Characterization of a Lytic Bacteriophage vB_EfaS_PHB08 Harboring Endolysin Lys08 against Enterococcus faecalis Biofilms
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultural Conditions
2.2. Phage Isolation and Purification
2.3. Host Range of the Phage
2.4. Physical Parameter of the Phage
2.5. One-Step Growth Curve
2.6. Lytic Activity
2.7. DNA Extraction and Analysis of Genome Sequence
2.8. Cloning, Expression and Activity Identification of the Endolysin
2.9. Antibiofilm by Phage and Its Endolysins
3. Results
3.1. Characteristics of Phage Specific for E. faecalis
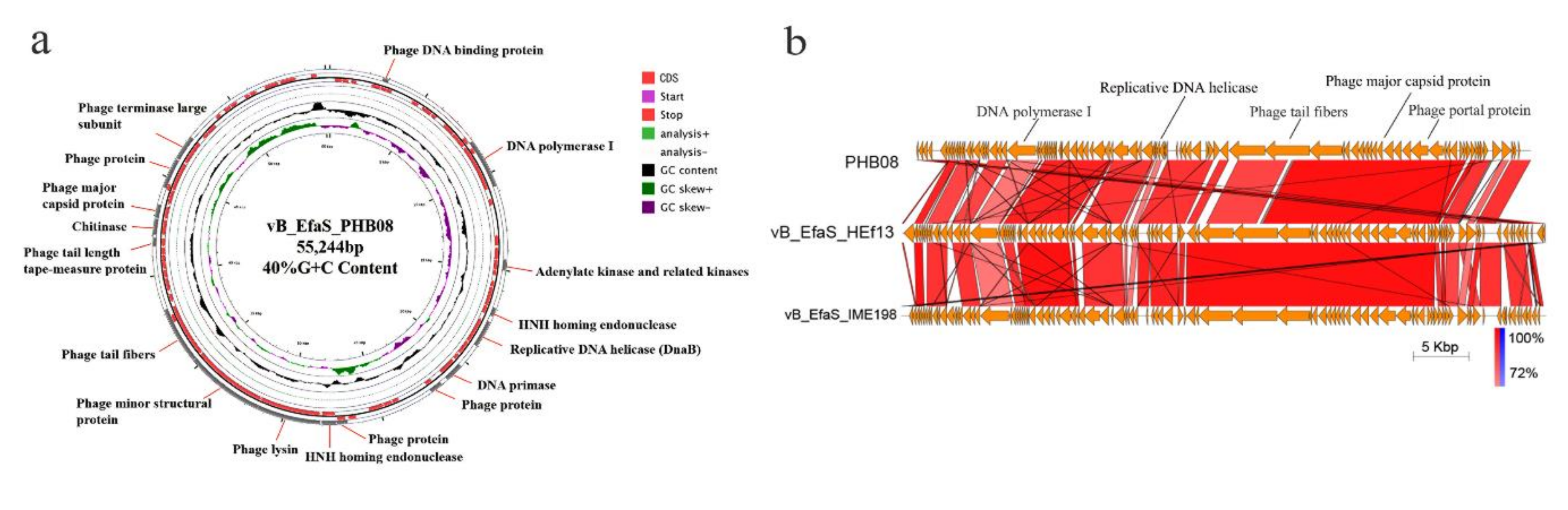
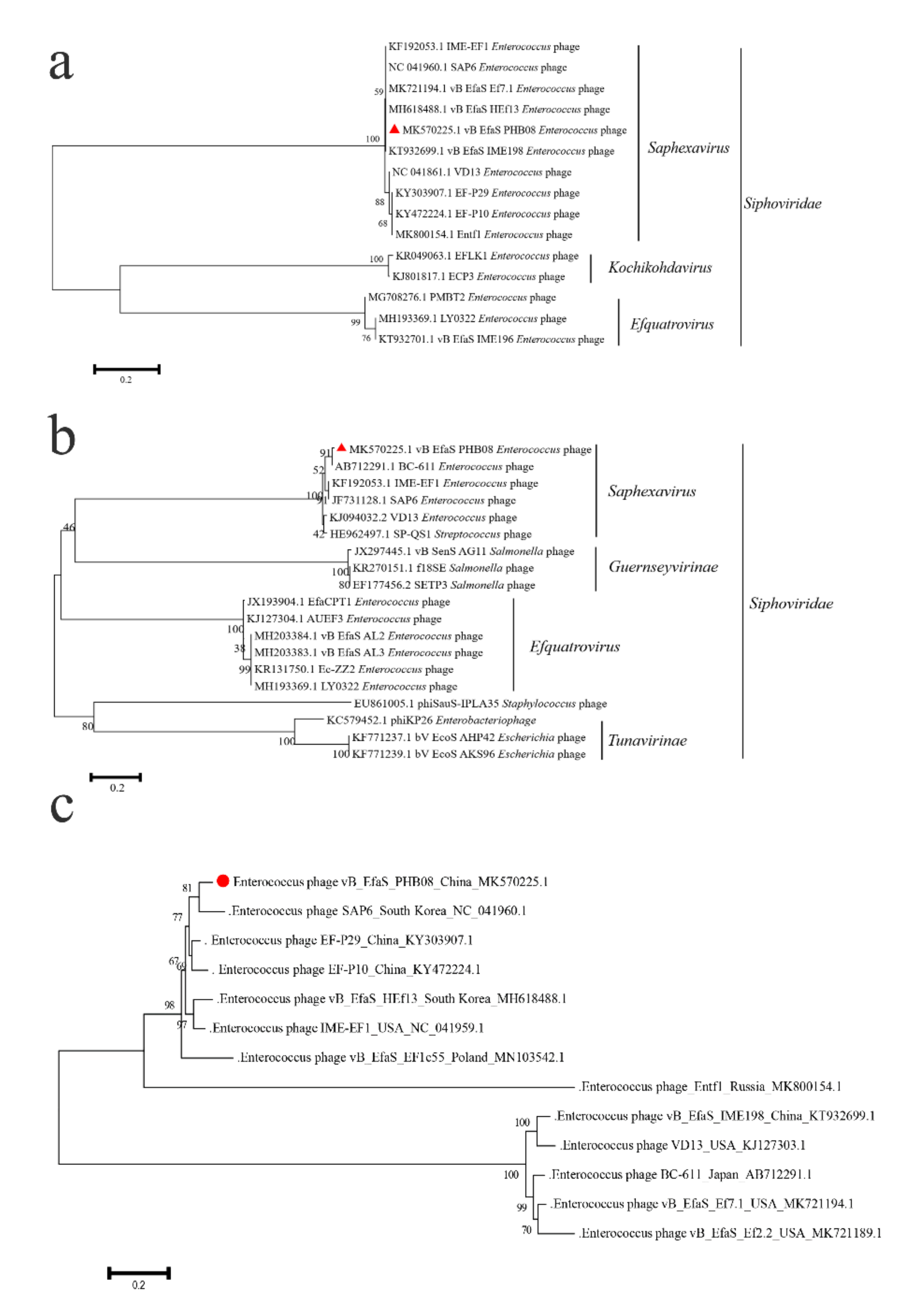
3.2. General Feature of Phage PHB08 Whole Genome Sequence
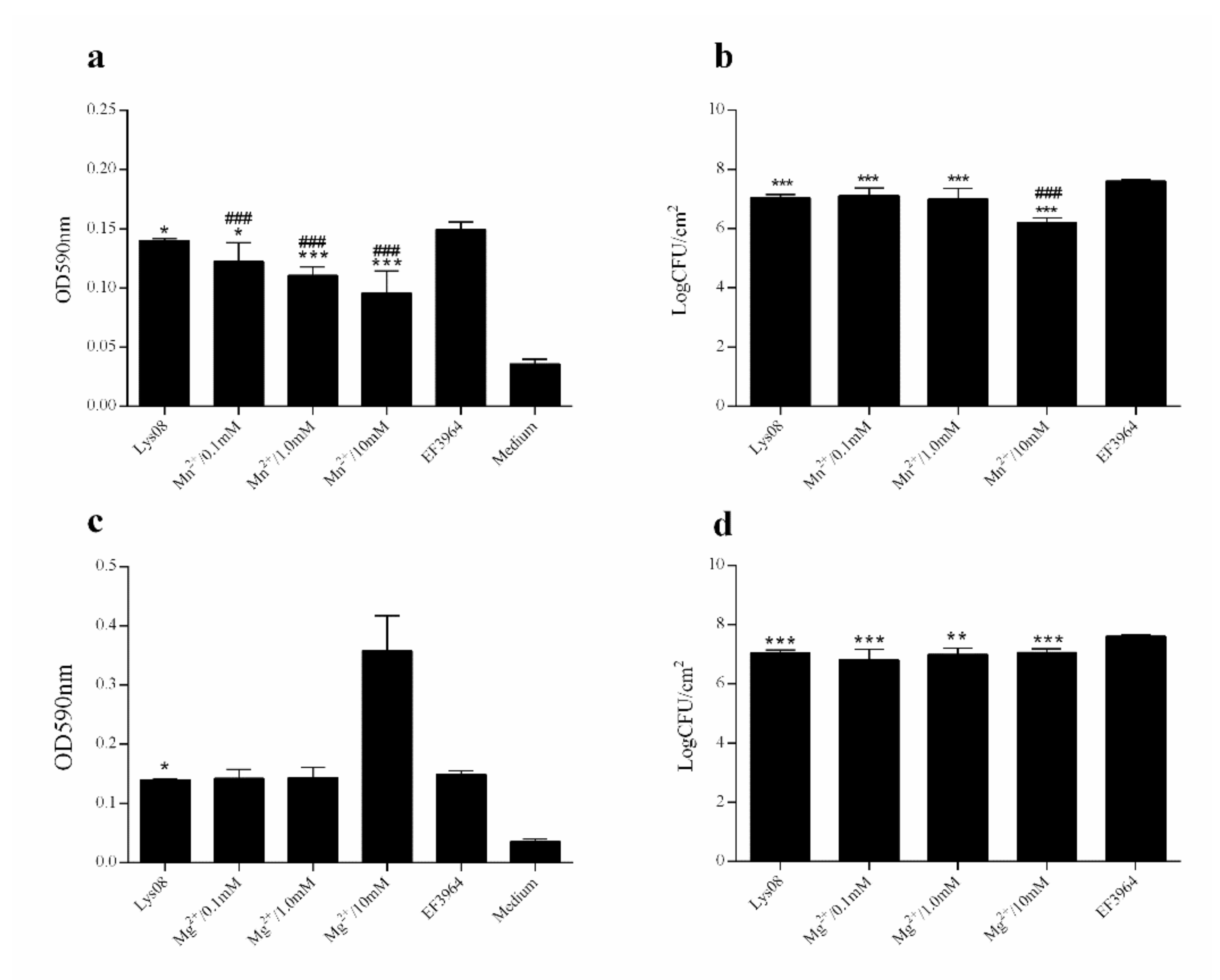
3.3. Antibiofilm Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yazgan, H.; Ozogul, Y.; Kuley, E. Antimicrobial influence of nanoemulsified lemon essential oil and pure lemon essential oil on food-borne pathogens and fish spoilage bacteria. Int. J. Food Microbiol. 2019, 306, 108266. [Google Scholar] [CrossRef] [PubMed]
- Dahl, A.; Bruun, N.E. Enterococcus faecalis infective endocarditis: Focus on clinical aspects. Expert. Rev. Cardiovasc. Ther. 2013, 11, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Stuart, C.H.; Schwartz, S.A.; Beeson, T.J.; Owatz, C.B. Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J. Endod. 2006, 32, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Tebruegge, M.; Pantazidou, A.; Clifford, V.; Gonis, G.; Ritz, N.; Connell, T.; Curtis, N. The age-related risk of co-existing meningitis in children with urinary tract infection. PLoS ONE 2011, 6, e26576. [Google Scholar] [CrossRef] [PubMed]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. Nhsn annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect. Control. Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Gjodsbol, K.; Christensen, J.J.; Karlsmark, T.; Jorgensen, B.; Klein, B.M.; Krogfelt, K.A. Multiple bacterial species reside in chronic wounds: A longitudinal study. Int. Wound J. 2006, 3, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Li, X.; Liu, P.; Luo, M.; Chen, S.; Su, K.; Zhang, Z.; He, Q.; Qiu, J.; Li, Y. Epidemiology of pathogens and antimicrobial resistanceof catheter-associated urinary tract infections in intensivecare units: A systematic review and meta-analysis. Am. J. Infect. Control. 2018, 46, e81–e90. [Google Scholar] [CrossRef]
- Zhang, C.; Du, J.; Peng, Z. Correlation between enterococcus faecalis and persistent intraradicular infection compared with primary intraradicular infection: A systematic review. J. Endod. 2015, 41, 1207–1213. [Google Scholar] [CrossRef]
- Vidana, R.; Sullivan, A.; Billstrom, H.; Ahlquist, M.; Lund, B. Enterococcus faecalis infection in root canals - host-derived or exogenous source? Lett. Appl. Microbiol. 2011, 52, 109–115. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006, 42 (Suppl. 1), S25–S34. [Google Scholar] [CrossRef]
- Nilsson, O. Vancomycin resistant enterococci in farm animals - occurrence and importance. Infect. Ecol. Epidemiol. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Baptiste, K. Vancomycin-resistant enterococci: A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microbial. Drug Resistance (Larchmont, N.Y.) 2018, 24, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.W. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 2000, 31, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Abamecha, A.; Wondafrash, B.; Abdissa, A. Antimicrobial resistance profile of enterococcus species isolated from intestinal tracts of hospitalized patients in jimma, ethiopia. BMC Res. Notes 2015, 8, 213. [Google Scholar] [CrossRef]
- Kaveh, M.; Bazargani, A.; Ramzi, M.; Ebrahim-Saraie, H.S.; Heidari, H. Colonization rate and risk factors of vancomycin-resistant enterococci among patients received hematopoietic stem cell transplantation in shiraz, southern iran. Int. J. Organ Transplant Med. 2016, 7, 197–205. [Google Scholar]
- Rizzotti, L.; Rossi, F.; Torriani, S. Biocide and antibiotic resistance of enterococcus faecalis and enterococcus faecium isolated from the swine meat chain. Food Microbiol. 2016, 60, 160–164. [Google Scholar] [CrossRef]
- van Harten, R.M.; Willems, R.J.L.; Martin, N.I.; Hendrickx, A.P.A. Multidrug-resistant enterococcal infections: New compounds, novel antimicrobial therapies? Trends Microbiol. 2017, 25, 467–479. [Google Scholar] [CrossRef]
- Tan, C.A.Z.; Antypas, H.; Kline, K.A. Overcoming the challenge of establishing biofilms in vivo: A roadmap for enterococci. Curr. Opin. Microbiol. 2020, 53, 9–18. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Hu, F.Z.; Gieseke, A.; Nistico, L.; Nguyen, D.; Hayes, J.; Forbes, M.; Greenberg, D.P.; Dice, B.; Burrows, A. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 2006, 296, 202–211. [Google Scholar] [CrossRef]
- Baldassarri, L.; Cecchini, R.; Bertuccini, L.; Ammendolia, M.G.; Iosi, F.; Arciola, C.R.; Montanaro, L.; Di Rosa, R.; Gherardi, G.; Dicuonzo, G.; et al. Enterococcus spp. Produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. 2001, 190, 113–120. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Ram, P.K.; Gupta, S.K.; Miller, M.A.; Mintz, E.D. Part i. Analysis of data gaps pertaining to salmonella enterica serotype typhi infections in low and medium human development index countries, 1984-2005. Epidemiol. Infect. 2008, 136, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.A.; Beyth, N.; Hazan, R. Targeting enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Gelman, D.; Beyth, S.; Lerer, V.; Adler, K.; Poradosu-Cohen, R.; Coppenhagen-Glazer, S.; Hazan, R. Combined bacteriophages and antibiotics as an efficient therapy against vre enterococcus faecalis in a mouse model. Res. Microbiol. 2018, 169, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Wroe, J.A.; Johnson, C.T.; Garcia, A.J. Bacteriophage delivering hydrogels reduce biofilm formation in vitro and infection in vivo. J. Biomed. Mater. Res. A 2020, 108, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Yuksel, F.N.; Buzrul, S.; Akcelik, M.; Akcelik, N. Inhibition and eradication of salmonella typhimurium biofilm using p22 bacteriophage, edta and nisin. Biofouling 2018, 34, 1046–1054. [Google Scholar] [CrossRef]
- Pennone, V.; Sanz-Gaitero, M.; O’Connor, P.; Coffey, A.; Jordan, K.; van Raaij, M.; McAuliffe, O. Inhibition of L. monocytogenes biofilm formation by the amidase domain of the phage vb_lmos_293 endolysin. Viruses 2019, 11, 722. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, X.; Wang, L.; Li, G.; Cong, C.; Li, R.; Cui, H.; Murtaza, B.; Xu, Y. The endolysin of the acinetobacter baumannii phage vb_abap_d2 shows broad antibacterial activity. Microbial. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P. A novel tail-associated o91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of shiga toxin-producing escherichia coli. Appl. Environ. Microb. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wen, Z.; Li, N.; Yang, W.; Wang, J.; Hu, L.; Dong, X.; Lu, J.; Li, J. Impact of relative humidity and collection media on mycobacteriophage d29 aerosol. Appl. Environ. Microb. 2012, 78, 1466–1472. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.B.; Sun, E.C.; Song, J.Y.; Yang, L.; Wu, B. Complete genome sequence of a novel t7-like bacteriophage from a pasteurella multocida capsular type a isolate. Curr. Microbiol. 2018, 75, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.B.; Sun, E.C.; Yang, L.; Song, J.Y.; Wu, B. Therapeutic application of bacteriophage phb02 and its putative depolymerase against pasteurella multocida capsular type a in mice. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cai, L.; Ma, R.; Xu, Y.; Tong, Y.; Huang, Y.; Jiao, N.; Zhang, R. A novel roseosiphophage isolated from the oligotrophic south china sea. Viruses 2017, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.H.; Li, J.Q. Application of a phage cocktail for control of salmonella in foods and reducing biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef]
- Huang, C.X.; Virk, S.M.; Shi, J.C.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.H.; Li, J.Q. Isolation, characterization, and application of bacteriophage lpse1 against salmonella enterica in ready to eat (rte) foods. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Adams, M.; Hartley, A.; Cox, L.J.F.M. Factors affecting the efficacy of washing procedures used in the production of prepared salads. Food Microbiol. 1989, 6, 69–77. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, L.; Yang, D.; Song, J.; Wang, C.; Sun, E.; Gu, C.; Chen, H.; Tong, Y.; Tao, P.; et al. Specific integration of temperate phage decreases the pathogenicity of host bacteria. Front. Cell Infect. Microbiol. 2020, 10, 14. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S. Soapdenovo-trans: De novo transcriptome assembly with short rna-seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, S.L.; Delcher, A.L.; Kasif, S.; White, O. Microbial gene identification using interpolated markov models. Nucleic Acids Res. 1998, 26, 544–548. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The trnascan-se, snoscan and snogps web servers for the detection of trnas and snornas. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. Aragorn, a program to detect trna genes and tmrna genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Diez-Martinez, R.; De Paz, H.D.; Garcia-Fernandez, E.; Bustamante, N.; Euler, C.W.; Fischetti, V.A.; Menendez, M.; Garcia, P. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against streptococcus pneumoniae. J. Antimicrob. Chemother. 2015, 70, 1763–1773. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar] [CrossRef] [PubMed]
- Milho, C.; Silva, M.; Alves, D.; Oliveira, H.; Sousa, C.; Pastrana, L.; Azeredo, J.; Sillankorva, S. Escherichia coli and salmonella enteritidis dual-species biofilms: Interspecies interactions and antibiofilm efficacy of phages. Sci. Rep. 2019, 9, 18183. [Google Scholar] [CrossRef]
- Schmelcher, M.; Waldherr, F.; Loessner, M. Listeria bacteriophage peptidoglycan hydrolases feature high thermoresistance and reveal increased activity after divalent metal cation substitution. Appl. Microbiol. Biotechnol. 2012, 93, 633–643. [Google Scholar] [CrossRef]
- Cha, Y.; Son, B.; Ryu, S. Effective removal of staphylococcal biofilms on various food contact surfaces by staphylococcus aureus phage endolysin lyscsa13. Food Microbiol. 2019, 84, 103245. [Google Scholar] [CrossRef]
- Aslam, S. Bacteriophage therapy as a treatment option for transplant infections. Curr. Opin. Infect. Dis. 2020, 33, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhang, Y.; Huang, C.; Wang, J.; Wang, X. Salmonellaan endolysin lysse24 by bacteriophage lpse1 confers specific bactericidal activity against multidrug-resistant strains. Microorganisms 2020, 8, 737. [Google Scholar] [CrossRef] [PubMed]
- Crotty, M.; Krekel, T.; Burnham, C.; Ritchie, D. New gram-positive agents: The next generation of oxazolidinones and lipoglycopeptides. J. Clin. Microbiol. 2016, 54, 2225–2232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henson, K.; Levine, M.; Wong, E.; Levine, D. Glycopeptide antibiotics: Evolving resistance, pharmacology and adverse event profile. Expert Rev. Anti-Infect. Ther. 2015, 13, 1265–1278. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Im, J.; Na, H.; Ryu, S.; Yun, C.; Han, S. Enterococcusthe novel phage vb_efas_hef13 has broad lytic activity against clinical isolates of. Front. Microbiol. 2019, 10, 2877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mi, Z.; Yin, X.; Fan, H.; An, X.; Zhang, Z.; Chen, J.; Tong, Y. Characterization of enterococcus faecalis phage ime-ef1 and its endolysin. PLoS ONE 2013, 8, e80435. [Google Scholar] [CrossRef]
- Cheng, M.; Zhang, Y.; Li, X.; Liang, J.; Hu, L.; Gong, P.; Zhang, L.; Cai, R.; Zhang, H.; Ge, J. Endolysin lysef-p10 shows potential as an alternative treatment strategy for multidrug-resistant enterococcus faecalis infections. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Cheng, M.; Liang, J.; Zhang, Y.; Hu, L.; Gong, P.; Cai, R.; Zhang, L.; Zhang, H.; Ge, J.; Ji, Y. The bacteriophage ef-p29 efficiently protects against lethal vancomycin-resistant enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Front. Microbiol. 2017, 8, 837. [Google Scholar] [CrossRef]
- Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Akechi, K.; Muraoka, A.; Wakiguchi, H.; Matsuzaki, S. Isolation and characterization of a novel enterococcus faecalis bacteriophage φef24c as a therapeutic candidate. FEMS Microbiol. Lett. 2008, 278, 200–206. [Google Scholar] [CrossRef]
- Guentzel, J.; Liang Lam, K.; Callan, M.; Emmons, S.; Dunham, V. Reduction of bacteria on spinach, lettuce, and surfaces in food service areas using neutral electrolyzed oxidizing water. Food Microbiol. 2008, 25, 36–41. [Google Scholar] [CrossRef]
- Mead, P.; Slutsker, L.; Dietz, V.; McCaig, L.; Bresee, J.; Shapiro, C.; Griffin, P.; Tauxe, R. Food-related illness and death in the united states. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Seo, H.; Seo, K.; Jeon, H.; Kim, D.; Kim, S.; Lim, S.; Lee, Y. Characteristics of high-level ciprofloxacin-resistant enterococcus faecalis and enterococcus faecium from retail chicken meat in korea. J. Food Prot. 2018, 81, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Jalowiecki, L.; Zur, J.; Chojniak, J.; Ejhed, H.; Plaza, G. Properties of antibiotic-resistant bacteria isolated from onsite wastewater treatment plant in relation to biofilm formation. Curr. Microbiol. 2018, 75, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Shida, T.; Hattori, H.; Ise, F.; Sekiguchi, J. Mutational analysis of catalytic sites of the cell wall lytic n-acetylmuramoyl-l-alanine amidases cwlc and cwlv. J. Biol. Chem. 2001, 276, 28140–28146. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Chen, Y.; Sun, E.; Hua, L.; Peng, Z.; Wu, B. Characterization of a Lytic Bacteriophage vB_EfaS_PHB08 Harboring Endolysin Lys08 against Enterococcus faecalis Biofilms. Microorganisms 2020, 8, 1332. https://doi.org/10.3390/microorganisms8091332
Yang D, Chen Y, Sun E, Hua L, Peng Z, Wu B. Characterization of a Lytic Bacteriophage vB_EfaS_PHB08 Harboring Endolysin Lys08 against Enterococcus faecalis Biofilms. Microorganisms. 2020; 8(9):1332. https://doi.org/10.3390/microorganisms8091332
Chicago/Turabian StyleYang, Dan, Yibao Chen, Erchao Sun, Lin Hua, Zhong Peng, and Bin Wu. 2020. "Characterization of a Lytic Bacteriophage vB_EfaS_PHB08 Harboring Endolysin Lys08 against Enterococcus faecalis Biofilms" Microorganisms 8, no. 9: 1332. https://doi.org/10.3390/microorganisms8091332
APA StyleYang, D., Chen, Y., Sun, E., Hua, L., Peng, Z., & Wu, B. (2020). Characterization of a Lytic Bacteriophage vB_EfaS_PHB08 Harboring Endolysin Lys08 against Enterococcus faecalis Biofilms. Microorganisms, 8(9), 1332. https://doi.org/10.3390/microorganisms8091332




