS000000000 | [14] |
| E325-705 | JACSWR000000000 | [14] |
| E325-750 | JACSWQ000000000 | [14] |
| E325-754 | JACSWP000000000 | [14] |
| E325-ad1 | JACSWO000000000 | [14] |
| E325-ad2 | JACSWN000000000 | [14] |
| Pantoea eucalypti | ||
| Pv21-6 | JACSXJ000000000 | [28] |
| Erwinia gerundensis | ||
| Eg13-2 | JACSXP000000000 | [28] |
| Eg13-3 | JACSXO000000000 | [28] |
| Eg31-2 | JACSXG000000000 | [28] |
| Eg31-5 | JACSXD000000000 | [28] |
| Rahnella aquatilis | ||
| Ra9-2 | JACSXQ000000000 | [28] |
| Pantoea sp. | ||
| Psp39-30 | JACSWU000000000 | [28] |
| Phage | Family | Genus | Species | Accession Number a | Propagation Host | Reference |
|---|---|---|---|---|---|---|
| ϕEa10-1 | Myoviridae | Kolesnikvirus | Erwinia virus Ea214 | NA | Ea17-1-1 | [29] |
| ϕEa10-2 | NA | Ea6-4 | [29] | |||
| ϕEa21-4 | NC_011811.1 | Ea6-4 | [29,30] | |||
| ϕEa45-1B | NA | Ea6-4 | [29] | |||
| ϕEa35-70 | Agricanvirus | Erwinia virus Ea35-70 | NC_023557.1 | Pa39-7 | [29,31] | |
| ϕEa10-7 | Podoviridae | Eracentumvirus | Erwinia virus Era103 | NA | Ea29-7 | [29] |
| ϕEa31-3 | NA | Ea29-7 | [29] | |||
| ϕEa46-1-A1 | NA | EaD7 | [29] | |||
| ϕEa46-1-A2 | NA | EaD7 | [29] | |||
| ϕEa9-2 | Johnsonvirus | Erwinia virus Ea9-2 | NC_023579.1 | Ea17-1-1 | [29] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gayder, S.; Parcey, M.; Nesbitt, D.; Castle, A.J.; Svircev, A.M. Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy. Microorganisms 2020, 8, 1449. https://doi.org/10.3390/microorganisms8091449
Gayder S, Parcey M, Nesbitt D, Castle AJ, Svircev AM. Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy. Microorganisms. 2020; 8(9):1449. https://doi.org/10.3390/microorganisms8091449
Chicago/Turabian StyleGayder, Steven, Michael Parcey, Darlene Nesbitt, Alan J. Castle, and Antonet M. Svircev. 2020. "Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy" Microorganisms 8, no. 9: 1449. https://doi.org/10.3390/microorganisms8091449
APA StyleGayder, S., Parcey, M., Nesbitt, D., Castle, A. J., & Svircev, A. M. (2020). Population Dynamics between Erwinia amylovora, Pantoea agglomerans and Bacteriophages: Exploiting Synergy and Competition to Improve Phage Cocktail Efficacy. Microorganisms, 8(9), 1449. https://doi.org/10.3390/microorganisms8091449





