Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence
Abstract
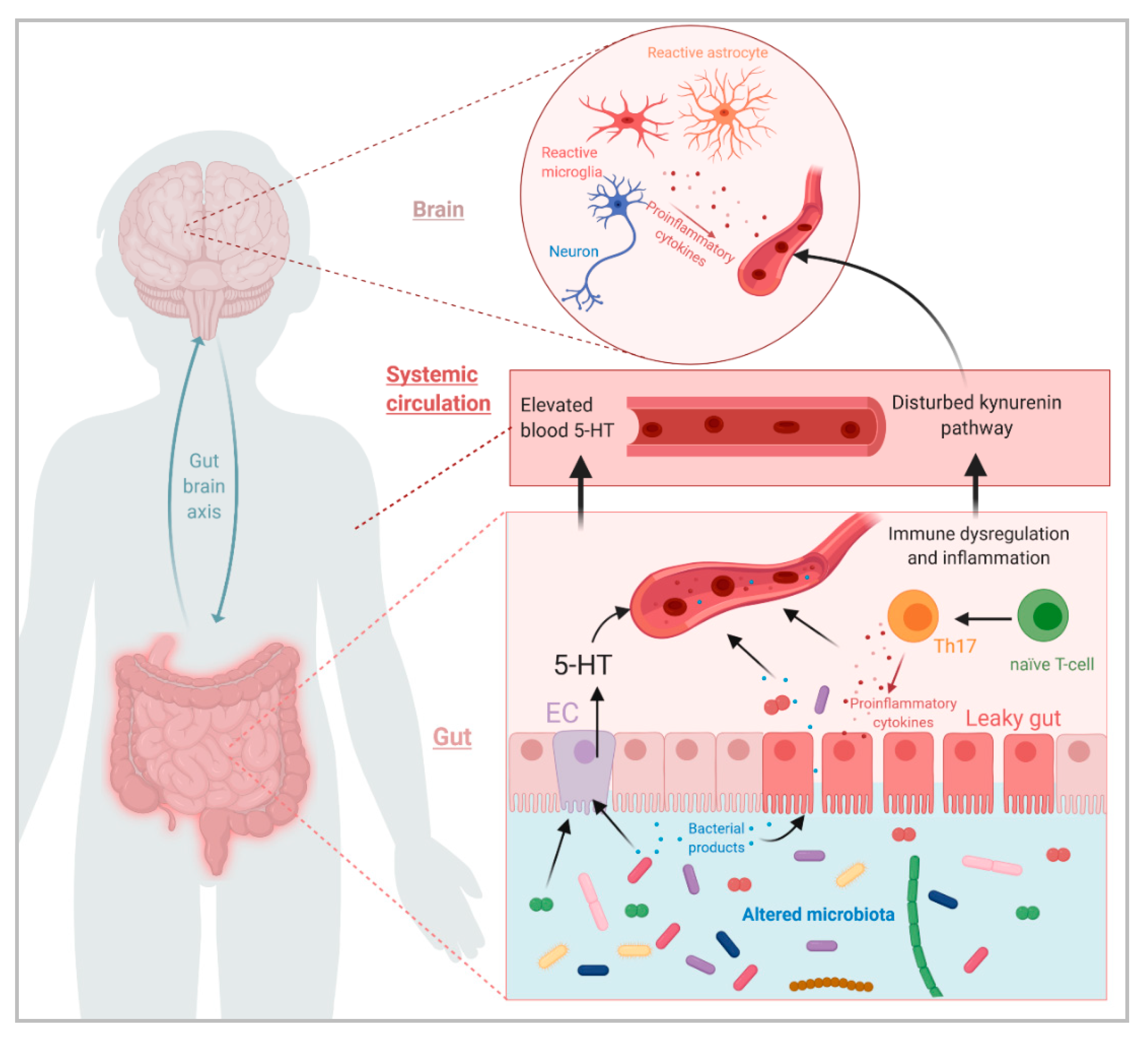
1. Introduction
2. Clinical and Preclinical Evidence for Involvement of the Gut Microbiota in Various Aspects of ASD
2.1. Dysbiosis and Changes in Bacterial Metabolites in ASD
2.1.1. Clinical Evidence
2.1.2. Preclinical Evidence
2.2. Influence of the Gut Microbiota on Immune System Dysregulation in ASD
2.2.1. Clinical Evidence
2.2.2. Preclinical Evidence
2.3. Influence of the Gut Microbiota on Dysregulation of Tryptophan Metabolism in ASD
2.3.1. Clinical Evidence
2.3.2. Preclinical Evidence
3. Clinical and Preclinical Interventions Targeting the Gut Microbiota
3.1. Probiotic Intervention Studies for ASD Symptoms
3.1.1. Clinical Studies
3.1.2. Preclinical Studies
3.2. FMT Studies
3.2.1. Clinical Studies
3.2.2. Preclinical Studies
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| BBB | Blood brain barrier |
| CMA | Cow’s milk allergy |
| EC | Enterochromaffin cells |
| 4-EPS | 4-Ethylphenylsulfate |
| FMT | Fecal microbiota transplantation |
| GABAergic | gamma-aminobutyric acid |
| GF | Germ free |
| GFAP | Glial fibrillary acidic protein |
| GI | Gastrointestinal |
| 5-HT | Serotonin |
| IFN | Interferon |
| ILC3 | Innate lymphoid cells |
| KA | Kynurenic acid |
| KYN | Kynurenin |
| LCL | Lymphoblastoid cell lines |
| LPS | Lipopolysaccharide |
| MHFD | Maternal high fat diet |
| MIA | Maternal immune activation |
| MPO | Myeloperoxidase |
| NMDA | N-methyl-D-aspartate |
| QA | Quinolinic acid |
| SCFAs | Short chain fatty acids |
| SFB | Segmented filamentous bacteria |
| SPF | Specific pathogen free |
| TD | Typically developing |
| Trp | Tryptophan |
| VPA | Valproic acid |
| WT | Wild-type |
Appendix A
| Model | Sex | Sample | Method | Difference in Microbiota Compared to Controls | Ref. |
|---|---|---|---|---|---|
| Shank3-/- | F/M | Feces | 16S rRNA seq RT-PCR | α-diversity: ↓ β-diversity: Modulated Phylum level: N.S Class level: ↓Bacilli Order level: ↓Lactobacillales, Rhodospirillales, Rickettsiales and Turicibacteriales Family level: ↑Veillonellaceae; ↓Lactobacillaceae, Bacteroidaceae, Acetobacteriaceae, mitochondria and Turicibacteriaceae; ↑Veillonellaceae Genus level: ↓Lactobacillus, Coprococcus, Bacteroides, Acetobacter, Turicibacter and Prevotella; ↑Veillonella in males ↓ in females Species level: ↓L. reuteri, L. brevis, L. ruminis in both male and female; ↓V. parvula and V. dispar in females, ↑V. dispar in males | [58] |
| N.S | Feces | 16S rRNA seq | No assessment of diversity Phylum level: ↑Actinobacteria and Firmicutes; ↓ Proteobacteria Absence of Verrucomicrobia; Presence of Deferribacteres, Chlamydiae and Tenericutes Class level: N.S Order level: ↑ Bifidobacteriales and Eggerthellales Family level: N.S Genus level: ↑Asaccharobacter, Eggerthella, Enterorhabdus and Paraeggerthella Species level: ↑ B. pseudolongum, Assacharobacter WCA-131-CoC-2, Eggerthella YY7918 and Enterorhabdus caecimuris. | [51] | |
| Shank3B-/- | M | Feces | 16S rRNA seq | α-diversity: No changes β-diversity: Modulated Bacterial modulation were not detailed except for: Species level: ↓ L. reuteri | [62] |
| NL3R451C | M | Feces | ARISA * 16S rRNA seq | α-diversity: No changes β-diversity: Modulated at 3 weeks of age (not at 9 weeks) Bacterial modulations were only detailed at OTU level: Species level (OTUs): ↑ OTUs from Lachnospiraceae family, ↓OTUs from Candidate phylum | [57] |
| BTBR | M | Feces and cecal content | 16S rRNA seq | α-diversity: ↓ β-diversity: Modulated in cecal content only Phylum level: ↑ Bacteroidetes in cecal content Class level: N.S Order level: N.S Family level: ↓ Enterobacteriaceae both cecal and fecal Genus level: N.S Species level: ↑ A. Muciniphila, Lactobacillus spp., Roseburia spp., C. leptum, Prevotella spp.; ↓ Clostridium cluster XI both cecal and fecal In cecal content only, ↑Methanobrevibacter spp.; ↓ C. coccoides and Clostridium cluster I In feces only, ↑ C. coccoides and Clostridium cluster I; ↓ Methanobrevibacter spp. | [59] |
| F/M | Feces | 16S rRNA seq | α-diversity: No changes β-diversity: Modulated Phylum level: ↑ Proteobacteria and TM7 in female Class level: N.S Order level: N.S Family level: N.S Genus level: ↑Bacteroides and Parabacteroides; ↓Dehalobacterium in both male and female. In females only, ↑Prevotella, Coprobacillus, Sutterella, Akkermansia, and unclassified genera of Desulfovibrionaceae and Enterobacteriaceae families; ↓ Oscillospira and unclassified members of TM7 and Rikenellaceae families In males only, ↑ Bacteroides, Parabacteroides, Lactobacillus, Coprobacillus and unclassified genus of the Helicobacteraceae family; ↓ Dehalobacterium, Ruminococcus and Desulfovibrio Species level: N.S | [52] | |
| M | Cecal content | 16S rRNA seq | α-diversity: ↓ β-diversity: Modulated Phylum level: ↑Verrucomicrobia, Bacteroidetes;↓ Firmicutes and Cyanobacteria Class level: N.S Order level: N.S Family level: N.S Genus level: ↑Akkermansia, Bacteroides, Bilophila, Enterorhabdus Intestinomonas and S24-7; ↓ Odoribacter, Parabacteroides, Rikenella, Blautia, Coprococcus, Bifidobacterium, Desulfovibrio, Lachnospiracae_Incertae Sedis and RC9 gut group Species level: N.S | [53] | |
| M | Feces | 16S rRNA seq | α-diversity: N.S β-diversity: Modulated Bacterial modulation were not detailed except for: Species levels: ↓ L. reuteri | [62] |
| Model | Sex | Sample | Method | Difference in Microbiota Compared to Controls | Ref. |
|---|---|---|---|---|---|
| MIA | M/F | Feces | 16S rRNA seq | α-diversity: No changes β-diversity: Modulated Bacterial modulations were only detailed at OTU level: ↑OTUs from the Alphaproteobacteria and Bacili classes, Bacteroidales order and Prevotellaceae, Lachnospiraceae and Porphyromonadaceae families ↓ OTUs from the Actinobacteria phylum, Gammaproteobacteria, Mollicutes and Erysipelotrichi classes and Ruminococcaceae, Erysipelotrichaceae and Aligenaceae families | [54] |
| M | Cecal content | 16S rRNA seq | No assessment of diversity Phylum level: N.S Class level: N.S Order level: N.S Family level: ↑ Ruminococcaceae, Porphyromonadaceae, Aoerococcaceae and Erysipelotrichaceae Genus level: ↑ Candidatus Species level: N.S | [94] | |
| N.S | Feces | 16S rRNA seq | α-diversity: ↓ β-diversity: Modulated Phylum level: ↑Bacteroidetes and Verrucomicrobia; ↓ Firmicutes Class level: N.S Order level: N.S Family level: N.S Genus level: ↑Prevotella, Prevotella_other, Akkermansia and a genus of S24-7 family; ↓Oscillospira, Ruminococcus, Bacteroides, Dehalobacterium, Desulfovibrio, Lactobacillus, and members of the Clostridiales order and Rikenellaceae, Lachnospiraceae and Ruminococcaceae families. Species level: ↑ F16 and OTUs from the Bacteroidales order, Clostridiaceae, Enterobacteriaceae and S24-7 familie and Akkermansia and Prevotella genera ↓ OTUs from the Clostridiales order, Ruminococcaceae and Rikenellaceae families and Ruminococcus, Bacteroides, Dehalobacterium, Desulfovibrio, Oscillospira and Odoribacter genera | [60] | |
| VPA | M/F | Feces | Total genomic DNA pyrosequencing | α-diversity: no changes β-diversity: no difference Phylum level: ↑ Firmicutes; ↓Bacteroidetes Class level: N.S Order level: N.S Family level: N.S Genus level: ↑ Uncultured genus of Erysipelotrichales, uncultured genera of the Bacteroidales and Desulfovibrionales orders Species level: N.S | [63] |
| M/F | Feces | 16SrDNA seq | α-diversity: ↓ β-diversity: Modulated Phylum level: only in males ↑ Bacteroidetes; only in female, ↑ Actinobacteria Class level: Only in males ↑Bacteroida, Alphaproteobacteria; ↓Coriobacteria Order level: N.S Family level: ↑ Eubacteriaceae, Rikenellaceae and Staphylococcaceae; ↓ Enterobacteriaceae Genus level: ↑ Anaerofustis, Proteus, Staphylococcus, and Allobaculum Only in females ↑, Bifidobacterium, Odoribacter and Candidatus Arthromitus Species level: ↑ Ruminococcus flavefaciens, OTUs from the Clostridiales order and the Ruminoccus and S24-7 genera. | [61] | |
| MHFD mice | M | Feces | 16SrDNA seq | α-diversity: ↓ β-diversity: Modulated No detail of the changes in bacterial taxa | [49] |
| N.S | Feces | 16SrDNA seq | α-diversity: ↓ β-diversity: Modulated Phylum level: ↑ Firmicutes, Verucomicrobia, ↓Bacteroidetes Class level: N.S Order level: N.S Family level: ↑Peptostreptococcaceae Genus level:↑Streptococcus, Akkermansia ↓ Lachnospiraceae_incertae_sedis Species level: N.S | [55] |
References
- Tchaconas, A.; Adesman, A. Autism Spectrum Disorders: A Pediatric Overview and Update. Curr. Opin. Pediatr. 2013, 25, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, L.D.; Rice, C.E.; Barger, B.; Soke, G.N.; Lee, L.-C.; Moody, E.; Edmondson-Pretzel, R.; Levy, S.E. DSM-5 Criteria for Autism Spectrum Disorder Maximizes Diagnostic Sensitivity and Specificity in Preschool Children. Soc. Psychiatry Psychiatr. Epidemiol. 2019, 54, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Autism Spectrum Disorders. Available online: https://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders (accessed on 6 July 2020).
- Maenner, M.J. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–23. [Google Scholar] [CrossRef]
- ASDEU. Findings. Available online: http://asdeu.eu/findings/ (accessed on 9 July 2020).
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K.; et al. Genetic Heritability and Shared Environmental Factors Among Twin Pairs with Autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Wiśniowiecka-Kowalnik, B.; Nowakowska, B.A. Genetics and Epigenetics of Autism Spectrum Disorder—Current Evidence in the Field. J. Appl. Genet. 2019, 60, 37–47. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The Familial Risk of Autism. JAMA 2014, 311, 1770–1777. [Google Scholar] [CrossRef]
- Bölte, S.; Girdler, S.; Marschik, P.B. The Contribution of Environmental Exposure to the Etiology of Autism Spectrum Disorder. Cell. Mol. Life Sci. CMLS 2019, 76, 1275–1297. [Google Scholar] [CrossRef]
- Gialloreti, L.E.; Mazzone, L.; Benvenuto, A.; Fasano, A.; Alcon, A.G.; Kraneveld, A.; Moavero, R.; Raz, R.; Riccio, M.P.; Siracusano, M.; et al. Risk and Protective Environmental Factors Associated with Autism Spectrum Disorder: Evidence-Based Principles and Recommendations. J. Clin. Med. 2019, 8, 217. [Google Scholar] [CrossRef]
- McElhanon, B.O.; McCracken, C.; Karpen, S.; Sharp, W.G. Gastrointestinal Symptoms in Autism Spectrum Disorder: A Meta-Analysis. Pediatrics 2014, 133, 872–883. [Google Scholar] [CrossRef]
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 218–230. [Google Scholar] [CrossRef]
- Sandler, R.H.; Finegold, S.M.; Bolte, E.R.; Buchanan, C.P.; Maxwell, A.P.; Väisänen, M.L.; Nelson, M.N.; Wexler, H.M. Short-Term Benefit from Oral Vancomycin Treatment of Regressive-Onset Autism. J. Child Neurol. 2000, 15, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Łukasik, J.; Patro-Gołąb, B.; Horvath, A.; Baron, R.; Szajewska, H.; Baron, R.; van der Vaart, I.B.; Gieruszczak-Białek, D.; Horvath, A.; The SAWANTI Working Group; et al. Early Life Exposure to Antibiotics and Autism Spectrum Disorders: A Systematic Review. J. Autism Dev. Disord. 2019, 49, 3866–3876. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Downes, J.; Summanen, P.H. Microbiology of Regressive Autism. Anaerobe 2012, 18, 260–262. [Google Scholar] [CrossRef]
- Xu, M.; Xu, X.; Li, J.; Li, F. Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Front. Psychiatry 2019, 10, 473. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Vázquez, L.; Van Riba, G.G.; Arija, V.; Canals, J. Composition of Gut Microbiota in Children with Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 792. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut Microbiota Changes in Children with Autism Spectrum Disorder: A Systematic Review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Bezawada, N.; Phang, T.H.; Hold, G.L.; Hansen, R. Autism Spectrum Disorder and the Gut Microbiota in Children: A Systematic Review. Ann. Nutr. Metab. 2020, 76, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium Prausnitzii: From Microbiology to Diagnostics and Prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing Study of Fecal Microflora of Autistic and Control Children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Parracho, H.M.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the Gut Microflora of Children with Autistic Spectrum Disorders and That of Healthy Children. J. Med. Microbiol. 2005, 54 Pt 10, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low Relative Abundances of the Mucolytic Bacterium Akkermansia Muciniphila and Bifidobacterium Spp. in Feces of Children with Autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased Abundance of Sutterella Spp. and Ruminococcus Torques in Feces of Children with Autism Spectrum Disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal Microbiota in Children with Autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.A.; Williams, K.; Kochel, R.; Powell, C.; Redel, C.; Versalovic, J.; Savidge, T. 217. The Role of the Microbiome in Complex Phenotypes of Pediatric Autism Spectrum Disorder. Biol. Psychiatry 2019, 85, S90. [Google Scholar] [CrossRef]
- Altieri, L.; Neri, C.; Sacco, R.; Curatolo, P.; Benvenuto, A.; Muratori, F.; Santocchi, E.; Bravaccio, C.; Lenti, C.; Saccani, M.; et al. Urinary P-Cresol Is Elevated in Small Children with Severe Autism Spectrum Disorder. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2011, 16, 252–260. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Cerullo, S.; Neri, C.; Urbani, A.; Tripi, G.; Malvy, J.; Barthelemy, C.; Bonnet-Brihault, F.; Persico, A.M. Urinary P-Cresol Is Elevated in Young French Children with Autism Spectrum Disorder: A Replication Study. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2014, 19, 463–470. [Google Scholar] [CrossRef]
- Kang, D.-W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howson, H.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in Fecal Microbial Metabolites and Microbiota of Children with Autism Spectrum Disorders. Anaerobe 2018, 49, 121–131. Available online: https://pubmed.ncbi.nlm.nih.gov/29274915/ (accessed on 6 July 2020). [CrossRef]
- Gevi, F.; Belardo, A.; Zolla, L. A Metabolomics Approach to Investigate Urine Levels of Neurotransmitters and Related Metabolites in Autistic Children. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165859. [Google Scholar] [CrossRef]
- Gabriele, S.; Sacco, R.; Altieri, L.; Neri, C.; Urbani, A.; Bravaccio, C.; Riccio, M.P.; Iovene, M.R.; Bombace, F.; De Magistris, L.; et al. Slow Intestinal Transit Contributes to Elevate Urinary P-Cresol Level in Italian Autistic Children. Autism Res. Off. J. Int. Soc. Autism Res. 2016, 9, 752–759. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The Role of Short-Chain Fatty Acids in Microbiota-Gut-Brain Communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal Flora and Gastrointestinal Status in Children with Autism--Comparisons to Typical Children and Correlation with Autism Severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Elevated Fecal Short Chain Fatty Acid and Ammonia Concentrations in Children with Autism Spectrum Disorder. Dig. Dis. Sci. 2012, 57, 2096–2102. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered Gut Microbiota and Short Chain Fatty Acids in Chinese Children with Autism Spectrum Disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef]
- Averina, O.V.; Kovtun, A.S.; Polyakova, S.I.; Savilova, A.M.; Rebrikov, D.V.; Danilenko, V.N. The Bacterial Neurometabolic Signature of the Gut Microbiota of Young Children with Autism Spectrum Disorders. J. Med. Microbiol. 2020, 69, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Cotrina, M.L.; Ferreiras, S.; Schneider, P. High Prevalence of Self-Reported Autism Spectrum Disorder in the Propionic Acidemia Registry. JIMD Rep. 2020, 51, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Valiente-Pallejà, A.; Torrell, H.; Muntané, G.; Cortés, M.J.; Martínez-Leal, R.; Abasolo, N.; Alonso, Y.; Vilella, E.; Martorell, L. Genetic and Clinical Evidence of Mitochondrial Dysfunction in Autism Spectrum Disorder and Intellectual Disability. Hum. Mol. Genet. 2018, 27, 891–900. [Google Scholar] [CrossRef]
- Frye, R.E.; Rose, S.; Chacko, J.; Wynne, R.; Bennuri, S.C.; Slattery, J.C.; Tippett, M.; Delhey, L.; Melnyk, S.; Kahler, S.G.; et al. Modulation of Mitochondrial Function by the Microbiome Metabolite Propionic Acid in Autism and Control Cell Lines. Transl. Psychiatry 2016, 6, e927. [Google Scholar] [CrossRef]
- Rose, S.; Bennuri, S.C.; Davis, J.E.; Wynne, R.; Slattery, J.C.; Tippett, M.; Delhey, L.; Melnyk, S.; Kahler, S.G.; MacFabe, D.F.; et al. Butyrate Enhances Mitochondrial Function during Oxidative Stress in Cell Lines from Boys with Autism. Transl. Psychiatry 2018, 8, 42. [Google Scholar] [CrossRef]
- Boccuto, L.; Lauri, M.; Sarasua, S.M.; Skinner, C.D.; Buccella, D.; Dwivedi, A.; Orteschi, D.; Collins, J.S.; Zollino, M.; Visconti, P.; et al. Prevalence of SHANK3 Variants in Patients with Different Subtypes of Autism Spectrum Disorders. Eur. J. Hum. Genet. EJHG 2013, 21, 310–316. [Google Scholar] [CrossRef]
- Peça, J.; Feliciano, C.; Ting, J.T.; Wang, W.; Wells, M.F.; Venkatraman, T.N.; Lascola, C.D.; Fu, Z.; Feng, G. Shank3 Mutant Mice Display Autistic-like Behaviours and Striatal Dysfunction. Nature 2011, 472, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, K.; Blundell, J.; Etherton, M.R.; Hammer, R.E.; Liu, X.; Powell, C.M.; Südhof, T.C. A Neuroligin-3 Mutation Implicated in Autism Increases Inhibitory Synaptic Transmission in Mice. Science 2007, 318, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Bruining, H.; Matsui, A.; Oguro-Ando, A.; Kahn, R.S.; Van’t Spijker, H.M.; Akkermans, G.; Stiedl, O.; van Engeland, H.; Koopmans, B.; van Lith, H.A.; et al. Genetic Mapping in Mice Reveals the Involvement of Pcdh9 in Long-Term Social and Object Recognition and Sensorimotor Development. Biol. Psychiatry 2015, 78, 485–495. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.G.; Kusek, G.K.; Yang, M.; Phoenix, J.L.; Bolivar, V.J.; Crawley, J.N. Autism-like Behavioral Phenotypes in BTBR T+tf/J Mice. Genes Brain Behav. 2008, 7, 152–163. [Google Scholar] [CrossRef]
- Ornoy, A. Valproic Acid in Pregnancy: How Much Are We Endangering the Embryo and Fetus? Reprod. Toxicol. Elmsford N 2009, 28, 1–10. [Google Scholar] [CrossRef]
- Malkova, N.V.; Yu, C.Z.; Hsiao, E.Y.; Moore, M.J.; Patterson, P.H. Maternal Immune Activation Yields Offspring Displaying Mouse Versions of the Three Core Symptoms of Autism. Brain. Behav. Immun. 2012, 26, 607–616. [Google Scholar] [CrossRef]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef]
- de Theije, C.G.M.; Wu, J.; Koelink, P.J.; Korte-Bouws, G.A.H.; Borre, Y.; Kas, M.J.H.; da Silva, S.L.; Korte, S.M.; Olivier, B.; Garssen, J.; et al. Autistic-like Behavioural and Neurochemical Changes in a Mouse Model of Food Allergy. Behav. Brain Res. 2014, 261, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Sauer, A.K.; Bockmann, J.; Steinestel, K.; Boeckers, T.M.; Grabrucker, A.M. Altered Intestinal Morphology and Microbiota Composition in the Autism Spectrum Disorders Associated SHANK3 Mouse Model. Int. J. Mol. Sci. 2019, 20, 2134. [Google Scholar] [CrossRef]
- Coretti, L.; Cristiano, C.; Florio, E.; Scala, G.; Lama, A.; Keller, S.; Cuomo, M.; Russo, R.; Pero, R.; Paciello, O.; et al. Sex-Related Alterations of Gut Microbiota Composition in the BTBR Mouse Model of Autism Spectrum Disorder. Sci. Rep. 2017, 7, 45356. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, A.V.; Joyce, S.A.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Pérez, A.; Peterson, V.; et al. Microbiota-Related Changes in Bile Acid & Tryptophan Metabolism Are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Sun, Y.; Wu, J.; Huang, S.; Jin, G.; Guo, Z.; Zhang, Y.; Liu, T.; Liu, X.; Cao, X.; et al. Maternal High Fat Diet Alters Gut Microbiota of Offspring and Exacerbates DSS-Induced Colitis in Adulthood. Front. Immunol. 2018, 9, 2608. [Google Scholar] [CrossRef] [PubMed]
- de Theije, C.G.M.; Koelink, P.J.; Korte-Bouws, G.A.H.; da Silva, S.L.; Korte, S.M.; Olivier, B.; Garssen, J.; Kraneveld, A.D. Intestinal Inflammation in a Murine Model of Autism Spectrum Disorders. Brain. Behav. Immun. 2014, 37, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Hosie, S.; Ellis, M.; Swaminathan, M.; Ramalhosa, F.; Seger, G.O.; Balasuriya, G.K.; Gillberg, C.; Råstam, M.; Churilov, L.; McKeown, S.J.; et al. Gastrointestinal Dysfunction in Patients and Mice Expressing the Autism-Associated R451C Mutation in Neuroligin-3. Autism Res. Off. J. Int. Soc. Autism Res. 2019, 12, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Tabouy, L.; Getselter, D.; Ziv, O.; Karpuj, M.; Tabouy, T.; Lukic, I.; Maayouf, R.; Werbner, N.; Ben-Amram, H.; Nuriel-Ohayon, M.; et al. Dysbiosis of Microbiome and Probiotic Treatment in a Genetic Model of Autism Spectrum Disorders. Brain. Behav. Immun. 2018, 73, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Newell, C.; Bomhof, M.R.; Reimer, R.A.; Hittel, D.S.; Rho, J.M.; Shearer, J. Ketogenic Diet Modifies the Gut Microbiota in a Murine Model of Autism Spectrum Disorder. Mol. Autism 2016, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Fu, Y.; Wang, Y.; Liao, L.; Xu, H.; Zhang, A.; Zhang, J.; Fan, L.; Ren, J.; Fang, B. Therapeutic Effects of the In Vitro Cultured Human Gut Microbiota as Transplants on Altering Gut Microbiota and Improving Symptoms Associated with Autism Spectrum Disorder. Microb. Ecol. 2020, 80, 475–486. [Google Scholar] [CrossRef]
- Liu, F.; Horton-Sparks, K.; Hull, V.; Li, R.W.; Martínez-Cerdeño, V. The Valproic Acid Rat Model of Autism Presents with Gut Bacterial Dysbiosis Similar to That in Human Autism. Mol. Autism 2018, 9, 61. [Google Scholar] [CrossRef]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259. [Google Scholar] [CrossRef]
- de Theije, C.G.M.; Wopereis, H.; Ramadan, M.; van Eijndthoven, T.; Lambert, J.; Knol, J.; Garssen, J.; Kraneveld, A.D.; Oozeer, R. Altered Gut Microbiota and Activity in a Murine Model of Autism Spectrum Disorders. Brain. Behav. Immun. 2014, 37, 197–206. [Google Scholar] [CrossRef]
- Babu, S.T.; Niu, X.; Raetz, M.; Savani, R.C.; Hooper, L.V.; Mirpuri, J. Maternal High-Fat Diet Results in Microbiota-Dependent Expansion of ILC3s in Mice Offspring. JCI Insight 2018, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Microbiota Is Essential for Social Development in the Mouse. Mol. Psychiatry 2014, 19, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of Phenol- and p-Cresol-Producing Intestinal Bacteria by Using Media Supplemented with Tyrosine and Its Metabolites. FEMS Microbiol. Ecol. 2018, 94, 125. [Google Scholar] [CrossRef]
- Bermudez-Martin, P.; Becker, J.; Fernandez, S.; Costa-Campos, R.; Barbosa, S.; Martinez-Gili, L.; Myridakis, A.; Dumas, M.-E.; Bruneau, A.; Cherbuy, C.; et al. The Microbial Metabolite p -Cresol Induces Autistic-like Behaviors in Mice by Remodeling of the Gut Microbiota. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hung, L.W.; Neuner, S.; Polepalli, J.S.; Beier, K.T.; Wright, M.; Walsh, J.J.; Lewis, E.M.; Luo, L.; Deisseroth, K.; Dölen, G.; et al. Gating of Social Reward by Oxytocin in the Ventral Tegmental Area. Science 2017, 357, 1406–1411. [Google Scholar] [CrossRef]
- Shultz, S.R.; MacFabe, D.F. Propionic Acid Animal Model of Autism. In Comprehensive Guide to Autism; Patel, V.B., Preedy, V.R., Martin, C.R., Eds.; Springer: New York, NY, USA, 2014; pp. 1755–1778. [Google Scholar] [CrossRef]
- Kamen, C.L.; Zevy, D.L.; Ward, J.M.; Bishnoi, I.R.; Kavaliers, M.; Ossenkopp, K.-P. Systemic Treatment with the Enteric Bacterial Fermentation Product, Propionic Acid, Reduces Acoustic Startle Response Magnitude in Rats in a Dose-Dependent Fashion: Contribution to a Rodent Model of ASD. Neurotox. Res. 2019, 35, 353–359. [Google Scholar] [CrossRef]
- Meeking, M.M.; MacFabe, D.F.; Mepham, J.R.; Foley, K.A.; Tichenoff, L.J.; Boon, F.H.; Kavaliers, M.; Ossenkopp, K.-P. Propionic Acid Induced Behavioural Effects of Relevance to Autism Spectrum Disorder Evaluated in the Hole Board Test with Rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 97, 109794. [Google Scholar] [CrossRef]
- Foley, K.A.; MacFabe, D.F.; Vaz, A.; Ossenkopp, K.-P.; Kavaliers, M. Sexually Dimorphic Effects of Prenatal Exposure to Propionic Acid and Lipopolysaccharide on Social Behavior in Neonatal, Adolescent, and Adult Rats: Implications for Autism Spectrum Disorders. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2014, 39, 68–78. [Google Scholar] [CrossRef]
- Kratsman, N.; Getselter, D.; Elliott, E. Sodium Butyrate Attenuates Social Behavior Deficits and Modifies the Transcription of Inhibitory/Excitatory Genes in the Frontal Cortex of an Autism Model. Neuropharmacology 2016, 102, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.; Van de Water, J. Elevated Plasma Cytokines in Autism Spectrum Disorders Provide Evidence of Immune Dysfunction and Are Associated with Impaired Behavioral Outcome. Brain. Behav. Immun. 2011, 25, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Croen, L.A.; Yoshida, C.K.; Heuer, L.; Hansen, R.; Zerbo, O.; DeLorenze, G.N.; Kharrazi, M.; Yolken, R.; Ashwood, P.; et al. Autism with Intellectual Disability Is Associated with Increased Levels of Maternal Cytokines and Chemokines During Gestation. Mol. Psychiatry 2017, 22, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, A.; Van de Water, J. The Role of the Immune System in Autism Spectrum Disorder. Neuropsychopharmacology 2017, 42, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the Microbiota and the Immune System. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Lee, G.R. The Balance of Th17 versus Treg Cells in Autoimmunity. Int. J. Mol. Sci. 2018, 19, 730. [Google Scholar] [CrossRef]
- Moaaz, M.; Youssry, S.; Elfatatry, A.; El Rahman, M.A. Th17/Treg Cells Imbalance and Their Related Cytokines (IL-17, IL-10 and TGF-β) in Children with Autism Spectrum Disorder. J. Neuroimmunol. 2019, 337, 577071. [Google Scholar] [CrossRef]
- Azhari, A.; Azizan, F.; Esposito, G. A Systematic Review of Gut-Immune-Brain Mechanisms in Autism Spectrum Disorder. Dev. Psychobiol. 2019, 61, 752–771. [Google Scholar] [CrossRef]
- Rose, D.R.; Yang, H.; Serena, G.; Sturgeon, C.; Ma, B.; Careaga, M.; Hughes, H.K.; Angkustsiri, K.; Rose, M.; Hertz-Picciotto, I.; et al. Differential Immune Responses and Microbiota Profiles in Children with Autism Spectrum Disorders and Co-Morbid Gastrointestinal Symptoms. Brain. Behav. Immun. 2018, 70, 354–368. [Google Scholar] [CrossRef]
- Rose, D.R.; Yang, H.; Careaga, M.; Angkustsiri, K.; Van de Water, J.; Ashwood, P. T Cell Populations in Children with Autism Spectrum Disorder and Co-Morbid Gastrointestinal Symptoms. Brain Behav. Immun.-Health 2020, 2, 100042. [Google Scholar] [CrossRef]
- Inoue, R.; Sakaue, Y.; Sawai, C.; Sawai, T.; Ozeki, M.; Romero-Pérez, G.A.; Tsukahara, T. A Preliminary Investigation on the Relationship between Gut Microbiota and Gene Expressions in Peripheral Mononuclear Cells of Infants with Autism Spectrum Disorders. Biosci. Biotechnol. Biochem. 2016, 80, 2450–2458. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of Type I Interferon Responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef]
- Vargas, D.L.; Nascimbene, C.; Krishnan, C.; Zimmerman, A.W.; Pardo, C.A. Neuroglial Activation and Neuroinflammation in the Brain of Patients with Autism. Ann. Neurol. 2005, 57, 67–81. [Google Scholar] [CrossRef]
- Morgan, J.T.; Chana, G.; Pardo, C.A.; Achim, C.; Semendeferi, K.; Buckwalter, J.; Courchesne, E.; Everall, I.P. Microglial Activation and Increased Microglial Density Observed in the Dorsolateral Prefrontal Cortex in Autism. Biol. Psychiatry 2010, 68, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sugihara, G.; Ouchi, Y.; Nakamura, K.; Futatsubashi, M.; Takebayashi, K.; Yoshihara, Y.; Omata, K.; Matsumoto, K.; Tsuchiya, K.J.; et al. Microglial Activation in Young Adults with Autism Spectrum Disorder. JAMA Psychiatry 2013, 70, 49–58. [Google Scholar] [CrossRef]
- Matta, S.M.; Hill-Yardin, E.L.; Crack, P.J. The Influence of Neuroinflammation in Autism Spectrum Disorder. Brain. Behav. Immun. 2019, 79, 75–90. [Google Scholar] [CrossRef]
- Erny, D.; de Angelis, A.L.H.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host Microbiota Constantly Control Maturation and Function of Microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Rothhammer, V.; Borucki, D.M.; Tjon, E.C.; Takenaka, M.C.; Chao, C.-C.; Ardura-Fabregat, A.; de Lima, K.A.; Gutiérrez-Vázquez, C.; Hewson, P.; Staszewski, O.; et al. Microglial Control of Astrocytes in Response to Microbial Metabolites. Nature 2018, 557, 724–728. [Google Scholar] [CrossRef]
- Wu, H.-J.; Wu, E. The Role of Gut Microbiota in Immune Homeostasis and Autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Saunders, J.M.; Moreno, J.L.; Ibi, D.; Sikaroodi, M.; Kang, D.J.; Muñoz-Moreno, R.; Dalmet, S.S.; García-Sastre, A.; Gillevet, P.M.; Dozmorov, M.G.; et al. Gut Microbiota Manipulation during the Prepubertal Period Shapes Behavioral Abnormalities in a Mouse Neurodevelopmental Disorder Model. Sci. Rep. 2020, 10, 4697. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H.; Yim, Y.S.; Ha, S.; Atarashi, K.; Tan, T.G.; Longman, R.S.; Honda, K.; Littman, D.R.; Choi, G.B.; et al. Maternal Gut Bacteria Promote Neurodevelopmental Abnormalities in Mouse Offspring. Nature 2017, 549, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Onore, C.E.; Careaga, M.; Babineau, B.A.; Schwartzer, J.J.; Berman, R.F.; Ashwood, P. Inflammatory Macrophage Phenotype in BTBR T+tf/J Mice. Front. Neurosci. 2013, 7, 158. [Google Scholar] [CrossRef] [PubMed]
- Lucchina, L.; Depino, A.M. Altered Peripheral and Central Inflammatory Responses in a Mouse Model of Autism. Autism Res. Off. J. Int. Soc. Autism Res. 2014, 7, 273–289. [Google Scholar] [CrossRef]
- Kazlauskas, N.; Seiffe, A.; Campolongo, M.; Zappala, C.; Depino, A.M. Sex-Specific Effects of Prenatal Valproic Acid Exposure on Sociability and Neuroinflammation: Relevance for Susceptibility and Resilience in Autism. Psychoneuroendocrinology 2019, 110, 104441. [Google Scholar] [CrossRef]
- Deckmann, I.; Schwingel, G.B.; Fontes-Dutra, M.; Bambini-Junior, V.; Gottfried, C. Neuroimmune Alterations in Autism: A Translational Analysis Focusing on the Animal Model of Autism Induced by Prenatal Exposure to Valproic Acid. Neuroimmunomodulation 2018, 25, 285–299. [Google Scholar] [CrossRef]
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary Metabolomics of Young Italian Autistic Children Supports Abnormal Tryptophan and Purine Metabolism. Mol. Autism 2016, 7, 47. [Google Scholar] [CrossRef]
- Bryn, V.; Verkerk, R.; Skjeldal, O.H.; Saugstad, O.D.; Ormstad, H. Kynurenine Pathway in Autism Spectrum Disorders in Children. Neuropsychobiology 2017, 76, 82–88. [Google Scholar] [CrossRef]
- Ormstad, H.; Bryn, V.; Verkerk, R.; Skjeldal, O.H.; Halvorsen, B.; Saugstad, O.D.; Isaksen, J.; Maes, M. Serum Tryptophan, Tryptophan Catabolites and Brain-Derived Neurotrophic Factor in Subgroups of Youngsters with Autism Spectrum Disorders. CNS Neurol. Disord. Drug Targets 2018, 17, 626–639. [Google Scholar] [CrossRef]
- Yang, C.-J.; Tan, H.-P.; Du, Y.-J. The Developmental Disruptions of Serotonin Signaling May Involved in Autism during Early Brain Development. Neuroscience 2014, 267, 1–10. [Google Scholar] [CrossRef]
- Muller, C.L.; Anacker, A.M.J.; Veenstra-VanderWeele, J. The Serotonin System in Autism Spectrum Disorder: From Biomarker to Animal Models. Neuroscience 2016, 321, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Gabriele, S.; Sacco, R.; Persico, A.M. Blood Serotonin Levels in Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2014, 24, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Gershon, M.D. The Bowel and beyond: The Enteric Nervous System in Neurological Disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Gillespie, C.H.; Anderson, G.M.; et al. Brief Report: Whole Blood Serotonin Levels and Gastrointestinal Symptoms in Autism Spectrum Disorder. J. Autism Dev. Disord. 2016, 46, 1124–1130. [Google Scholar] [CrossRef]
- Kato-Kataoka, A.; Nishida, K.; Takada, M.; Suda, K.; Kawai, M.; Shimizu, K.; Kushiro, A.; Hoshi, R.; Watanabe, O.; Igarashi, T.; et al. Fermented Milk Containing Lactobacillus Casei Strain Shirota Prevents the Onset of Physical Symptoms in Medical Students under Academic Examination Stress. Benef. Microbes 2016, 7, 153–156. [Google Scholar] [CrossRef]
- Riezzo, G.; Chimienti, G.; Orlando, A.; D’Attoma, B.; Clemente, C.; Russo, F. Effects of Long-Term Administration of Lactobacillus Reuteri DSM-17938 on Circulating Levels of 5-HT and BDNF in Adults with Functional Constipation. Benef. Microbes 2019, 10, 137–147. [Google Scholar] [CrossRef]
- Kazemi, A.; Noorbala, A.A.; Azam, K.; Eskandari, M.H.; Djafarian, K. Effect of Probiotic and Prebiotic vs Placebo on Psychological Outcomes in Patients with Major Depressive Disorder: A Randomized Clinical Trial. Clin. Nutr. Edinb. Scotl. 2019, 38, 522–528. [Google Scholar] [CrossRef]
- Rudzki, L.; Ostrowska, L.; Pawlak, D.; Małus, A.; Pawlak, K.; Waszkiewicz, N.; Szulc, A. Probiotic Lactobacillus Plantarum 299v Decreases Kynurenine Concentration and Improves Cognitive Functions in Patients with Major Depression: A Double-Blind, Randomized, Placebo Controlled Study. Psychoneuroendocrinology 2019, 100, 213–222. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The Microbiome-Gut-Brain Axis during Early Life Regulates the Hippocampal Serotonergic System in a Sex-Dependent Manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut Microbes Promote Colonic Serotonin Production through an Effect of Short-Chain Fatty Acids on Enterochromaffin Cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 1395–1403. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Horder, J.; Petrinovic, M.M.; Mendez, M.A.; Bruns, A.; Takumi, T.; Spooren, W.; Barker, G.J.; Künnecke, B.; Murphy, D.G. Glutamate and GABA in Autism Spectrum Disorder-a Translational Magnetic Resonance Spectroscopy Study in Man and Rodent Models. Transl. Psychiatry 2018, 8, 106. [Google Scholar] [CrossRef]
- Puts, N.A.J.; Wodka, E.L.; Harris, A.D.; Crocetti, D.; Tommerdahl, M.; Mostofsky, S.H.; Edden, R.A.E. Reduced GABA and Altered Somatosensory Function in Children with Autism Spectrum Disorder. Autism Res. Off. J. Int. Soc. Autism Res. 2017, 10, 608–619. [Google Scholar] [CrossRef]
- Coghlan, S.; Horder, J.; Inkster, B.; Mendez, M.A.; Murphy, D.G.; Nutt, D.J. GABA System Dysfunction in Autism and Related Disorders: From Synapse to Symptoms. Neurosci. Biobehav. Rev. 2012, 36, 2044–2055. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P. Neurotransmitter Modulation by the Gut Microbiota. Brain Res. 2018, 1693 Pt B, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Parracho, H.M.R.T.; Gibson, G.R.; Knott, F.; Bosscher, D.; Kleerebezem, M.; McCartney, A.L. A Double-Blind, Placebo-Controlled, Crossover-Designed Probiotic Feeding Study in Children Diagnosed with Autistic Spectrum Disorders. Int. J. Probiotics Prebiotics 2010, 5, 69–74. [Google Scholar]
- Arnold, L.E.; Luna, R.A.; Williams, K.; Chan, J.; Parker, R.A.; Wu, Q.; Hollway, J.A.; Jeffs, A.; Lu, F.; Coury, D.L.; et al. Probiotics for Gastrointestinal Symptoms and Quality of Life in Autism: A Placebo-Controlled Pilot Trial. J. Child Adolesc. Psychopharmacol. 2019, 29, 659–669. [Google Scholar] [CrossRef]
- Sanctuary, M.R.; Kain, J.N.; Chen, S.Y.; Kalanetra, K.; Lemay, D.G.; Rose, D.R.; Yang, H.T.; Tancredi, D.J.; German, J.B.; Slupsky, C.M.; et al. Pilot Study of Probiotic/Colostrum Supplementation on Gut Function in Children with Autism and Gastrointestinal Symptoms. PLoS ONE 2019, 14, e0210064. [Google Scholar] [CrossRef]
- Grossi, E.; Melli, S.; Dunca, D.; Terruzzi, V. Unexpected Improvement in Core Autism Spectrum Disorder Symptoms after Long-Term Treatment with Probiotics. SAGE Open Med. Case Rep. 2016, 4. [Google Scholar] [CrossRef]
- Shaaban, S.Y.; El Gendy, Y.G.; Mehanna, N.S.; El-Senousy, W.M.; El-Feki, H.S.A.; Saad, K.; El-Asheer, O.M. The Role of Probiotics in Children with Autism Spectrum Disorder: A Prospective, Open-Label Study. Nutr. Neurosci. 2018, 21, 676–681. [Google Scholar] [CrossRef]
- Liu, Y.-W.; Liong, M.T.; Chung, Y.-C.E.; Huang, H.-Y.; Peng, W.-S.; Cheng, Y.-F.; Lin, Y.-S.; Wu, Y.-Y.; Tsai, Y.-C. Effects of Lactobacillus Plantarum PS128 on Children with Autism Spectrum Disorder in Taiwan: A Randomized, Double-Blind, Placebo-Controlled Trial. Nutrients 2019, 11, 820. [Google Scholar] [CrossRef]
- Liu, J.; Wan, G.-B.; Huang, M.-S.; Agyapong, G.; Zou, T.; Zhang, X.-Y.; Liu, Y.-W.; Song, Y.-Q.; Tsai, Y.-C.; Kong, X.-J. Probiotic Therapy for Treating Behavioral and Gastrointestinal Symptoms in Autism Spectrum Disorder: A Systematic Review of Clinical Trials. Curr. Med. Sci. 2019, 39, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Scientists, Doctors, Companies–Gemma. Available online: https://www.gemma-project.eu/information/scientists-doctors-companies/ (accessed on 8 July 2020).
- Kong, X.-J.; Liu, J.; Li, J.; Kwong, K.; Koh, M.; Sukijthamapan, P.; Guo, J.J.; Sun, Z.J.; Song, Y. Probiotics and Oxytocin Nasal Spray as Neuro-Social-Behavioral Interventions for Patients with Autism Spectrum Disorders: A Pilot Randomized Controlled Trial Protocol. Pilot Feasibility Stud. 2020, 6, 20. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Gasbarrini, A. Fecal Microbiota Transplantation for the Treatment of Clostridium Difficile Infection: A Systematic Review. J. Clin. Gastroenterol. 2014, 48, 693–702. [Google Scholar] [CrossRef]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal Microbiota Transplantation Induces Remission in Patients with Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015, 149, 102–109. [Google Scholar] [CrossRef]
- El-Salhy, M.; Hatlebakk, J.G.; Gilja, O.H.; Kristoffersen, A.B.; Hausken, T. Efficacy of Faecal Microbiota Transplantation for Patients with Irritable Bowel Syndrome in a Randomised, Double-Blind, Placebo-Controlled Study. Gut 2020, 69, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojärvi, J.; Kootte, R.S.; Bartelsman, J.F.W.M.; Dallinga–Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of Intestinal Microbiota from Lean Donors Increases Insulin Sensitivity in Individuals With Metabolic Syndrome. Gastroenterology 2012, 143, 913–916. [Google Scholar] [CrossRef]
- Zhang, Z.; Mocanu, V.; Cai, C.; Dang, J.; Slater, L.; Deehan, E.C.; Walter, J.; Madsen, K.L. Impact of Fecal Microbiota Transplantation on Obesity and Metabolic Syndrome—A Systematic Review. Nutrients 2019, 11, 2291. [Google Scholar] [CrossRef] [PubMed]
- Vendrik, K.E.W.; Ooijevaar, R.E.; de Jong, P.R.C.; Laman, J.D.; van Oosten, B.W.; van Hilten, J.J.; Ducarmon, Q.R.; Keller, J.J.; Kuijper, E.J.; Contarino, M.F. Fecal Microbiota Transplantation in Neurological Disorders. Front. Cell. Infect. Microbiol. 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy Alters Gut Ecosystem and Improves Gastrointestinal and Autism Symptoms: An Open-Label Study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-Term Benefit of Microbiota Transfer Therapy on Autism Symptoms and Gut Microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–16187. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Yu, L.; Tian, F.; Zhai, Q.; Fan, L.; Chen, W. Gut Microbiota: A Target for Heavy Metal Toxicity and a Probiotic Protective Strategy. Sci. Total Environ. 2020, 742, 140429. [Google Scholar] [CrossRef] [PubMed]
- Obrenovich, M.E.; Shamberger, R.J.; Lonsdale, D. Altered Heavy Metals and Transketolase Found in Autistic Spectrum Disorder. Biol. Trace Elem. Res. 2011, 144, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Cen, S.; Jiang, J.; Zhao, J.; Zhang, H.; Chen, W. Disturbance of Trace Element and Gut Microbiota Profiles as Indicators of Autism Spectrum Disorder: A Pilot Study of Chinese Children. Environ. Res. 2019, 171, 501–509. [Google Scholar] [CrossRef]
- Belardo, A.; Gevi, F.; Zolla, L. The Concomitant Lower Concentrations of Vitamins B6, B9 and B12 May Cause Methylation Deficiency in Autistic Children. J. Nutr. Biochem. 2019, 70, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roussin, L.; Prince, N.; Perez-Pardo, P.; Kraneveld, A.D.; Rabot, S.; Naudon, L. Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence. Microorganisms 2020, 8, 1369. https://doi.org/10.3390/microorganisms8091369
Roussin L, Prince N, Perez-Pardo P, Kraneveld AD, Rabot S, Naudon L. Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence. Microorganisms. 2020; 8(9):1369. https://doi.org/10.3390/microorganisms8091369
Chicago/Turabian StyleRoussin, Léa, Naika Prince, Paula Perez-Pardo, Aletta D. Kraneveld, Sylvie Rabot, and Laurent Naudon. 2020. "Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence" Microorganisms 8, no. 9: 1369. https://doi.org/10.3390/microorganisms8091369
APA StyleRoussin, L., Prince, N., Perez-Pardo, P., Kraneveld, A. D., Rabot, S., & Naudon, L. (2020). Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence. Microorganisms, 8(9), 1369. https://doi.org/10.3390/microorganisms8091369





