Francisella tularensis Subspecies holarctica and Tularemia in Germany
Abstract
1. Introduction
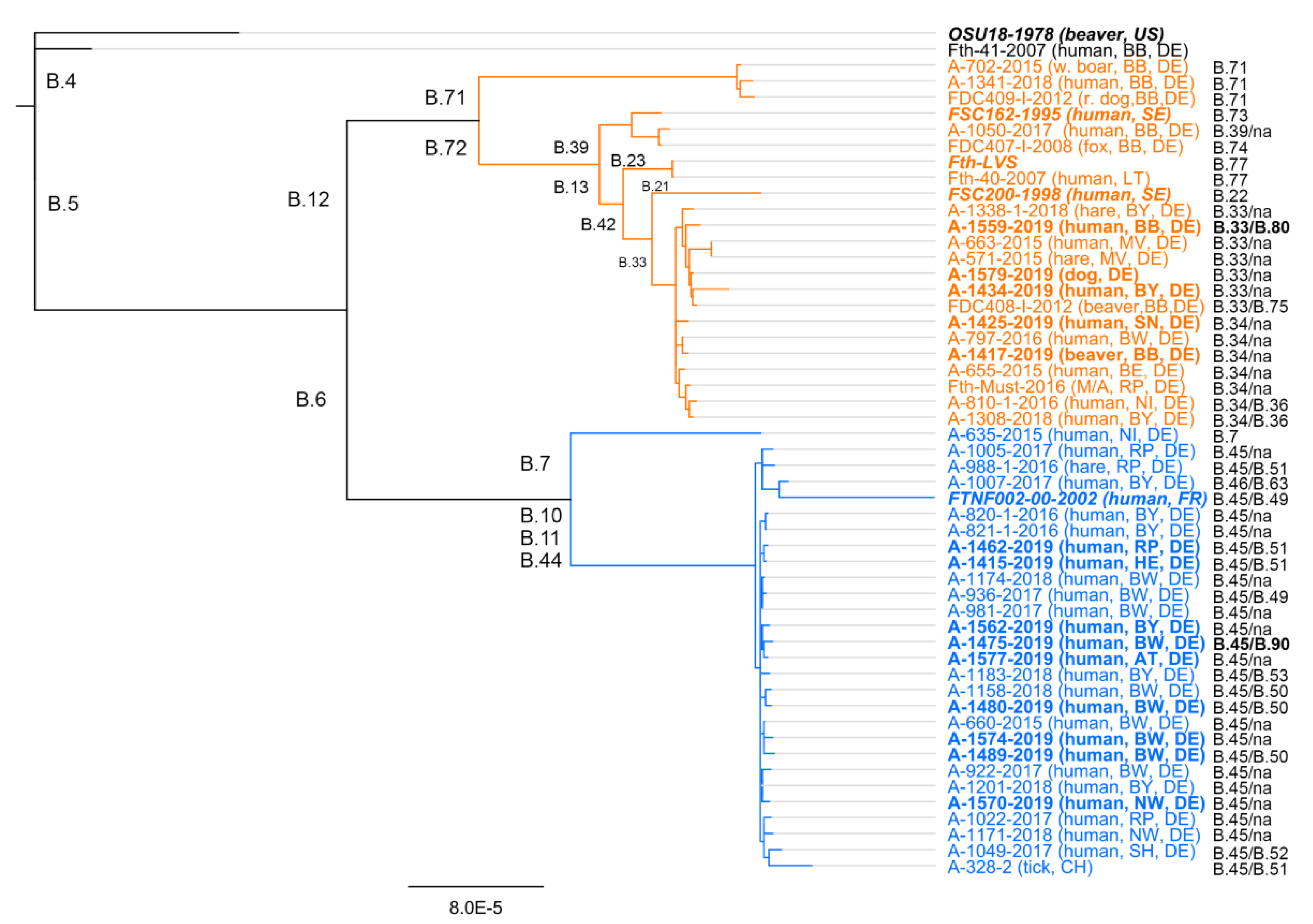
2. Francisella tularensis Subsp. holarctica—Genetic Diversity and Geographic Distribution
3. Tularemia in Germany
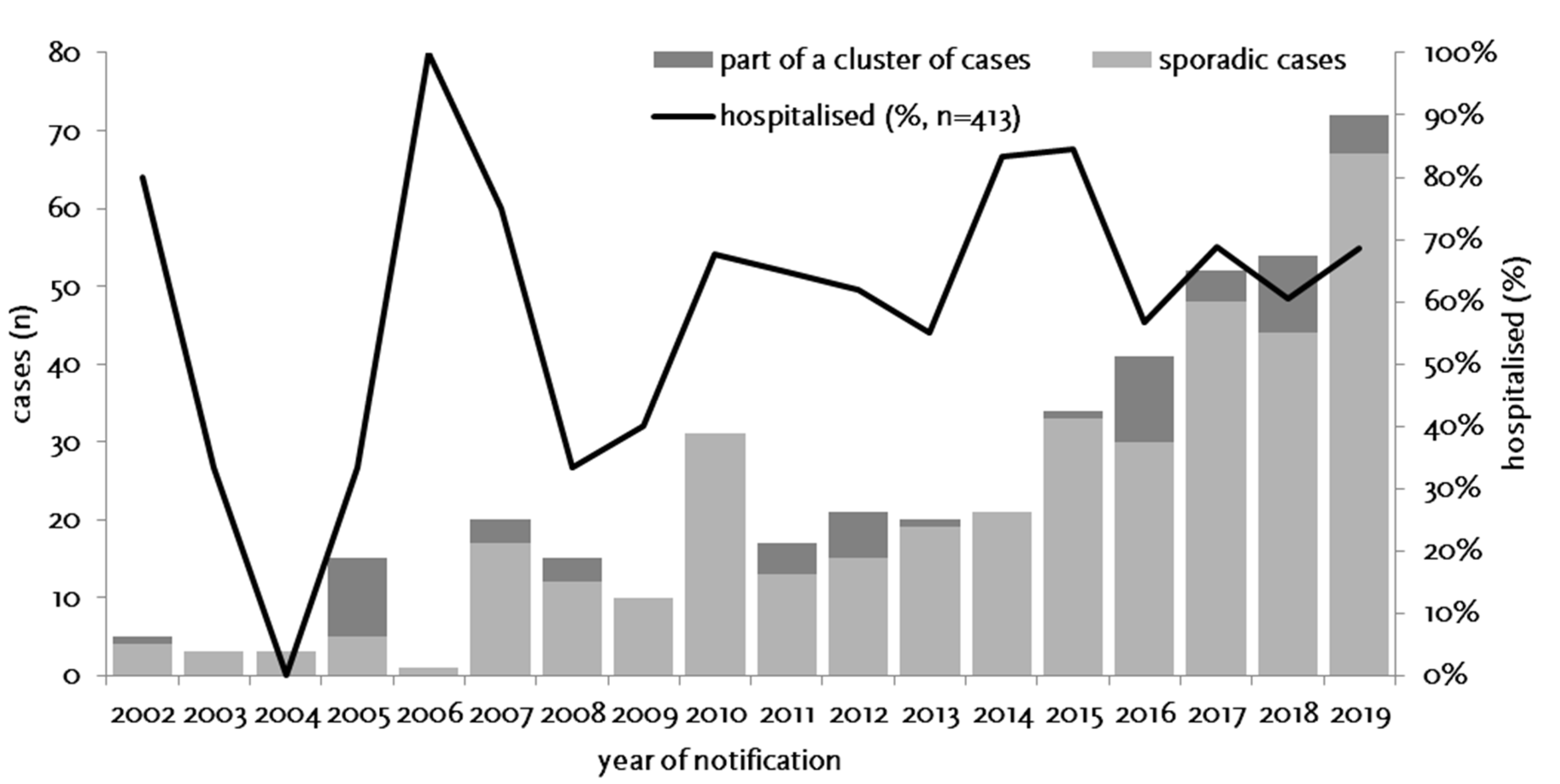
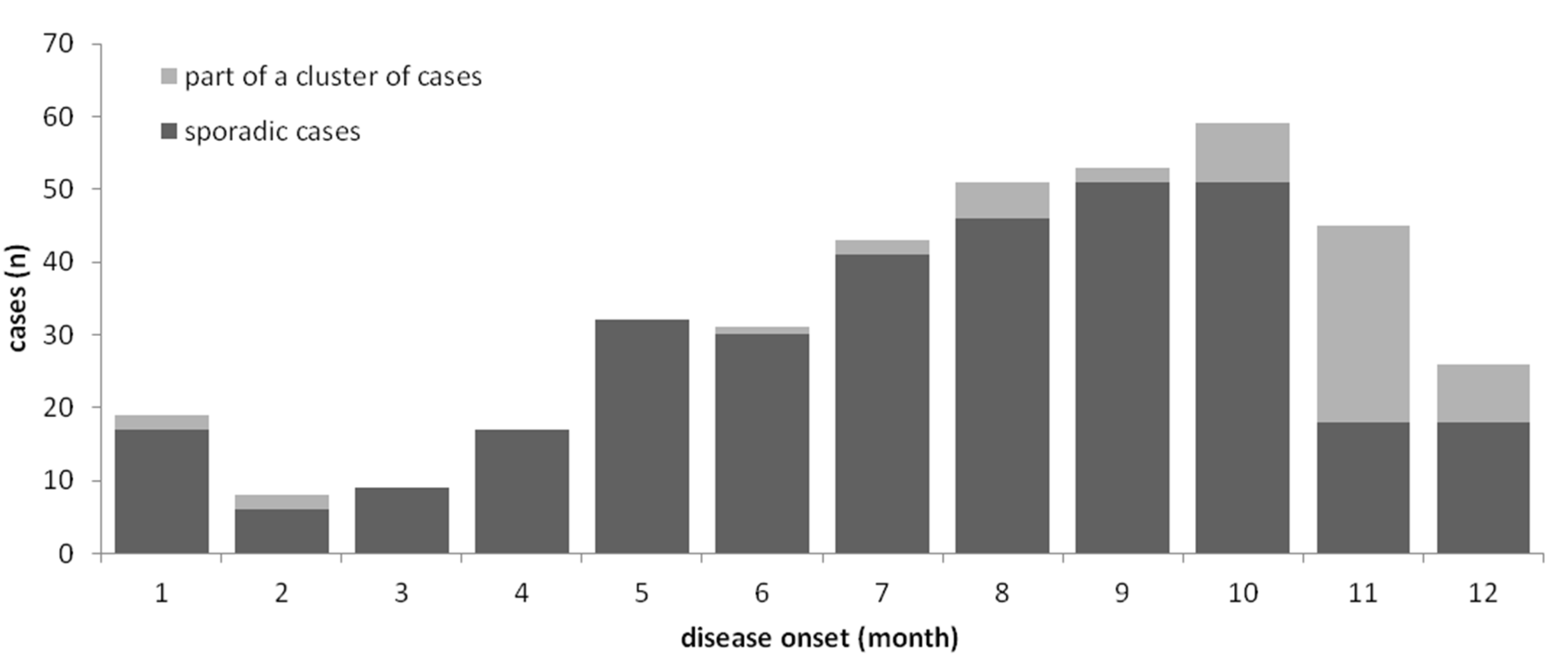
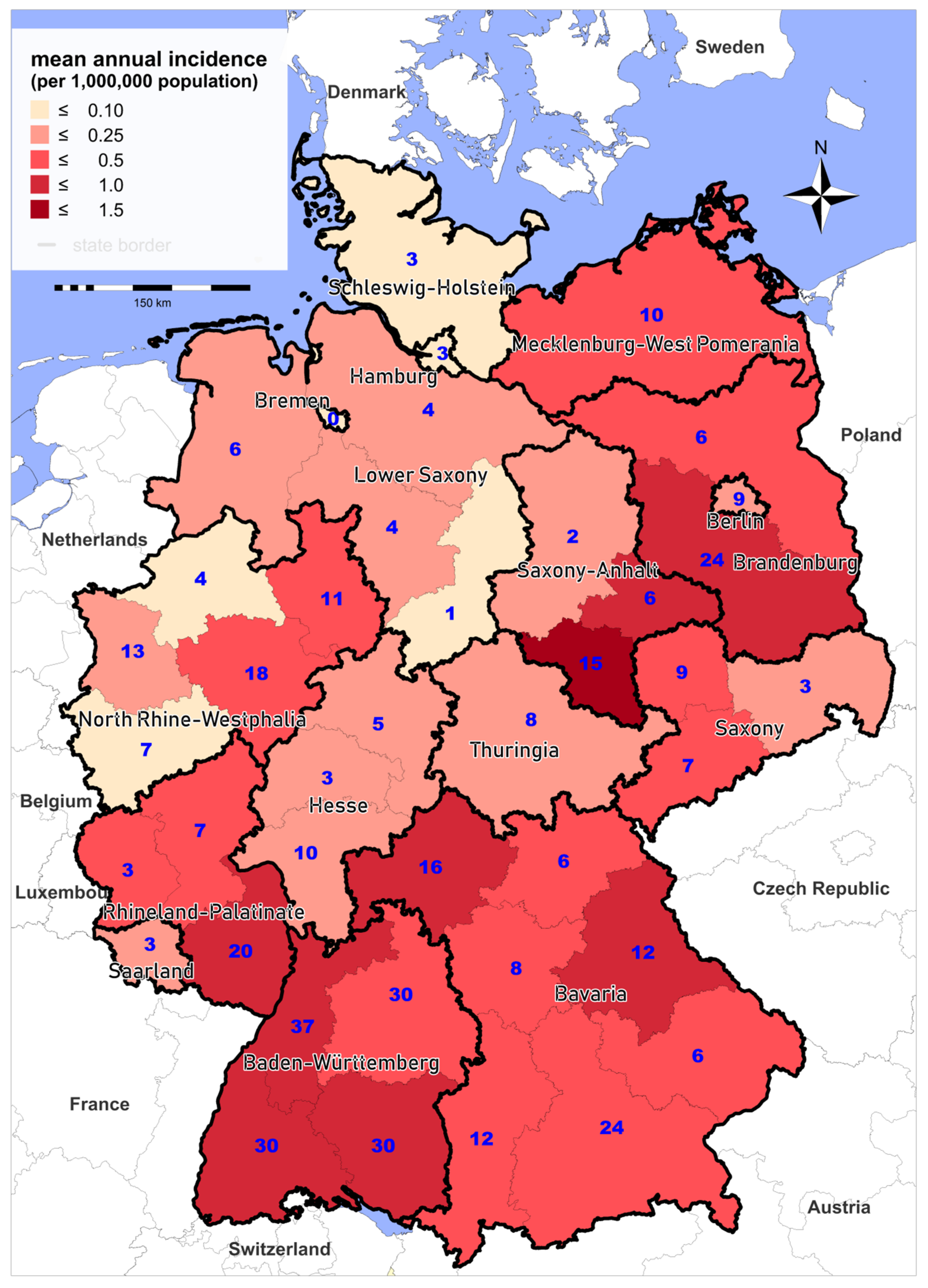
3.1. Epidemiology of Notified Cases
3.2. Outbreaks of Tularemia in Humans
3.3. Clinical Aspects and Diagnosis
4. Francisella sp. Strain W12-1067 (F-W12)—What Is Known
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moreau, G.B.; Mann, B.J. Adherence and uptake ofFrancisellainto host cells. Virulence 2013, 4, 826–832. [Google Scholar] [CrossRef]
- Clemens, D.L.; Lee, B.-Y.; Horwitz, M.A. Francisella tularensis Enters Macrophages via a Novel Process Involving Pseudopod Loops. Infect. Immun. 2005, 73, 5892–5902. [Google Scholar] [CrossRef] [PubMed]
- Santic, M.; Molmeret, M.; Klose, K.E.; Abu Kwaik, Y. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006, 14, 37–44. [Google Scholar] [CrossRef]
- Ellis, J.; Oyston, P.C.F.; Green, M.; Titball, R.W. Tularemia. Clin. Microbiol. Rev. 2002, 15, 631–646. [Google Scholar] [CrossRef]
- Maurin, M.; Gyuranecz, M. Tularaemia: Clinical aspects in Europe. Lancet Infect. Dis. 2016, 16, 113–124. [Google Scholar] [CrossRef]
- Oyston, P.C.; Griffiths, R. Francisellavirulence: Significant advances, ongoing challenges and unmet needs. Expert Rev. Vaccines 2009, 8, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, F.; Hoffmann, D.; Jahn, K.; Heuner, K.; Jacob, D.; Vogt, M.; Bent, S.; Grunow, R.; Zanger, P. Oropharyngeal Tularemia from Freshly Pressed Grape Must. N. Engl. J. Med. 2018, 379, 197–199. [Google Scholar] [CrossRef]
- Jacob, D.; Köppen, K.; Radonić, A.; Haldemann, B.; Zanger, P.; Heuner, K.; Grunow, R. Molecular identification of the source of an uncommon tularaemia outbreak, Germany, autumn 2016. Eurosurveillance 2019, 24, 1800419. [Google Scholar] [CrossRef] [PubMed]
- Wetzstein, N.; Kärcher, I.; Küpper-Tetzel, C.P.; Kann, G.; Hogardt, M.; Jozsa, K.; Jacob, D.; Grunow, R.; Just-Nübling, G.; Wolf, T.; et al. Clinical characteristics in a sentinel case as well as in a cluster of tularemia patients associated with grape harvest. Int. J. Infect. Dis. 2019, 84, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Jenzora, A.; Jansen, A.; Ranisch, H.; Lierz, M.; Wichmann, O.; Grunow, R. Seroprevalence study of Francisella tularensis among hunters in Germany. FEMS Immunol. Med. Microbiol. 2008, 53, 183–189. [Google Scholar] [CrossRef]
- Yeşilyurt, M.; Kılıç, S.; Celebi, B.; Gül, S. Tularemia: Are hunters really a risk group? Mikrobiyoloji Bul. 2012, 46, 153–155. [Google Scholar]
- Hofstetter, I.; Eckert, J.; Splettstoesser, W.; Hauri, A.M. Tularaemia outbreak in hare hunters in the Darmstadt-Dieburg district, Germany. Wkly. Releases 2006, 11, 2878. [Google Scholar] [CrossRef] [PubMed]
- Hauri, A.M.; Hofstetter, I.; Seibold, E.; Kaysser, P.; Eckert, J.; Neubauer, H.; Splettstoesser, W.D. Investigating an Airborne Tularemia Outbreak, Germany. Emerg. Infect. Dis. 2010, 16, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.A.; Murua, C.; Jones, J.M.; Mohler, K.; Zhang, Y.; Wiggins, L.; Kwit, N.A.; Respicio-Kingry, L.; Kingry, L.C.; Petersen, J.M.; et al. Francisella tularensis Transmission by Solid Organ Transplantation, 20171. Emerg. Infect. Dis. 2019, 25, 767–775. [Google Scholar] [CrossRef]
- Jackson, J.; McGregor, A.; Cooley, L.; Ng, J.; Brown, M.; Ong, C.W.; Darcy, C.; Sintchenko, V. Francisella tularensis Subspecies holarctica, Tasmania, Australia, 2011. Emerg. Infect. Dis. 2012, 18, 1484–1486. [Google Scholar] [CrossRef]
- Eden, J.-S.; Rose, K.; Ng, J.; Shi, M.; Wang, Q.; Sintchenko, V.; Holmes, E.C. Francisella tularensis ssp. holarctica in Ringtail Possums, Australia. Emerg. Infect. Dis. 2017, 23, 1198–1201. [Google Scholar] [CrossRef]
- Dwibedi, C.; Birdsell, D.; Lärkeryd, A.; Myrtennäs, K.; Öhrman, C.; Nilsson, E.; Karlsson, E.; Hochhalter, C.; Rivera, A.; Maltinsky, S.; et al. Long-range dispersal moved Francisella tularensis into Western Europe from the East. Microb. Genom. 2016, 2, e000100. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.; Svensson, K.; Lindgren, P.; Byström, M.; Sjödin, A.; Forsman, M.; Johansson, A. The phylogeographic pattern of Francisella tularensis in Sweden indicates a Scandinavian origin of Eurosiberian tularaemia. Environ. Microbiol. 2012, 15, 634–645. [Google Scholar] [CrossRef]
- Vogler, A.J.; Birdsell, D.; Price, L.B.; Bowers, J.R.; Beckstrom-Sternberg, S.M.; Auerbach, R.K.; Beckstrom-Sternberg, J.S.; Johansson, A.; Clare, A.; Buchhagen, J.L.; et al. Phylogeography of Francisella tularensis: Global Expansion of a Highly Fit Clone. J. Bacteriol. 2009, 191, 2474–2484. [Google Scholar] [CrossRef] [PubMed]
- Hestvik, G.; Warns-Petit, E.; Smith, L.A.; Fox, N.J.; Uhlhorn, H.; Artois, M.; Hannant, D.; Hutchings, M.R.; Mattsson, R.; Yon, L.; et al. The status of tularemia in Europe in a one-health context: A review. Epidemiol. Infect. 2014, 143, 2137–2160. [Google Scholar] [CrossRef] [PubMed]
- Faber, M.S.; Heuner, K.; Jacob, D.; Grunow, R. Tularemia in Germany—A Re-emerging Zoonosis. Front. Microbiol. 2018, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Appelt, S.; Köppen, K.; Radonić, A.; Drechsel, O.; Jacob, D.; Grunow, R.; Heuner, K. Genetic Diversity and Spatial Segregation of Francisella tularensis Subspecies holarctica in Germany. Front. Microbiol. 2019, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Gyuranecz, M.; Birdsell, D.N.; Splettstoesser, W.; Seibold, E.; Beckstrom-Sternberg, S.M.; Makrai, L.; Fodor, L.; Fabbi, M.; Vicari, N.; Johansson, A.; et al. Phylogeography of Francisella tularensis subsp. holarctica, Europe. Emerg. Infect. Dis. 2012, 18, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Origgi, F.C.; Frey, J.; Pilo, P. Characterisation of a new group of Francisella tularensis subsp. holarctica in Switzerland with altered antimicrobial susceptibilities, 1996 to 2013. Eurosurveillance 2014, 19, 20858. [Google Scholar] [CrossRef]
- Svensson, K.; Bäck, E.; Eliasson, H.; Berglund, L.; Granberg, M.; Karlsson, L.; Larsson, P.; Forsman, M.; Johansson, A. Landscape Epidemiology of Tularemia Outbreaks in Sweden. Emerg. Infect. Dis. 2009, 15, 1937–1947. [Google Scholar] [CrossRef]
- Wittwer, M.; Altpeter, E.; Pilo, P.; Gygli, S.M.; Beuret, C.; Foucault, F.; Ackermann-Gäumann, R.; Karrer, U.; Jacob, D.; Grunow, R.; et al. Population Genomics of Francisella tularensis subsp. holarctica and its Implication on the Eco-Epidemiology of Tularemia in Switzerland. Front. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- I Kudelina, R.; Olsufiev, N.G. Sensitivity to macrolide antibiotics and lincomycin in Francisella tularensis holarctica. J. Hyg. Epidemiol. Microbiol. Immunol. 1980, 24, 84–91. [Google Scholar]
- Kudelina, R.I. Change in the properties of the causative agent of tularemia due to erythromycin. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1973, 50, 98–101. [Google Scholar]
- Origgi, F.C.; Pilo, P. Francisella Tularensis Clades B.FTN002-00 and B.13 Are Associated With Distinct Pathology in the European Brown Hare (Lepus europaeus). Vet. Pathol. 2016, 53, 1220–1232. [Google Scholar] [CrossRef]
- Rydzewski, K.; Schulz, T.; Brzuszkiewicz, E.; Holland, G.; Lück, C.; Fleischer, J.; Grunow, R.; Heuner, K. Genome sequence and phenotypic analysis of a first German Francisella sp. isolate (W12-1067) not belonging to the species Francisella tularensis. BMC Microbiol. 2014, 14, 169. [Google Scholar] [CrossRef]
- Johansson, A.; Petersen, J.M. Genotyping of Francisella tularensis, the Causative Agent of Tularemia. J. AOAC Int. 2010, 93, 1930–1943. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.; Granberg, M.; Karlsson, L.; Neubauerova, V.; Forsman, M.; Johansson, A. A Real-Time PCR Array for Hierarchical Identification of Francisella Isolates. PLoS ONE 2009, 4, e8360. [Google Scholar] [CrossRef] [PubMed]
- Vogler, A.; Birdsell, D.; Wagner, D.; Keim, P. An optimized, multiplexed multi-locus variable-number tandem repeat analysis system for genotyping Francisella tularensis. Lett. Appl. Microbiol. 2009, 48, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Lärkeryd, A.; Widerström, M.; Mörtberg, S.; Myrtännäs, K.; Öhrman, C.; Birdsell, D.; Keim, P.; Wagner, D.M.; Forsman, M.; et al. An Outbreak of Respiratory Tularemia Caused by Diverse Clones of Francisella tularensis. Clin. Infect. Dis. 2014, 59, 1546–1553. [Google Scholar] [CrossRef] [PubMed]
- Antwerpen, M.H.; Prior, K.; Mellmann, A.; Hoppner, S.; Splettstoesser, W.D.; Harmsen, D. Rapid High Resolution Genotyping of Francisella tularensis by Whole Genome Sequence Comparison of Annotated Genes (“MLST+”). PLoS ONE 2015, 10, e0123298. [Google Scholar] [CrossRef]
- Schulze, C.; Heuner, K.; Myrtennäs, K.; Karlsson, E.; Jacob, D.; Kutzer, P.; Große, K.; Forsman, M.; Grunow, R. High and novel genetic diversity of Francisella tularensis in Germany and indication of environmental persistence. Epidemiol. Infect. 2016, 144, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Borde, J.P.; Zange, S.; Antwerpen, M.H.; Georgi, E.; Von Buttlar, H.; Kern, W.; Rieg, S. Five cases of vector-borne Francisella tularensis holarctica infections in south-western Germany and genetic diversity. Ticks Tick Borne Dis. 2017, 8, 808–812. [Google Scholar] [CrossRef]
- Tomaso, H.; Hotzel, H.; Otto, P.; Myrtennäs, K.; Forsman, M. Antibiotic susceptibility in vitro of Francisella tularensis subsp. holarctica isolates from Germany. J. Antimicrob. Chemother. 2017, 72, 2539–2543. [Google Scholar] [CrossRef]
- Tomaso, H.; Otto, P.; Peters, M.; Süss, J.; Karger, A.; Schamoni, H.; Zuchantke, E.; Hotzel, H. Francisella tularensis and other bacteria in hares and ticks in North Rhine-Westphalia (Germany). Ticks Tick Borne Dis. 2018, 9, 325–329. [Google Scholar] [CrossRef]
- Kevin, M.; Girault, G.; Caspar, Y.; Cherfa, M.A.; Mendy, C.; Tomaso, H.; Gavier-Widen, D.; Escudero, R.; Maurin, M.; Durand, B.; et al. Phylogeography and Genetic Diversity of Francisella tularensis subsp. holarctica in France (1947–2018). Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Gehringer, H.; Schacht, E.; Maylaender, N.; Zeman, E.; Kaysser, P.; Oehme, R.; Pluta, S.; Splettstoesser, W.D. Presence of an emerging subclone of Francisella tularensis holarctica in Ixodes ricinus ticks from south-western Germany. Ticks Tick Borne Dis. 2013, 4, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.; Hotzel, H.; Otto, P.H.; Karger, A.; Bettin, B.; Bocklisch, H.; Braune, S.; Eskens, U.; Hörmansdorfer, S.; Konrad, R.; et al. German Francisella tularensis isolates from European brown hares (Lepus europaeus) reveal genetic and phenotypic diversity. BMC Microbiol. 2013, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Kaysser, P.; Seibold, E.; Mätz-Rensing, K.; Pfeffer, M.; Essbauer, S.; Splettstoesser, W.D. Re-emergence of tularemia in Germany: Presence of Francisella tularensis in different rodent species in endemic areas. BMC Infect. Dis. 2008, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, A.; Schulze, C.; Kutzer, P.; Probst, C.; Hlinak, A.; Ochs, A.; Grunow, R. Tularaemia seroprevalence of captured and wild animals in Germany: The fox (Vulpes vulpes) as a biological indicator. Epidemiol. Infect. 2012, 141, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.H.; Chaignat, V.; Klimpel, D.; Diller, R.; Melzer, F.; Müller, W.; Tomaso, H. Serological Investigation of Wild Boars (Sus scrofa) and Red Foxes (Vulpes vulpes) As Indicator Animals for Circulation of Francisella tularensis in Germany. Vector Borne Zoonotic Dis. 2014, 14, 46–51. [Google Scholar] [CrossRef]
- Porsch-Özcürümez, M.; Kischel, N.; Priebe, H.; Splettstösser, M.; Finke, E.-J.; Grunow, R. Comparison of Enzyme-Linked Immunosorbent Assay, Western Blotting, Microagglutination, Indirect Immunofluorescence Assay, and Flow Cytometry for Serological Diagnosis of Tularemia. Clin. Diagn. Lab. Immunol. 2004, 11, 1008–1015. [Google Scholar] [CrossRef]
- Splettstoesser, W.D.; Piechotowski, I.; Buckendahl, A.; Frangoulidis, D.; Kaysser, P.; Kratzer, W.; Kimmig, P.; Seibold, E.; Brockmann, S.O. Tularemia in Germany: The tip of the iceberg? Epidemiol. Infect. 2008, 137, 736–743. [Google Scholar] [CrossRef]
- Straube, E.; Hess, C. Tularemia—Report of 2 cases without detectable contact with animals. Z. Gesamte Hyg. 1986, 32, 580–581. [Google Scholar]
- Schätzle, W.; Schwenk, R. Three cases of tularaemia in southern Baden-Wuerttemberg, Germany, November 2007. Eurosurveillance 2008, 13, 1–2. [Google Scholar] [CrossRef]
- Schubert, A.; Splettstoesser, W.; Bätzing-Feigenbaum, J. Tularaemia in Berlin—Two independent cases in travellers returning from central Anatolia, Turkey, February 2011. Eurosurveillance 2011, 16, 16. [Google Scholar]
- Kohlmann, R.; Geis, G.; Gatermann, S. Die Tularämie in Deutschland. Dtsch. Med. Wochenschr. 2014, 139, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Boone, I.; Hassler, D.; Nguyen, T.; Splettstoesser, W.; Wagner-Wiening, C.; Pfaff, G. Tularaemia in southwest Germany: Three cases of tick-borne transmission. Ticks Tick Borne Dis. 2015, 6, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Lübbert, C.; Taege, C.; Seufferlein, T.; Grunow, R. Prolongierter Verlauf einer durch Zeckenstich übertragenen ulzeroglandulären Tularämie bei einer 20-jährigen Patientin. Dtsch. Med. Wochenschr. 2009, 134, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.; Kohlmann, R.; Müller, W.; Julich, S.; Geis, G.; Gatermann, S.G.; Peters, M.; Wolf, P.J.; Karlsson, E.; Forsman, M.; et al. Hare-to-Human Transmission of Francisella tularensis subsp. holarctica, Germany. Emerg. Infect. Dis. 2015, 21, 153–155. [Google Scholar] [CrossRef]
- Regier, Y.; Komma, K.; Weigel, M.; Kraiczy, P.; Laisi, A.; Pulliainen, A.T.; Hain, T.; Kempf, V.A. Combination of microbiome analysis and serodiagnostics to assess the risk of pathogen transmission by ticks to humans and animals in central Germany. Parasites Vectors 2019, 12, 11. [Google Scholar] [CrossRef]
- Seemann, R.; Kleinschmidt, M.C.; Trampuz, A.; Mardian, S. Ulceroglandular tularemia after contact with a wild boar: Risk of infection for medical personnel by aerosol inhalation during lymph node resection. Unfallchirurg 2020, 123, 740–743. [Google Scholar] [CrossRef]
- Hestvik, G.; Uhlhorn, H.; Koene, M.; Åkerström, S.; Malmsten, A.; Dahl, F.; Åhlén, P.-A.; Dalin, A.-M.; Gavier-Widén, D. Francisella tularensis in Swedish predators and scavengers. Epidemiol. Infect. 2019, 147, e293. [Google Scholar] [CrossRef]
- Ecke, F.; Johansson, A.; Forsman, M.; Khalil, H.; Magnusson, M.; Hörnfeldt, B. Selective Predation by Owls on Infected Bank Voles (Myodes glareolus) as a Possible Sentinel of Tularemia Outbreaks. Vector Borne Zoonotic Dis. 2020, 20, 630–632. [Google Scholar] [CrossRef]
- Tsokana, C.N.; Sokos, C.; Giannakopoulos, A.; Birtsas, P.; Valiakos, G.; Spyrou, V.; Athanasiou, L.V.; Burriel, A.R.; Billinis, C. European Brown hare (Lepus europaeus) as a source of emerging and re-emerging pathogens of Public Health importance: A review. Veter Med. Sci. 2020, 6, 550–564. [Google Scholar] [CrossRef]
- Sutor, A.; Schwarz, S.; Conraths, F.J. The biological potential of the raccoon dog (Nyctereutes procyonoides, Gray 1834) as an invasive species in Europe—New risks for disease spread? Acta Thériol. 2013, 59, 49–59. [Google Scholar] [CrossRef]
- Gyuranecz, M.; Rigó, K.; Dán, Á; Földvári, G.; Makrai, L.; Denes, B.; Fodor, L.; Majoros, G.; Tirják, L.; Erdélyi, K. Investigation of the Ecology of Francisella tularensis During an Inter-Epizootic Period. Vector Borne Zoonotic Dis. 2011, 11, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Cronhjort, S.; Wilhelmsson, P.; Karlsson, L.; Thelaus, J.; Sjödin, A.; Pawełczyk, O.; Forsberg, P.; Lindgren, P.-E. The Tick-Borne Diseases STING study: Real-time PCR analysis of three emerging tick-borne pathogens in ticks that have bitten humans in different regions of Sweden and the Aland islands, Finland. Infect. Ecol. Epidemiol. 2019, 9, 1683935. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zellner, B.; Huntley, J.F. Ticks and Tularemia: Do We Know What We Don’t Know? Front. Microbiol. 2019, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Dryselius, R.; Hjertqvist, M.; Mäkitalo, S.; Lindblom, A.; Lilja, T.; Eklöf, D.; Lindström, A. Large outbreak of tularaemia, central Sweden, July to September 2019. Eurosurveillance 2019, 24, 1900603. [Google Scholar] [CrossRef]
- Ma, Y.; Bring, A.; Kalantari, Z.; Destouni, G. Potential for Hydroclimatically Driven Shifts in Infectious Disease Outbreaks: The Case of Tularemia in High-Latitude Regions. Int. J. Environ. Res. Public Health 2019, 16, 3717. [Google Scholar] [CrossRef]
- Grunow, R.; Splettstoesser, W.; McDonald, S.; Otterbein, C.; O’Brien, T.; Morgan, C.; Aldrich, J.; Hofer, E.; Finke, E.-J.; Meyer, H. Detection of Francisella tularensis in Biological Specimens Using a Capture Enzyme-Linked Immunosorbent Assay, an Immunochromatographic Handheld Assay, and a PCR. Clin. Diagn. Lab. Immunol. 2000, 7, 86–90. [Google Scholar] [CrossRef]
- Grunow, R.; Splettstößer, W.; Hirsch, F.W.; Kleemann, D.; Finke, E.-J. Differential diagnosis of tularaemia. Dtsch. Med. Wochenschr. 2001, 126, 408–413. [Google Scholar] [CrossRef]
- Grunow, R.; Ippolito, G.; Jacob, D.; Sauer, U.; Rohleder, A.; Di Caro, A.; Iacovino, R. Benefits of a European project on diagnostics of highly pathogenic agents and assessment of potential “dual use” issues. Front. Public Health 2014, 2, 199. [Google Scholar] [CrossRef]
- Schmitt, P.; Splettstösser, W.; Porsch-Ozcürümez, M.; Finke, E.J.; Grunow, R. A novel screening ELISA and a confirmatory Western blot useful for diagnosis and epidemiological studies of tularemia. Epidemiol. Infect. 2005, 133, 759–766. [Google Scholar] [CrossRef]
- Splettstoesser, W.D.; Tomaso, H.; Dahouk, S.; Neubauer, H.; Schuff-Werner, P. Diagnostic Procedures in Tularaemia with Special Focus on Molecular and Immunological Techniques. J. Veter Med. Ser. B 2005, 52, 249–261. [Google Scholar] [CrossRef]
- Seibold, E.; Maier, T.; Kostrzewa, M.; Zeman, E.; Splettstoesser, W. Identification of Francisella tularensis by Whole-Cell Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry: Fast, Reliable, Robust, and Cost-Effective Differentiation on Species and Subspecies Levels. J. Clin. Microbiol. 2010, 48, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Wahab, T.; Edvinsson, B.; Peterzon, A.; Boskani, T.; Farhadi, L.; Barduhn, A.; Grunow, R.; Sandström, G. Identification and subtyping of Francisella by pyrosequencing and signature matching of 16S rDNA fragments. Lett. Appl. Microbiol. 2011, 53, 592–595. [Google Scholar] [CrossRef] [PubMed]
- Euler, M.; Wang, Y.; Otto, P.; Tomaso, H.; Escudero, R.; Anda, P.; Hufert, F.T.; Weidmann, M. Recombinase Polymerase Amplification Assay for Rapid Detection of Francisella tularensis. J. Clin. Microbiol. 2012, 50, 2234–2238. [Google Scholar] [CrossRef] [PubMed]
- Georgi, E.; Schacht, E.; Scholz, H.C.; Splettstoesser, W.D. Standardized broth microdilution antimicrobial susceptibility testing of Francisella tularensis subsp. holarctica strains from Europe and rare Francisella species. J. Antimicrob. Chemother. 2012, 67, 2429–2433. [Google Scholar] [CrossRef] [PubMed]
- Sting, R.; Runge, M.; Eisenberg, T.; Braune, S.; Müller, W.; Otto, P. Comparison of bacterial culture and polymerase chain reaction (PCR) for the detection of F. tularensis subsp. holarctica in wild animals. Berl. Munch. Tierarztl. Wochenschr. 2013, 126, 285–290. [Google Scholar]
- Becker, S.; Lochau, P.; Jacob, D.; Heuner, K.; Grunow, R. Successful re-evaluation of broth medium T for the growth of Francisella tularensis ssp. and other higly pathogenic bacteria. J. Microbiol. Methods 2016, 121, 5–7. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Petersen, J.M.; Gallegos-Graves, L.V.; Hodge, D.; Pillai, S.; Kuske, C.R. Whole-Genome Relationships among Francisella Bacteria of Diverse Origins Define New Species and Provide Specific Regions for Detection. Appl. Environ. Microbiol. 2016, 83. [Google Scholar] [CrossRef]
- Qu, P.-H.; Chen, S.; Scholz, H.C.; Busse, H.-J.; Gu, Q.; Kämpfer, P.; Foster, J.T.; Glaeser, S.P.; Chen, C.; Yang, Z.-C. Francisella guangzhouensis sp. nov., isolated from air-conditioning systems. Int. J. Syst. Evol. Microbiol. 2013, 63, 3628–3635. [Google Scholar] [CrossRef]
- Qu, P.-H.; Li, Y.; Salam, N.; Chen, S.; Liu, L.; Gu, Q.; Fang, B.-Z.; Xiao, M.; Li, M.; Chen, C.; et al. Allofrancisella inopinata gen. nov., sp. nov. and Allofrancisella frigidaquae sp. nov., isolated from water-cooling systems, and transfer of Francisella guangzhouensis Qu et al. 2013 to the new genus as Allofrancisella guangzhouensis comb. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4832–4838. [Google Scholar] [CrossRef]
- Köppen, K.; Chen, F.; Rydzewski, K.; Einenkel, R.; Böttcher, T.; Morguet, C.; Grunow, R.; Eisenreich, W.; Heuner, K. Screen for fitness and virulence factors of Francisella sp. strain W12-1067 using amoebae. Int. J. Med. Microbiol. 2019, 309, 151341. [Google Scholar] [CrossRef]
- Chen, F.; Köppen, K.; Rydzewski, K.; Einenkel, R.; Morguet, C.; Vu, D.T.; Eisenreich, W.; Heuner, K. Myo-Inositol as a carbon substrate in Francisella and insights into the metabolism of Francisella sp. strain W12-1067. Int. J. Med. Microbiol. 2020, 310, 151426. [Google Scholar] [CrossRef] [PubMed]
- Tlapák, H.; Köppen, K.; Rydzewski, K.; Grunow, R.; Heuner, K. Construction of a New Phage Integration Vector pFIV-Val for Use in Different Francisella Species. Front. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Gyuranecz, M.; Reiczigel, J.; Krisztalovics, K.; Monse, L.; Szabóné, G.K.; Szilágyi, A.; Szépe, B.; Makrai, L.; Magyar, T.; Bhide, M.; et al. Factors Influencing Emergence of Tularemia, Hungary, 1984–2010. Emerg. Infect. Dis. 2012, 18, 1379–1381. [Google Scholar] [CrossRef] [PubMed]
| Form | n | % |
|---|---|---|
| Glandular (lymphadenitis and not meeting criteria for other forms) | 129 | 29.7 |
| Ulceroglandular (lymphadenitis + skin ulcer) | 68 | 15.6 |
| Pneumonic (dyspnoea or pneumonia) | 53 | 12.2 |
| Intestinal (diarrhea, vomiting, or abdominal pain) | 20 | 4.6 |
| Oropharyngeal (lymphadenitis AND tonsillitis, pharyngitis, stomatitis) | 23 | 5.3 |
| Oculoglandular (lymphadenitis + conjunctivitis) | 8 | 1.8 |
| Combination (meeting criteria of >1 form) | 52 | 12.0 |
| Typhoidal | 5 | 1.1 |
| Other (symptoms not meeting any of the above criteria, e.g., “only fever”) | 77 | 17.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Appelt, S.; Faber, M.; Köppen, K.; Jacob, D.; Grunow, R.; Heuner, K. Francisella tularensis Subspecies holarctica and Tularemia in Germany. Microorganisms 2020, 8, 1448. https://doi.org/10.3390/microorganisms8091448
Appelt S, Faber M, Köppen K, Jacob D, Grunow R, Heuner K. Francisella tularensis Subspecies holarctica and Tularemia in Germany. Microorganisms. 2020; 8(9):1448. https://doi.org/10.3390/microorganisms8091448
Chicago/Turabian StyleAppelt, Sandra, Mirko Faber, Kristin Köppen, Daniela Jacob, Roland Grunow, and Klaus Heuner. 2020. "Francisella tularensis Subspecies holarctica and Tularemia in Germany" Microorganisms 8, no. 9: 1448. https://doi.org/10.3390/microorganisms8091448
APA StyleAppelt, S., Faber, M., Köppen, K., Jacob, D., Grunow, R., & Heuner, K. (2020). Francisella tularensis Subspecies holarctica and Tularemia in Germany. Microorganisms, 8(9), 1448. https://doi.org/10.3390/microorganisms8091448





