Four New Genes of Cyanobacterium Synechococcus elongatus PCC 7942 Are Responsible for Sensitivity to 2-Nonanone
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Transposon Mutagenesis of Synechococcus
2.3. Selection of Synechococcus Mutants Resistant to 2-Nonanone Action
2.4. Identification of Transposon Insertion Sites
2.5. Construction of Knockout-Mutants by Site-Directed Mutagenesis
2.6. Light Microscopy
2.7. Data Analysis
3. Results
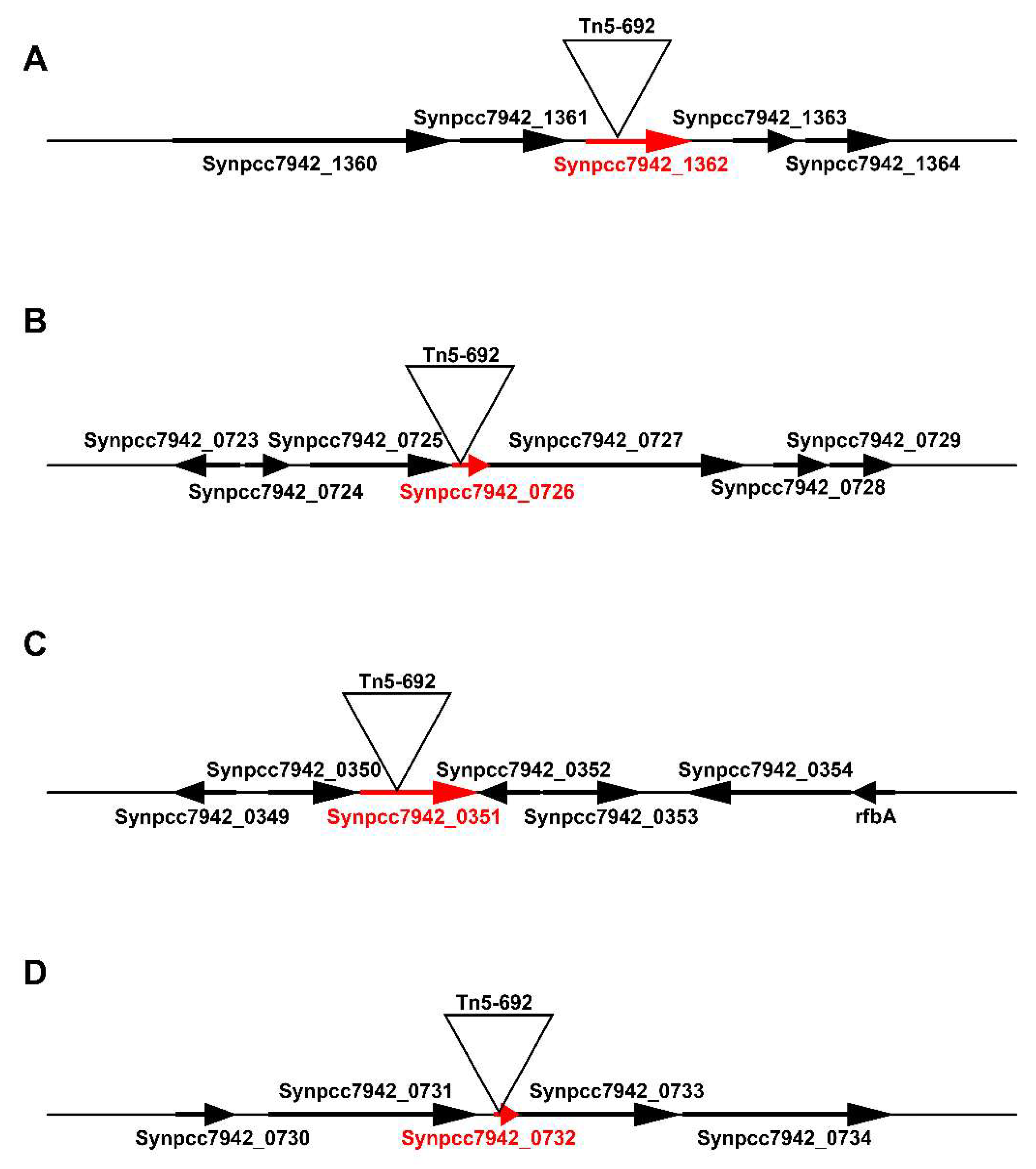
3.1. Localization of Tn5-692 Transposon Insertions in the Resistant Mutants of Synechococcus
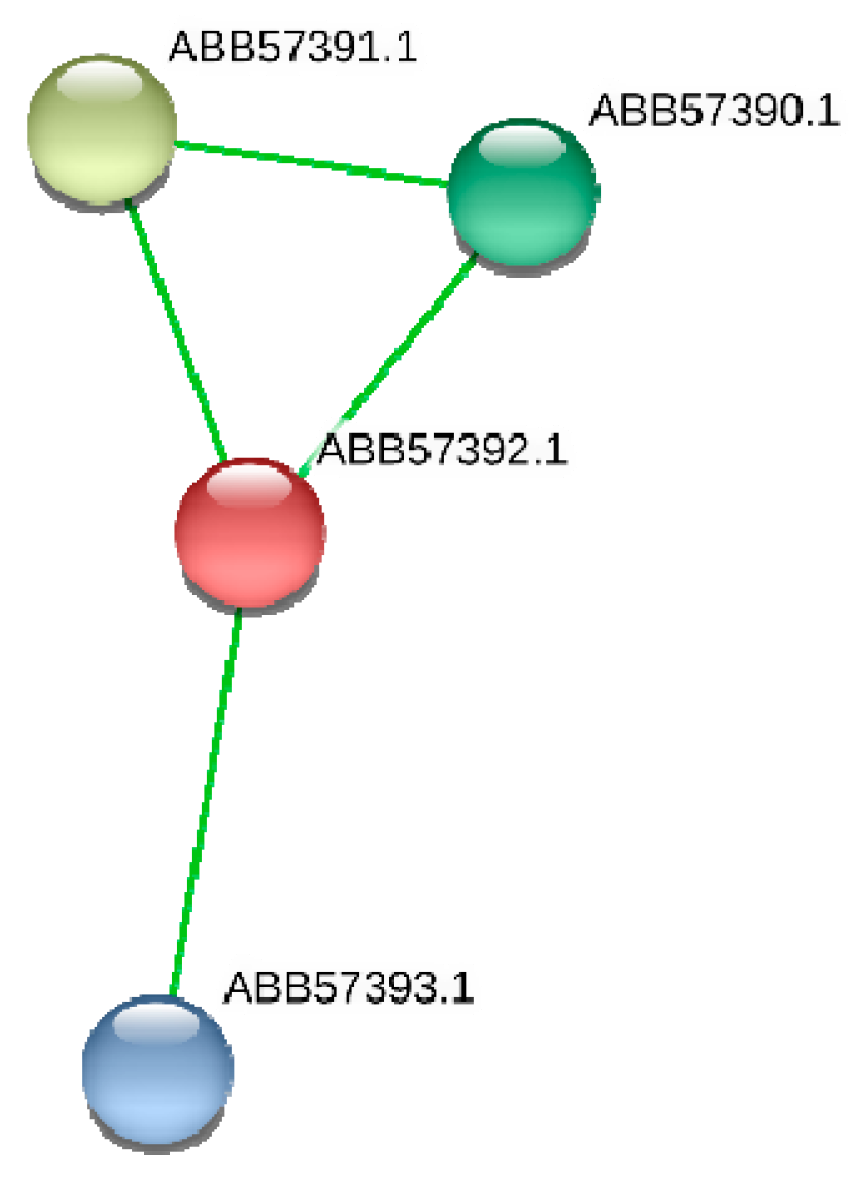
3.1.1. The Synpcc7942_1362 Gene Was Inactivated by Tn5-692 in the Mutant NR401(Tn)
3.1.2. In the Mutant NR365(Tn) the Transposon Was Inserted in the Synpcc7942_0351 Gene
3.1.3. In the Mutant NR359(Tn) the Tn5-692 Transposon Was Inserted in the Synpcc7942_0732 Gene
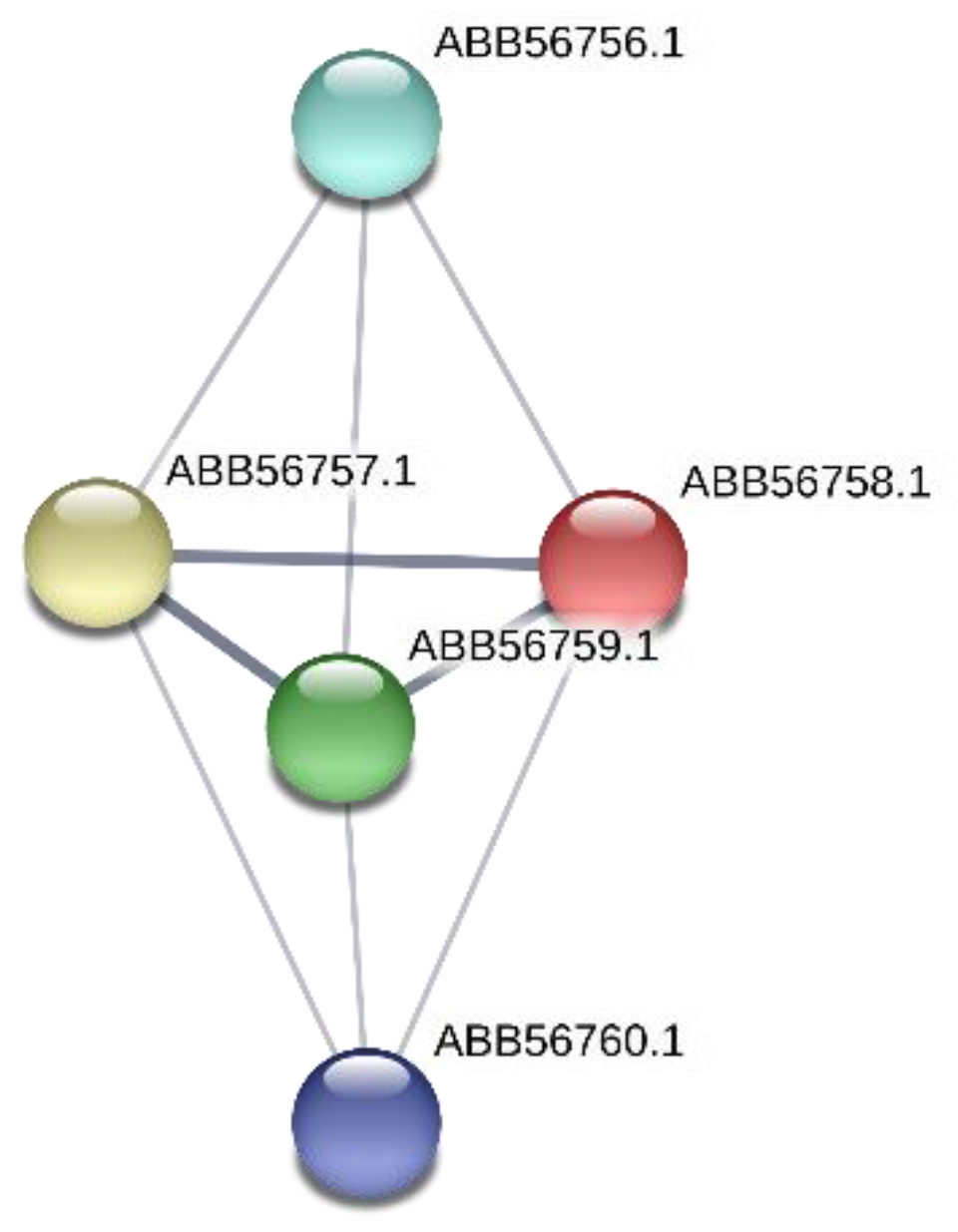
3.1.4. The Gene Synpcc7942_0726 Was Inactivated by Tn5-692 in the Mutant NR385(Tn)
3.2. Directional Inactivation of Synpcc7942_1362 and Synpcc7942_0726 Genes
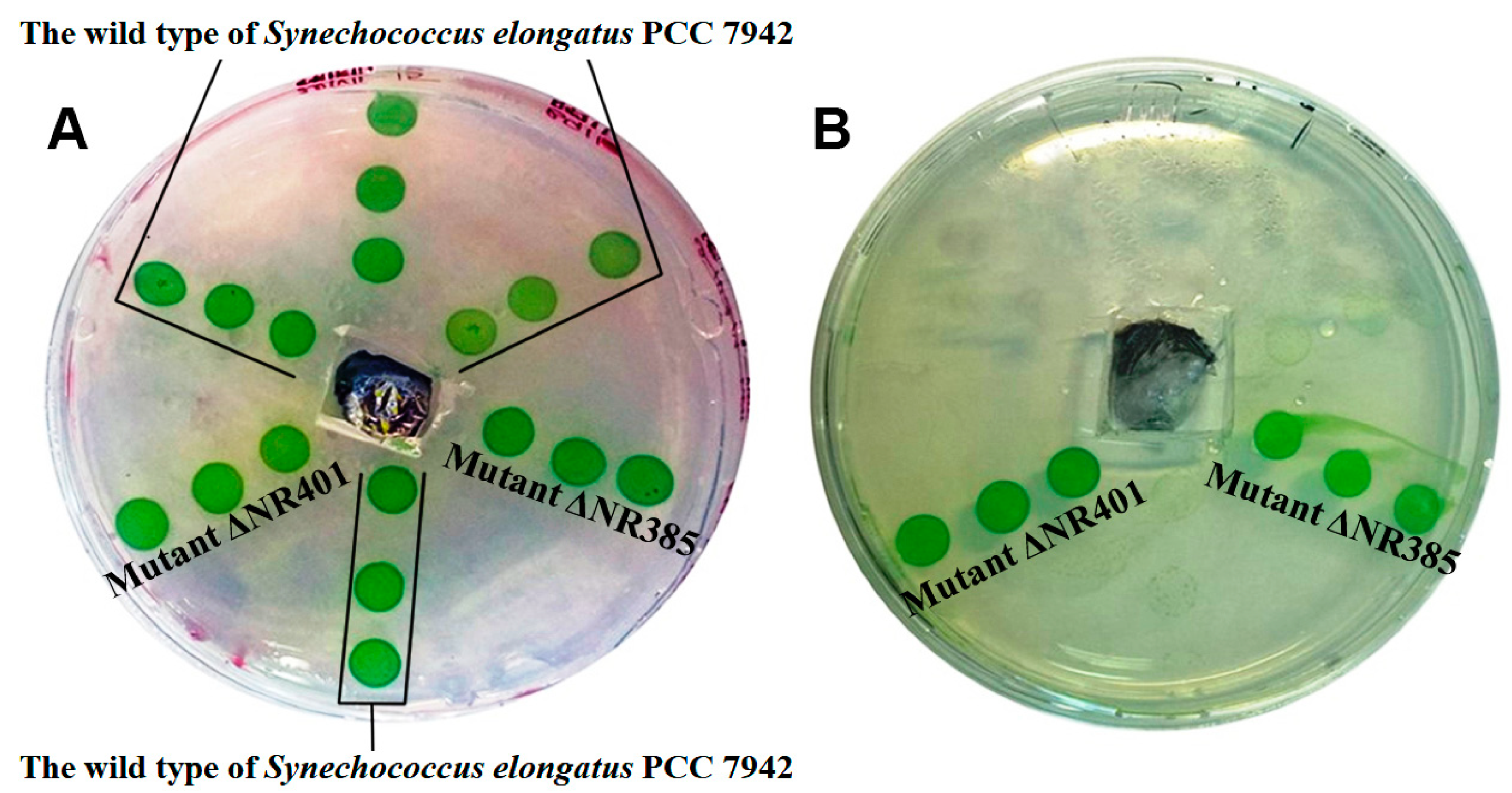
3.3. Morphological Characterization of ΔNR401 and ΔNR385 Mutants
4. Discussion
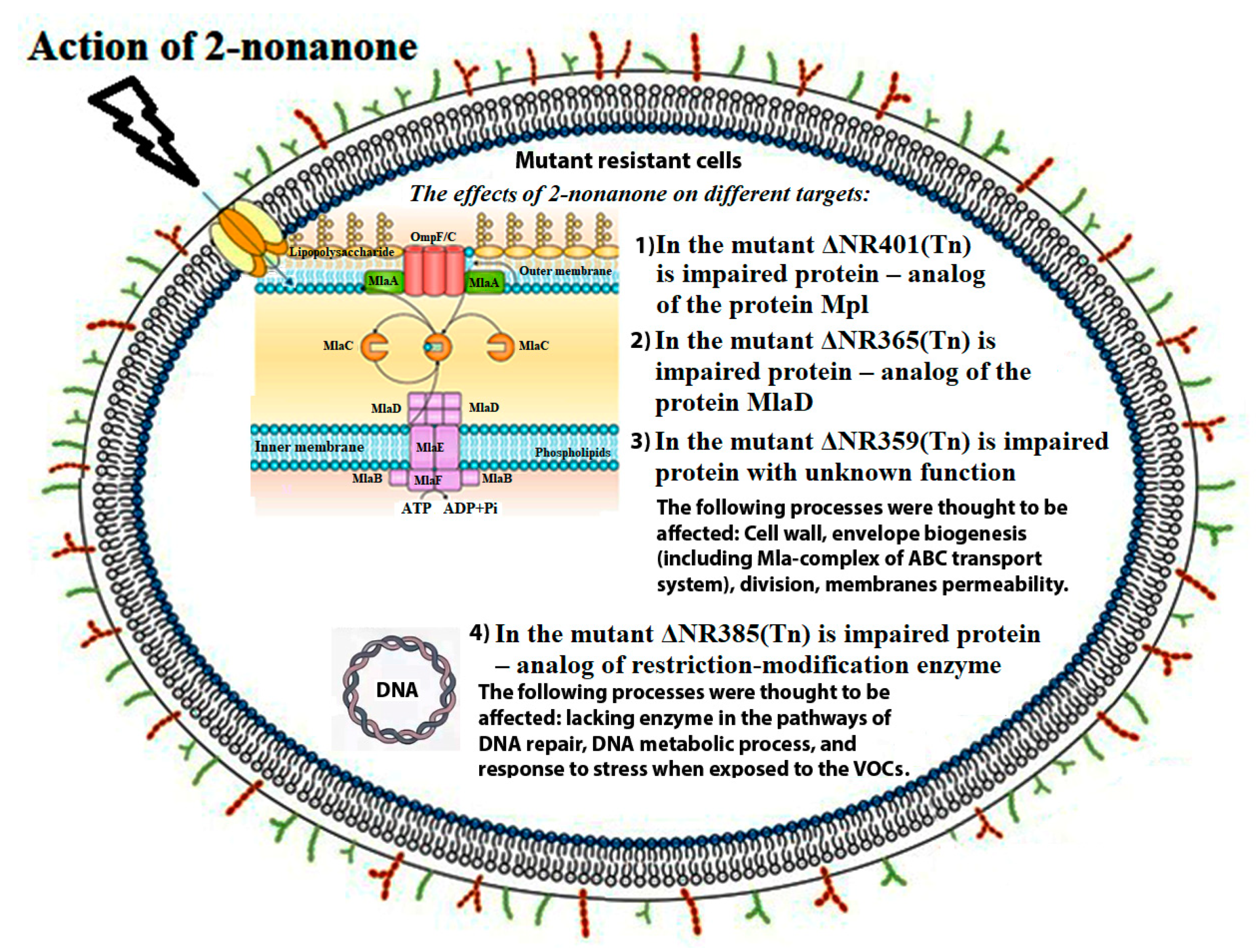
4.1. Three Genes (Synpcc7942_1362, Synpcc7942_0351, Synpcc7942_0732) That Encode Proteins Involved in the Formation, Maintenance, and Functionality of Cyanobacterial Cell Wall
4.2. The Gene Synpcc7942_0726 Encodes Protein Involved in DNA Metabolism
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Piechulla, B.; Degenhardt, J. The emerging importance of microbial volatile organic compounds. Plant Cell Environ. 2014, 37, 811–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemfack, M.C.; Gohlke, B.O.; Toguem, S.M.T.; Preissner, S.; Piechulla, B.; Preissner, R. mVOC 2.0: A database of microbial volatiles. Nucl. Acids Res. 2018, 46, D1261–D1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veselova, M.A.; Plyuta, V.A.; Khmel, I.A. Volatile compounds of bacteria: Structure, biosynthesis, biological activity. Microbiology 2019, 88, 261–274. [Google Scholar] [CrossRef]
- Koksharova, O.A. Cyanobacterial VOCs as allelopathic tools. In Bacterial Volatile Compounds as Mediators of Airborne Interactions; Ryu, C., Weisskopf, L., Piechulla, B., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2020. [Google Scholar] [CrossRef]
- Kai, M.; Haustein, M.; Molina, F.; Petri, A.; Scholz, B.; Piechulla, B. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 2009, 81, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Effmert, U.; Kalderas, J.; Warnke, R.; Piechulla, B. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 2012, 38, 665–703. [Google Scholar] [CrossRef]
- Popova, A.A.; Koksharova, O.A.; Lipasova, V.A.; Zaitseva, J.V.; Katkova-Zhukotskaya, O.A.; Eremina, S.I.; Mironov, A.S.; Chernin, L.S.; Khmel, I.A. Inhibitory and toxic Effects of Volatiles emitted by Strains of Pseudomonas and Serratia on Growth and Survival of selected Microorganisms, Caenorhabditis elegans and Drosophila melanogaster. BioMed. Res. Int. 2014, 2014, 125704. [Google Scholar] [CrossRef] [Green Version]
- Schulz-Bohm, K.; Gerards, S.; Hundscheid, M.; Melenhorst, J.; de Boer, W.; Garbeva, P. Calling from distance: Attraction of soil bacteria by plant root volatiles. ISMEJ 2018, 12, 1252–1262. [Google Scholar] [CrossRef] [Green Version]
- Giorgio, A.; De Stradis, A.; Lo Cantore, P.; Iacobellis, N.S. Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 1056. [Google Scholar] [CrossRef] [Green Version]
- Means, G.E.; Feeney, R.E. Chemical Modification of Proteins; Holden-Day, Inc.: San Francisco, CA, USA, 1971; pp. 214–217. [Google Scholar]
- Arulmurugan, S.; Kavitha, H.P.; Venkatraman, B.R. Biological activities of schiff base and its complexes: A review. RASAYAN J. Chem. 2010, 3, 385–410. [Google Scholar]
- Damodaran, S.; Kinsella, J.E. Stabilization of Proteins by Solvents. Effect of pH and anions on the positive cooperativity of 2-nonanone binding to bovine serum albumin. JBC 1980, 255, 8503–8508. [Google Scholar]
- Melkina, O.E.; Khmel, I.A.; Plyuta, V.A.; Koksharova, O.A.; Zavilgelsky, G.B. Ketones 2-heptanone, 2-nonanone, and 2-undecanone inhibit DnaK-dependent refolding of heat inactivated bacterial luciferases in Escherichia coli cells lacking small chaperon IbpB. Appl. Microbiol. Biotechnol. 2017, 101, 5765–5771. [Google Scholar] [CrossRef] [PubMed]
- Plyuta, V.A.; Popova, A.A.; Koksharova, O.A.; Kuznetsov, A.E.; Khmel, I.A. The ability of natural ketones to interact with bacterial quorum sensing systems. Mol. Genet. Microbiol. Virol. 2014, 4, 167–171. [Google Scholar] [CrossRef]
- Plyuta, V.; Lipasova, V.; Popova, A.; Koksharova, O.; Kuznetsov, A.; Szegedi, E.; Chernin, L.; Khmel, I. Influence of volatile organic compounds emitted by Pseudomonas and Serratia strains on Agrobacterium tumefaciens biofilms. APMIS 2016, 124, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Voronova, E.N.; Konyukhov, I.V.; Koksharova, O.A.; Popova, A.A.; Pogosyan, S.I.; Khmel, I.A.; Rubin, A.B. Inhibition of cyanobacterial photosynthetic activity by natural ketones. J. Phycol. 2019, 55, 840–857. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1974; pp. 14–66. [Google Scholar]
- Koksharova, O.A.; Wolk, C.P. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 2002, 184, 5524–5528. [Google Scholar] [CrossRef] [Green Version]
- Elhai, J.; Vepritskiy, A.; Muro-Pastor, A.M.; Flores, E.; Wolk, C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997, 179, 1998–2005. [Google Scholar] [CrossRef] [Green Version]
- Elhai, J.; Wolk, C.P. A versatile class of positive selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 1988, 68, 119–138. [Google Scholar] [CrossRef]
- Karaushu, E.V.; Lazebnaya, I.V.; Kravzova, T.R.; Vorobey, N.A.; Lazebny, O.E.; Kiriziy, D.A.; Olkhovich, O.P.; Taran, N.Y.; Kots, S.Y.; Popova, A.A.; et al. Biochemical and Molecular Phylogenetic Study of Agriculturally Useful Association of a Nitrogen-Fixing Cyanobacterium and Nodule Sinorhizobium with Medicago sativa L. BioMed. Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Koksharova, O.; Schubert, M.; Shestakov, S.; Cerff, R. Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 1998, 36, 183–194. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; pp. 16–26. [Google Scholar]
- Mengin-Lecreulx, D.; van Heijenoort, J.; Park, J.T. Identification of the rnpl gene encoding UDP-N-acetylmuramate: L-alanyl-yD-glutamyl-meso-diamilnopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 1996, 178, 5347–5352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bramhill, D. Bacterial Cell Division. Annu. Rev. Cell Dev. Biol. 1997, 13, 395–424. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.N. Peptidoglycan Synthesis and Cell Division. In Microbial Biochemistry, 2nd ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 17–22. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Babykin, M.M. Cyanobacterial cell division: Genetics and comparative genomics of cyanobacterial cell division. Russ. J. Genet. 2011, 47, 255–261. [Google Scholar] [CrossRef]
- Bisson-Filho, A.W.; Hsu, Y.P.; Squyres, G.R.; Kuru, E.; Wu, F.; Jukes, C.; Sun, Y.; Dekker, C.; Holden, S.; VanNieuwenhze, M.S.; et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 2017, 355, 739–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorelova, O.A.; Baulina, O.I.; Rasmussen, U.; Koksharova, O.A. The pleiotropic effects of ftn2 and ftn6 mutations in cyanobacterium Synechococcus sp. PCC 7942: An ultrastructural study. Protoplasma 2013, 250, 931–942. [Google Scholar] [CrossRef]
- Malinverni, J.C.; Silhavy, T.J. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 2009, 106, 8009–8014. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.W.; Hall, S.C.L.; Laxton, C.S.; Sridhar, P.; Mahadi, A.H.; Hatton, C.; Knowles, T.J. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 2019, 4, 1692–1705. [Google Scholar] [CrossRef]
- Avrani, S.; Wurtzel, O.; Sharon, I.; Sorek, R.; Lindell, D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 2011, 474, 604–608. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Riemann, L.; Marston, M.F.; Middelboe, M. Antagonistic coevolution of marine planktonic viruses and their hosts. Annu. Rev. Mar. Sci. 2014, 6, 393–414. [Google Scholar] [CrossRef]
- Silva, B.J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [Green Version]
- Zborowsky, S.; Lindell, D. Resistance in marine cyanobacteria differs against specialist and generalist cyanophages. Proc. Natl. Acad. Sci. USA 2019, 116, 16899–16908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, N.; Nishioka, T.; Takabayashi, J.; Furuichi, T. Plant volatiles regulate the activities of Ca2+-permeable channels and promote cytoplasmic calcium transients in Arabidopsis leaf cells. Plant Signal. Behav. 2009, 4, 294–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trombetta, D.; Castelli, F.; Sarpietro, M.G.; Venuti, V.; Cristani, M.; Daniele, C.; Saija, A.; Mazzanti, G.; Bisignano, G. Mechanisms of Antibacterial Action of Three Monoterpenes. Antimicrob. Agents Chemother. 2005, 49, 2474–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tromelin, A.; Andriot, I.; Guichard, E. Protein-flavour interactions. In Flavour in Food; Voilley, A., Etiévant, P.X., Eds.; Woodhead Publishing Limited: Cambridge, UK, 2006; pp. 172–207. [Google Scholar] [CrossRef]
- Kinch, L.N.; Ginalski, K.; Rychlewski, L.; Grishin, N.V. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005, 33, 3598–3605. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Babu, M.M.; Aravind, L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle 2006, 5, 775–782. [Google Scholar] [CrossRef] [Green Version]
- MacKay, C.; Déclais, A.C.; Lundin, C.; Agostinho, A.; Deans, A.J.; MacArtney, T.J.; Hofmann, K.; Gartner, A.; West, S.C.; Helleday, T.; et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 2010, 142, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Sharples, G.J.; Curtis, F.A.; McGlynn, P.; Bolt, E.L. Holliday junction binding and resolution by the Rap structure-specific endonuclease of phage lambda. J. Mol. Biol. 2004, 340, 739–751. [Google Scholar] [CrossRef]
- Alpha, C.J.; Campos, M.; Jacobs-Wagner, C.; Strobel, S. Mycofumigation by the Volatile Organic Compound-Producing Fungus Muscodor albus Induces Bacterial Cell Death through DNA Damage. Appl. Environ. Microbiol. 2015, 81, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.L.; Alpha-Cobb, C.J.; Hiller, D.A.; Berro, J.; Strobel, S.A. Mycofumigation Through Production of the Volatile DNA Methylating Agent N-methyl-N-nitrosoisobutyramide by Fungi in the Genus Muscodor. J. Biol. Chem. 2017, 292, 7358–7371. [Google Scholar] [CrossRef] [Green Version]
- Yung, P.Y.; Grasso, L.L.; Mohidin, A.F.; Acerbi, E.; Hinks, J.; Seviour, T.; Marsili, E.; Lauro, F.M. Global transcriptomic responses of Escherichia coli K-12 to volatile organic compounds. Sci. Rep. 2016, 6, 19899. [Google Scholar] [CrossRef]
- Bishop, R.E. Phospholipid middle management. Nat. Microbiol. 2019, 4, 1608–1609. [Google Scholar] [CrossRef] [PubMed]
- Pei, G.; Chen, L.; Wang, J.; Qiao, J.; Zhang, W. Protein Network Signatures Associated with Exogenous Biofuels Treatments in Cyanobacterium Synechocystis sp. PCC 6803. Front. Bioeng. Biotechnol. 2014, 2, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]



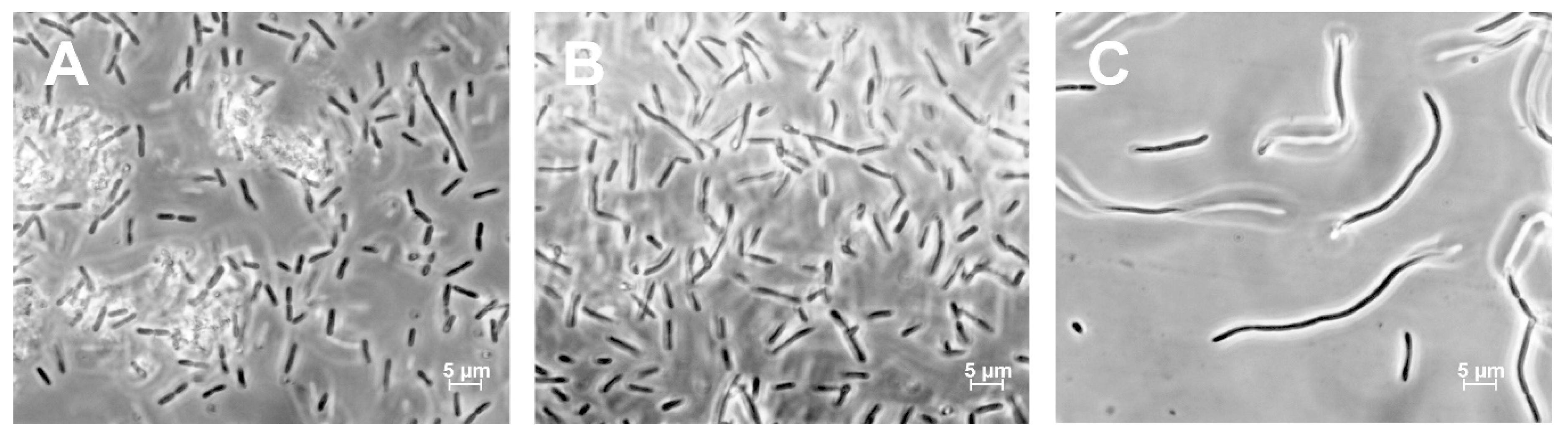
| Strain | Characterization and/or Derivation | Reference or Origin |
|---|---|---|
| Synechococcus elongatus | ||
| PCC 7942 | The wild type | Laboratory collection |
| NR401(Tn) | Sm R Sp R Em R; Tn5-692 mutant | This work |
| NR385(Tn) | Sm R Sp REm R; Tn5-692 mutant | This work |
| NR365(Tn) | Sm R Sp R Em R; Tn5-692 mutant | This work |
| NR359(Tn) | Sm R Sp R Em R; Tn5-692 mutant | This work |
| ΔNR401 | Km R; PCC 7942::pΔNR401, transconjugant | This work |
| ΔNR385 | Km R; PCC 7942::pΔNR385, transconjugant | This work |
| Escherichia coli | ||
| XL1-Blue | Host for routine cloning | Collection of the Genetics Department, Moscow State University |
| HB101 692 | HB101 harboring pRL692, Sm R Sp R Em R | Laboratory collection |
| HB101 443 | HB101 harboring pRL443, Ap R Tc R | Laboratory collection |
| Plasmids | ||
| pRL692 | Carrying the mobile element Tn5-692, Sm R Sp R Em R | [19] |
| pRL443 | Conjugal plasmid, derivative of RP4, Ap R Tc R; Km S | [20] |
| pRL498 | Km R; plasmid vector for the direct selection | [21] |
| pJet1.2/blunt | pMB1-ori bla eco47 IR::MCS; Ap R | Thermo Fisher Scientific |
| pJet1.2-385 | Ap R; pJet1.2::ΔSynpcc7942_0726 | This work |
| pΔNR401 | Km R; pRL498::ΔSynpcc7942_1362 | This work |
| pΔNR385 | Km R; pRL498::ΔSynpcc7942_0726 | This work |
| Primer Name | Sequence |
|---|---|
| pW-TnR:946 U21 | 5′-CTGCTGGCCATTGAGGACACC-3′ |
| pW-TnR:923 L21 | 5′-CGGGAAACTCCTGAGCCAACT-3′ |
| TN692 R-155 | 5′-GGCGTTGACATCACTCTG-3′ |
| TN692 F-5485 | 5′-GTCTAGCTATCGCCATGTAAGC-3′ |
| NR401-F | 5′-CCGAATTCGATGCTGTTAGAGG-3′ |
| NR401-R | 5′-CCGAATTCGCTTCCAGCTCGAG-3′ |
| NR385-F | 5′-CCGAATTCCTCTGGAAGACG-3′ |
| NR385-R | 5′-CCGAATTCGCGTCTTGCATC-3′ |
| NR365-F | 5′-CCGAATTCGAGAAGGCAGTG-3′ |
| NR365-R | 5′-CCGAATTCGAGATCCGTGAC-3′ |
| NR359-F | 5′-CCGAATTCGAAGACTTGCAAGC-3′ |
| NR359-R | 5′-CCGAATTCGACGGTACTGGATG-3′ |
| Mutant | Gene | Protein | Protein Function | Pathway |
|---|---|---|---|---|
| ΔNR401(Tn) | Synpcc7942_1362 | conserved hypothetical protein (ABB57392.1) | murein-peptide ligase (Mpl, UDP-N-acetylmuramate: L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase) | Biogenesis of cell wall |
| ΔNR365(Tn) | Synpcc7942_0351 | putative ABC transport system substrate-binding proteins MlaD (ABB56383.1) | Protein is similar to ABC transporters providing the resistance to organic solvents | Phospholipid transport pathway that maintains lipid asymmetry in the outer membrane |
| ΔNR359(Tn) | Synpcc7942_0732 | a small conserved hypothetical protein (ABB56764.1) of 69 amino acids with an unknown function | Gene belonging to phage gene cluster in the Synechococcus genome and this protein is involved in interactions with phage proteins | Hypothetically cell membrane modification |
| ΔNR385(Tn) | Synpcc7942_0726 | hypothetical protein (ABB56758.1) containing the VRR-NUC domain | Protein contains a Viral replication and repair (VRR) nuclease (VRR nuc) domain. This functional unit is present in the PD-(D/E)XK nuclease superfamily and repair enzymes | DNA metabolism |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koksharova, O.A.; Popova, A.A.; Plyuta, V.A.; Khmel, I.A. Four New Genes of Cyanobacterium Synechococcus elongatus PCC 7942 Are Responsible for Sensitivity to 2-Nonanone. Microorganisms 2020, 8, 1234. https://doi.org/10.3390/microorganisms8081234
Koksharova OA, Popova AA, Plyuta VA, Khmel IA. Four New Genes of Cyanobacterium Synechococcus elongatus PCC 7942 Are Responsible for Sensitivity to 2-Nonanone. Microorganisms. 2020; 8(8):1234. https://doi.org/10.3390/microorganisms8081234
Chicago/Turabian StyleKoksharova, Olga A., Alexandra A. Popova, Vladimir A. Plyuta, and Inessa A. Khmel. 2020. "Four New Genes of Cyanobacterium Synechococcus elongatus PCC 7942 Are Responsible for Sensitivity to 2-Nonanone" Microorganisms 8, no. 8: 1234. https://doi.org/10.3390/microorganisms8081234
APA StyleKoksharova, O. A., Popova, A. A., Plyuta, V. A., & Khmel, I. A. (2020). Four New Genes of Cyanobacterium Synechococcus elongatus PCC 7942 Are Responsible for Sensitivity to 2-Nonanone. Microorganisms, 8(8), 1234. https://doi.org/10.3390/microorganisms8081234






