Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems
Abstract
1. Introduction
2. Materials and Methods
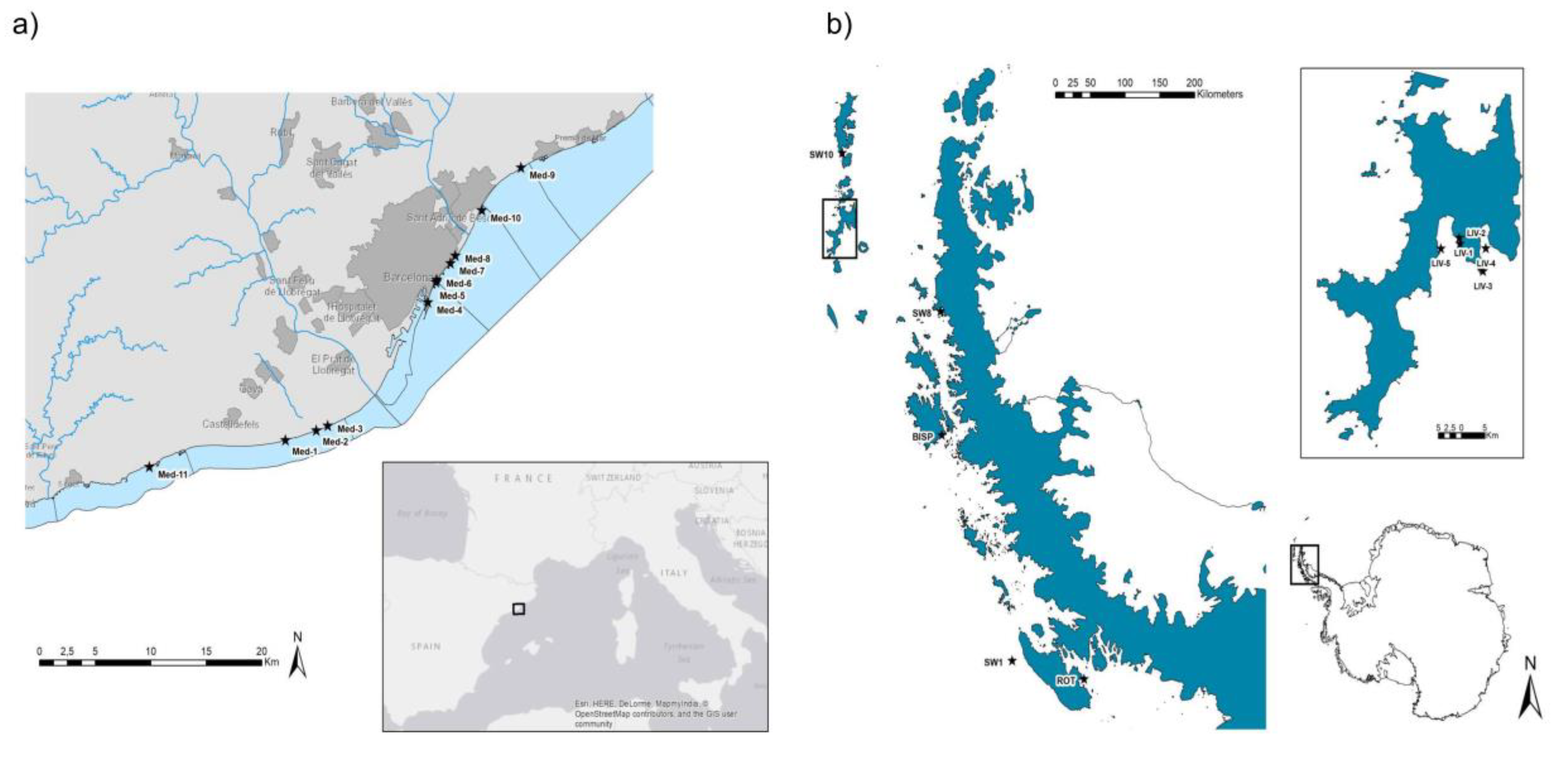
2.1. Samples
2.2. Phage Isolation from Seawater Samples
2.3. Phage Isolation from Induced Antarctic Bacterial Communities
2.4. Phage DNA Extraction
2.5. Amplification of ARGs in the DNA Extracted from Phage Particles
2.6. Bacterial and Viral Indicators
2.7. Propagation Cultures for Phages in Fish and Shellfish Samples
3. Results
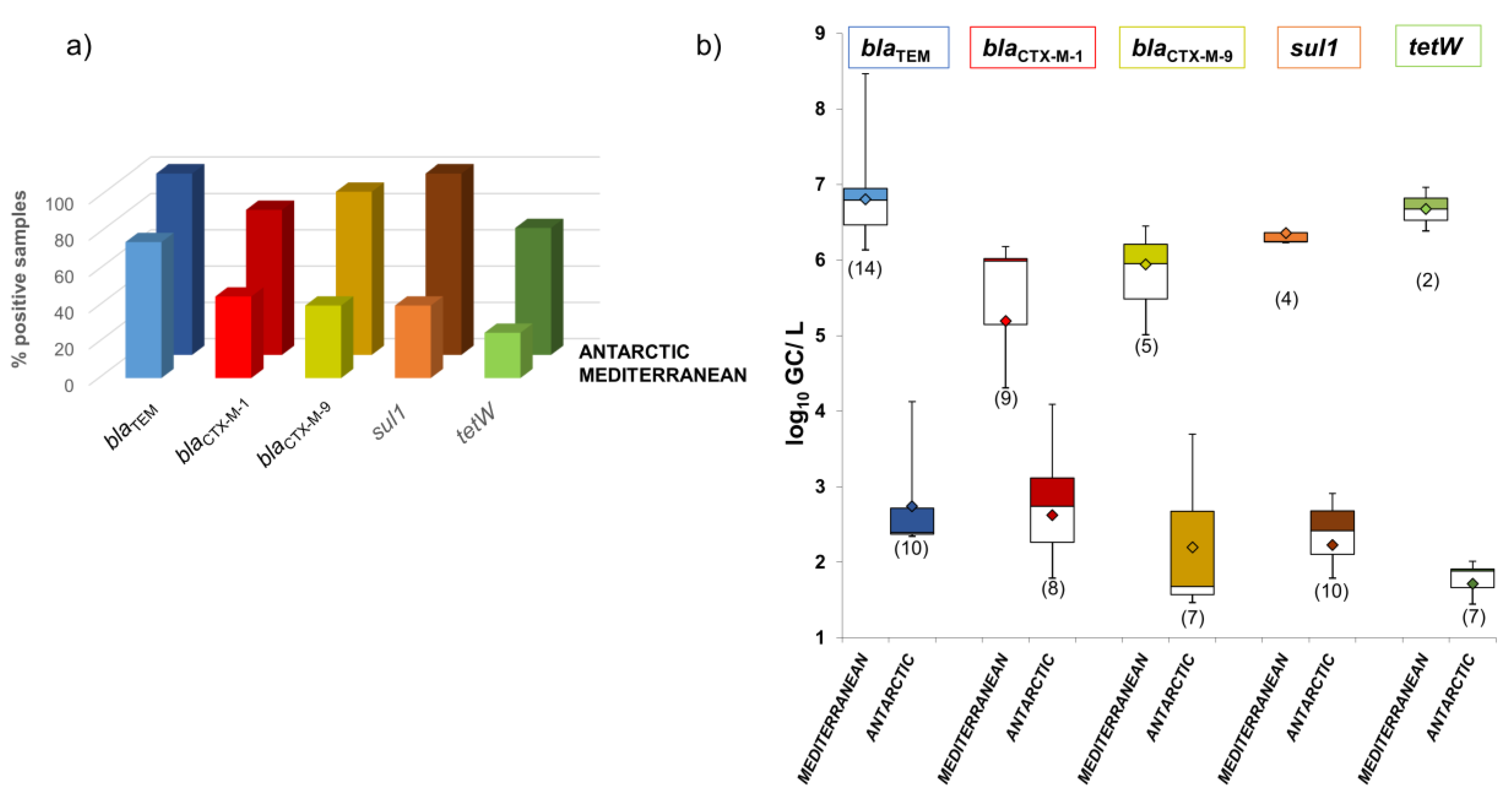
3.1. Comparison of ARGs in the Phage DNA Fraction from Mediterranean and Antarctic Seawater Samples
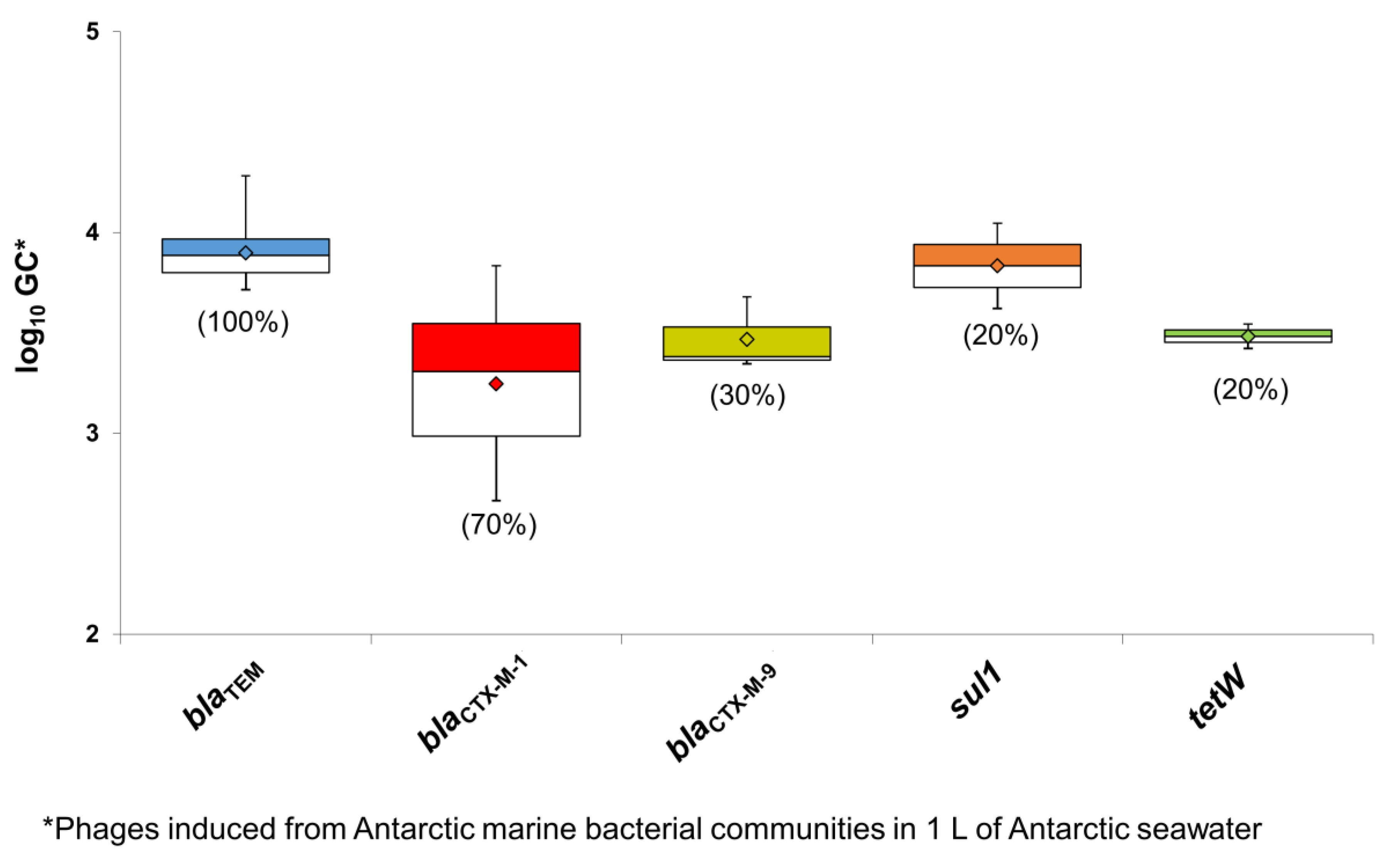
3.2. ARGs in Phage Particles Induced from Antarctic Bacterial Communities
3.3. Consequences of Anthropogenic Pollution in Mediterranean Environments
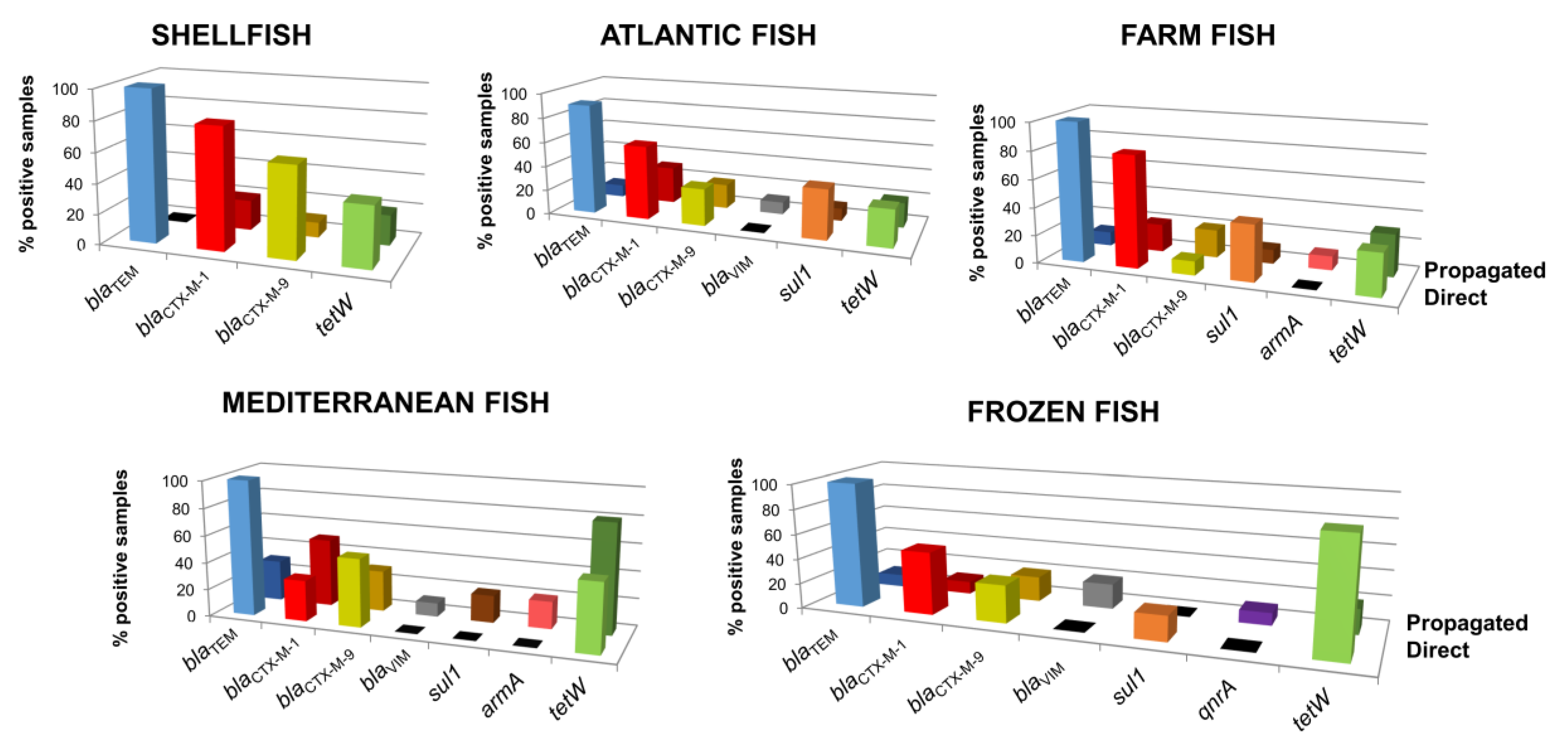
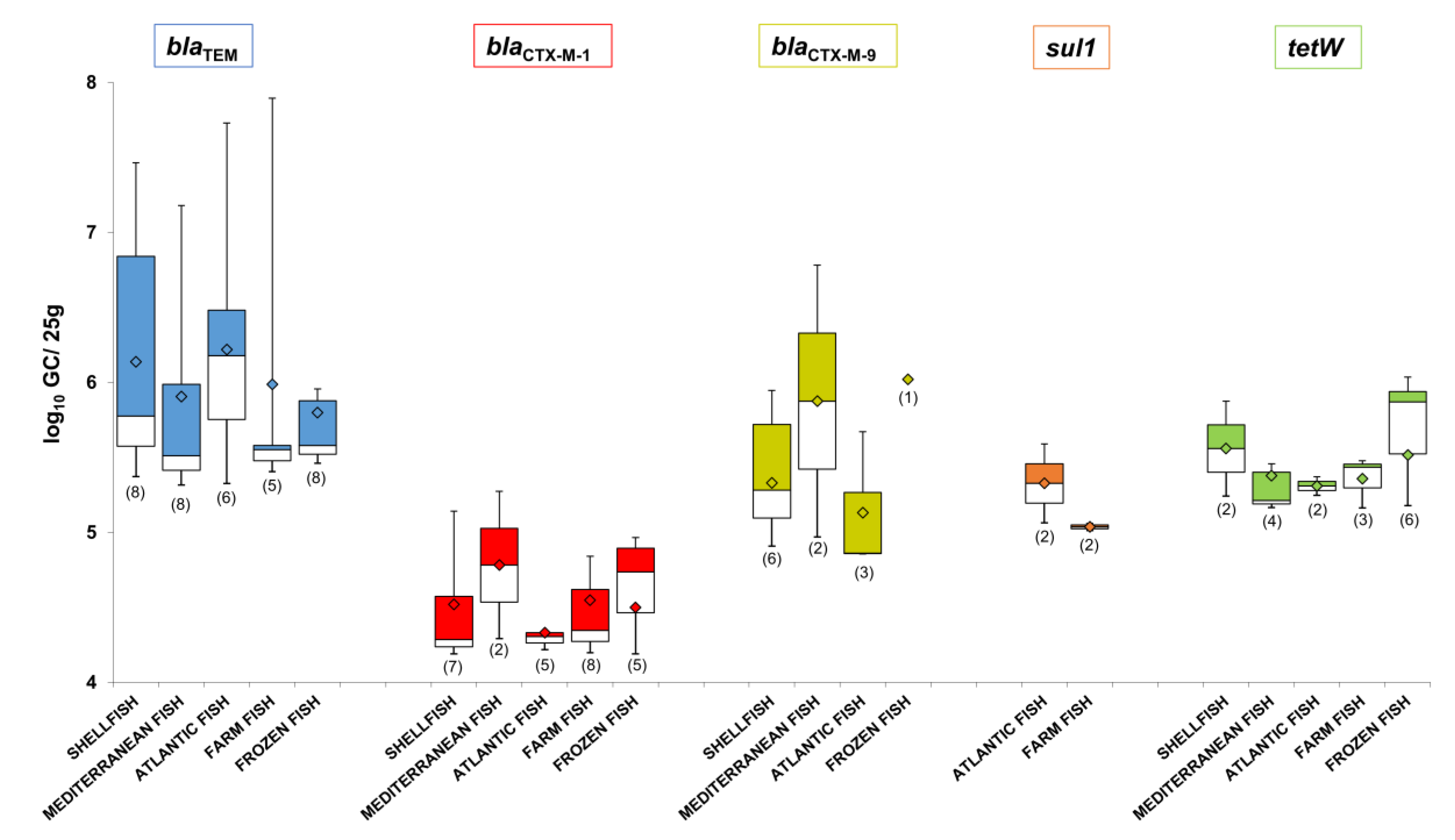
3.4. Prevalence of ARGs in Phage DNA Fraction Isolated from Shellfish and Fish Samples
3.5. Abundance of ARGs in the Phage DNA Fraction Isolated from Shellfish and Fish Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 8 January 2019).
- Ventola, C.L. Antibiotic Resistance Crisis. Int. J. Med. Dev. Ctries. 2019, 40, 561–564. [Google Scholar] [CrossRef]
- García, J.; García-Galán, M.J.; Day, J.W.; Boopathy, R.; White, J.R.; Wallace, S.; Hunter, R.G. A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour. Technol. 2020, 307, 123228. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Méndez, J.; Martín-Díaz, J.; Muniesa, M. The occurrence of antibiotic resistance genes in a Mediterranean river and their persistence in the riverbed sediment. Environ. Pollut. 2017, 223. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.V.; Gammon, K.; Day, M.J. Antibiotic resistance among bacteria isolated from seawater and penguin fecal samples collected near Palmer Station, Antarctica. Can. J. Microbiol. 2009, 55, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Balcazar, J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014, 10, e1004219. [Google Scholar] [CrossRef]
- Lekunberri, I.; Subirats, J.; Borrego, C.M.; Balcázar, J.L. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017, 220, 981–984. [Google Scholar] [CrossRef]
- Keen, E.C. A century of phage research: Bacteriophages and the shaping of modern biology. BioEssays 2015, 37, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Calero-Cáceres, W.; Muniesa, M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016, 95, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penadés, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef] [PubMed]
- Shousha, A.; Awaiwanont, N.; Sofka, D.; Smulders, F.J.M.; Paulsen, P.; Szostak, M.P.; Humphrey, T.; Hilbert, F. Bacteriophages Isolated from Chicken Meat and the Horizontal Transfer of Antimicrobial Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 4600–4606. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Melgarejo, A.; Colomer-Lluch, M.; Stoll, C.; Lucena, F.; Jofre, J.; Muniesa, M. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ. Sci. Technol. 2014, 48. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Calero-Cáceres, W.; Jebri, S.; Hmaied, F.; Muniesa, M.; Jofre, J. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 2014, 73, 167–175. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Ye, M.; Balcázar, J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019, 27, 570–577. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Calero-Cáceres, W.; Espinal, P.; Rodríguez-Navarro, J.; Miró, E.; González-López, J.J.; Cornejo, T.; Hurtado, J.C.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in phage particles isolated from human feces and induced from clinical bacterial isolates. Int. J. Antimicrob. Agents 2018, 51, 434–442. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Rodriguez Oyarzun, L.; Cornejo-Sánchez, T.; Martín-Gómez, M.T.; Gartner, S.; de Gracia, J.; Rovira, S.; Alvarez, A.; Jofre, J.; González-López, J.J.; et al. Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients. Front. Microbiol. 2018, 9, 856. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Imamovic, L.; Jofre, J.; Muniesa, M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 2011, 55, 4908–4911. [Google Scholar] [CrossRef] [PubMed]
- Larrañaga, O.; Brown-Jaque, M.; Quirós, P.; Gómez-Gómez, C.; Blanch, A.R.; Rodríguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 13281. [Google Scholar] [CrossRef] [PubMed]
- Poulos, B.T.; John, S.G.; Sullivan, M.B. Iron chloride flocculation of bacteriophages from seawater. Methods Mol. Biol. 2018, 1681, 49–57. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; p. 999. [Google Scholar]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages Methods and Protocols Volume 2: Molecular and Applied Aspects; Springer: Berlin, Germany, 2009; Volume 502, ISBN 978-1-60327-564-4. [Google Scholar]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef]
- Pereira, A.M.P.T.; Silva, L.J.G.; Meisel, L.M.; Pena, A. Fluoroquinolones and Tetracycline Antibiotics in a Portuguese Aquaculture System and Aquatic Surroundings: Occurrence and Environmental Impact. J. Toxicol. Environ. Health Part A Curr. Issues 2015, 78, 959–975. [Google Scholar] [CrossRef]
- Galimand, M.; Sabtcheva, S.; Lambert, T. Worldwide Disseminated armA Aminoglycoside Resistance Methylase Gene Is Borne by Composite Transposon Tn1548. Am. Soc. Microbiol. 2005, 49, 2949–2953. [Google Scholar] [CrossRef]
- ISO. Water Quality—Detection and Enumeration of Intestinal Enterococci—Part 2: Membrane Filtration Method; ISO 7899-2:2000; ISO: London, UK, 2000. [Google Scholar]
- Jofre, J. Indicators of Waterborne Enteric Viruses. In Perspectives in Medical Virology, Volume 17; Elsevier: Amsterdam, The Netherlands, 2007; pp. 227–249. ISBN 9780444521576. [Google Scholar]
- ISO. Water Quality. Detection and Enumeration of Bacteriophages—Part 2: Enumeration of Somatic Coliphages; ISO 10705-2; ISO: London, UK, 2000. [Google Scholar]
- Imamovic, L.; Misiakou, M.-A.; van der Helm, E.; Panagiotou, G.; Muniesa, M.; Sommer, M.O.A. Complete Genome Sequence of Escherichia coli Strain WG5. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Jofre, J.; Lucena, F.; Blanch, A.; Muniesa, M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water 2016, 8, 199. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.; Stedt, J.; Bonnedahl, J.; Molin, Y.; Drobni, M.; Calisto-Ulloa, N.; Gomez-Fuentes, C.; Astorga-España, M.S.; González-Acuña, D.; Waldenström, J.; et al. Human-associated extended-spectrum β-lactamase in the Antarctic. Appl. Environ. Microbiol. 2012, 78, 2056–2058. [Google Scholar] [CrossRef] [PubMed]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; González-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Wang, C.; Gao, H.; Li, R.; Jin, S.; Zhang, W.; Zong, H. The occurrence of sulfonamide and quinolone resistance genes at the Fildes Peninsula in Antarctica. Mar. Pollut. Bull. 2019, 149. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Tin, T.; Fleming, Z.L.; Hughes, K.A.; Ainley, D.G.; Convey, P.; Moreno, C.A.; Pfeiffer, S.; Scott, J.; Snape, I. Impacts of local human activities on the Antarctic environment. Antarct. Sci. 2009, 21, 3–33. [Google Scholar] [CrossRef]
- Brum, J.R.; Hurwitz, B.L.; Schofield, O.; Ducklow, H.W.; Sullivan, M.B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016, 10, 437–449. [Google Scholar] [CrossRef]
- Lindell, D.; Sullivan, M.B.; Johnson, Z.I.; Tolonen, A.C.; Rohwer, F.; Chisholm, S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 11013–11018. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcázar, J.L. Bacteriophages as a reservoir of extended-spectrum β-lactamase and fluoroquinolone resistance genes in the environment. Clin. Microbiol. Infect. 2014, 20, O456–O459. [Google Scholar] [CrossRef]
- Tamminen, M.; Karkman, A.; Lõhmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 2011, 45, 386–391. [Google Scholar] [CrossRef]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- Anonymous Commission Regulations (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, L338/1. Available online: https://eur-lex.europa.eu/eli/reg/2005/2073/oj (accessed on 23 August 2020).
- Anonymous Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 Concerning the Management of Bathing Water Quality. Off. J. Eur. Union 2006, L64/37. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006L0007&rid=1 (accessed on 23 August 2020).
- Lachmayr, K.L.; Kerkhof, L.J.; Dirienzo, A.G.; Cavanaugh, C.M.; Ford, T.E. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, H.; Schwartz, T.; Bischoff, P.; Kirchen, S.; Obst, U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 2004, 56, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J. Antimicrob. Chemother. 2014, 69, 1265–1274. [Google Scholar] [CrossRef]
- Quirós, P.; Colomer-Lluch, M.; Martínez-Castillo, A.; Miró, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014, 58, 606–609. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
| Microorganism | Mediterranean Seawater * | Fish | |||||
|---|---|---|---|---|---|---|---|
| Shellfish | Mediterranean | Atlantic | Farm | Frozen | |||
| n | 20 | 10 | 10 | 10 | 10 | 10 | |
| Total aerobic bacteria AmpR | % positive samples | 100 | 90 | 80 | 70 | 100 | 40 |
| Average | 2.4 × 103 | 4.6 × 107 | 5.1 × 104 | 5.7 × 106 | 3.8 × 106 | 1.0 × 108 | |
| SD | 5.6 × 103 | 4.9 × 107 | 5.9 × 104 | 9.4 × 106 | 8.3 × 106 | 1.2 × 108 | |
| E. coli | % positive samples | 85 | 0 | 10 | 10 | 0 | 0 |
| Average | 6.9 × 102 | 0 | 3.7 × 101 | 7.5 × 101 | 0 | 0 | |
| SD | 2.1 × 103 | 0 | − | − | 0 | 0 | |
| E. coli AmpR | % positive samples | 60 | 0 | 0 | 0 | 0 | 0 |
| Average | 6.8 × 102 | 0 | 0 | 0 | 0 | 0 | |
| SD | 1.9 × 103 | 0 | 0 | 0 | 0 | 0 | |
| Enterococcus spp. | % positive samples | 90 | 30 | 10 | 40 | 20 | 10 |
| Average | 1.0 × 102 | 1.4 × 102 | 7.5 × 101 | 7.3 × 102 | 1.9 × 102 | 1.1 × 102 | |
| SD | 1.5 × 102 | 1.2 × 102 | − | 1.2 × 103 | 5.3 × 101 | − | |
| Somatic coliphages | % positive samples | 40 | 70 | 0 | 30 | 10 | 20 |
| Average | 1.0 × 103 | 1.1 × 103 | 0 | 9.2 × 101 | 7.5 × 101 | 6.6 × 102 | |
| SD | 1.3 × 103 | 9.9 × 102 | 0 | 5.2 × 101 | - | 8.2 × 102 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms 2020, 8, 1293. https://doi.org/10.3390/microorganisms8091293
Blanco-Picazo P, Roscales G, Toribio-Avedillo D, Gómez-Gómez C, Avila C, Ballesté E, Muniesa M, Rodríguez-Rubio L. Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms. 2020; 8(9):1293. https://doi.org/10.3390/microorganisms8091293
Chicago/Turabian StyleBlanco-Picazo, Pedro, Gabriel Roscales, Daniel Toribio-Avedillo, Clara Gómez-Gómez, Conxita Avila, Elisenda Ballesté, Maite Muniesa, and Lorena Rodríguez-Rubio. 2020. "Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems" Microorganisms 8, no. 9: 1293. https://doi.org/10.3390/microorganisms8091293
APA StyleBlanco-Picazo, P., Roscales, G., Toribio-Avedillo, D., Gómez-Gómez, C., Avila, C., Ballesté, E., Muniesa, M., & Rodríguez-Rubio, L. (2020). Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms, 8(9), 1293. https://doi.org/10.3390/microorganisms8091293






