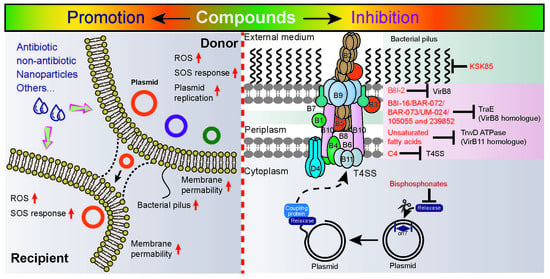
Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Horizontal Transfer of Antibiotic Resistance Genes
3. Exogenous Compounds Promote Horizontal Genes Transfer
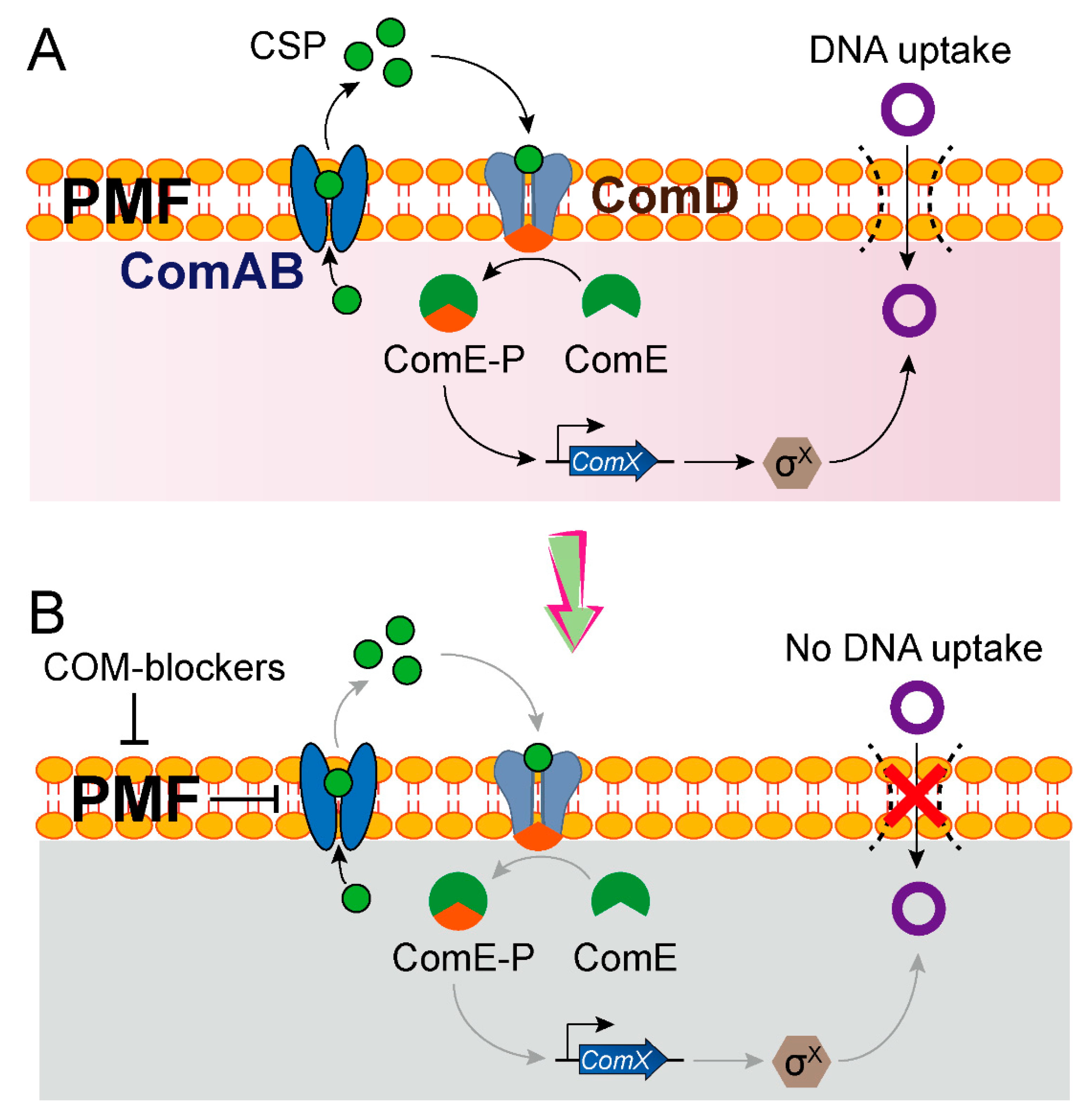
4. Exogenous Compounds Inhibit Plasmid Transfer
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupferschmidt, K. Resistance fighter. Science 2016, 352, 758–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review on Antimicrobial Resistance: London, UK, 2016. [Google Scholar]
- Harrison, E.; Brockhurst, M.A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012, 20, 262–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grohmann, E.; Muth, G.; Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 277–301. [Google Scholar] [CrossRef] [Green Version]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm animals and aquaculture: Significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Gogarten, J.P.; Doolittle, W.F.; Lawrence, J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002, 19, 2226–2238. [Google Scholar] [CrossRef] [Green Version]
- Huang, J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays 2013, 35, 868–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, R.A.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef]
- Kohler, V.; Keller, W.; Grohmann, E. Regulation of Gram-positive conjugation. Front. Microbiol. 2019, 10, 1134. [Google Scholar] [CrossRef] [Green Version]
- Guglielmini, J.; de la Cruz, F.; Rocha, E.P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013, 30, 315–331. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 2009, 62, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [Green Version]
- Tamminen, M.; Virta, M.; Fani, R.; Fondi, M. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol. Biol. Evol. 2012, 29, 1225–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.P.; Mullany, P. A modular master on the move: The Tn916 family of mobile genetic elements. Trends Microbiol. 2009, 17, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Harms, K.; Fricke, W.F.; Johnsen, P.J.; da Silva, G.J.; Nielsen, K.M. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012, 8, e1002837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claverys, J.P.; Martin, B.; Polard, P. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol. Rev. 2009, 33, 643–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Sturød, K.; Salvadori, G.; Junges, R.; Petersen, F.C. Antibiotics alter the window of competence for natural transformation in streptococci. Mol. Oral Microbiol. 2018, 33, 378–387. [Google Scholar] [CrossRef]
- Ferrándiz, M.J.; Fenoll, A.; Liñares, J.; De La Campa, A.G. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 840–847. [Google Scholar] [CrossRef] [Green Version]
- Bowler, L.D.; Zhang, Q.Y.; Riou, J.Y.; Spratt, B.G. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: Natural events and laboratory simulation. J. Bacteriol. 1994, 176, 333–337. [Google Scholar] [CrossRef] [Green Version]
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013, 499, 219–222. [Google Scholar] [CrossRef] [Green Version]
- Rolain, J.M.; Fancello, L.; Desnues, C.; Raoult, D. Bacteriophages as vehicles of the resistome in cystic fibrosis. J. Antimicrob. Chemother. 2011, 66, 2444–2447. [Google Scholar] [CrossRef] [Green Version]
- Colomer-Lluch, M.; Calero-Cáceres, W.; Jebri, S.; Hmaied, F.; Muniesa, M.; Jofre, J. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 2014, 73, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Quirós, P.; Colomer-Lluch, M.; Martínez-Castillo, A.; Miró, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014, 58, 606–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soler, N.; Forterre, P. Vesiduction: The fourth way of HGT. Environ. Microbiol. 2020, 22, 2457–2460. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.W.; Vesk, M.; Work, E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 1966, 92, 1206–1217. [Google Scholar] [CrossRef] [Green Version]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Erdmann, S.; Tschitschko, B.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat. Microbiol. 2017, 2, 1446–1455. [Google Scholar] [CrossRef]
- Biller, S.J.; Schubotz, F.; Roggensack, S.E.; Thompson, A.W.; Summons, R.E.; Chisholm, S.W. Bacterial vesicles in marine ecosystems. Science 2014, 343, 183–186. [Google Scholar] [CrossRef]
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019, 43, 273–303. [Google Scholar] [CrossRef] [PubMed]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudin, M.; Gauliard, E.; Schouten, S.; Houel-Renault, L.; Lenormand, P.; Marguet, E.; Forterre, P. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 2013, 5, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Haghi, F.; Lohrasbi, V.; Zeighami, H. High incidence of virulence determinants, aminoglycoside and vancomycin resistance in enterococci isolated from hospitalized patients in Northwest Iran. BMC Infect. Dis. 2019, 19, 744. [Google Scholar] [CrossRef] [Green Version]
- Kneis, D.; Berendonk, T.U.; Heß, S. High prevalence of colistin resistance genes in German municipal wastewater. Sci. Total Environ. 2019, 694, 133454. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Zhao, R.; Feng, J.; Liu, J.; Fu, W.; Li, X.; Li, B. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar]
- Møller, T.S.B.; Liu, G.; Boysen, A.; Thomsen, L.E.; Lüthje, F.L.; Mortensen, S.; Møller-Jensen, J.; Olsen, J.E. Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front. Microbiol. 2017, 8, 2365. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zeng, J.; Wang, L.; Lan, K.; Shunmei, E.; Wang, L.; Xiao, Q.; Luo, Q.; Huang, X.; Huang, B.; et al. Antibiotics promote Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting quorum sensing. Antimicrob. Agents Chemother. 2017, 61, e01284-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gu, A.Z.; He, M.; Li, D.; Chen, J. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 2017, 51, 570–580. [Google Scholar] [CrossRef]
- Cen, T.; Zhang, X.; Xie, S.; Li, D. Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environ. Int. 2020, 138, 105544. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Mao, L.; Li, J.; Yuan, Z.; Bond, P.L.; Guo, J. Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 2019, 13, 509–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Wang, Y.; Li, J.; Mao, L.; Nguyen, S.H.; Duarte, T.; Coin, L.; Bond, P.; Yuan, Z.; Guo, J. Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int. 2018, 121, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Y.; Zhang, S.; Bond, P.; Yuan, Z.; Guo, J. Triclosan at environmental concentrations can enhance the spread of extracellular antibiotic resistance genes through transformation. Sci. Total Environ. 2020, 713, 136621. [Google Scholar] [CrossRef]
- Quesada-González, D.; Merkoçi, A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018, 47, 4697–4709. [Google Scholar] [CrossRef]
- Xie, J.; Gong, L.; Zhu, S.; Yong, Y.; Gu, Z.; Zhao, Y. Emerging strategies of nanomaterial-mediated tumor radiosensitization. Adv. Mater. 2019, 31, e1802244. [Google Scholar] [CrossRef]
- Zhu, M.; Nie, G.; Meng, H.; Xia, T.; Nel, A.; Zhao, Y. Physicochemical properties determine nanomaterial cellular uptake, transport, and fate. Acc. Chem. Res. 2013, 46, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Yu, Y.; Chen, Z.; Jin, M.; Yang, D.; Zhao, Z.; Wang, J.; Shen, Z.; Wang, X.; Qian, D.; et al. Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. USA 2012, 109, 4944–4949. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Mao, D.; Mu, Q.; Luo, Y. Enhanced horizontal transfer of antibiotic resistance genes in freshwater microcosms induced by an ionic liquid. PLoS ONE 2015, 10, e0126784. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, Q.; Mao, D.; Cui, Y.; Luo, Y. The horizontal transfer of antibiotic resistance genes is enhanced by ionic liquid with different structure of varying alkyl chain length. Front. Microbiol. 2015, 6, 864. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Wang, Q.; Lu, Q.; Mu, Q.; Mao, D. An ionic liquid facilitates the proliferation of antibiotic resistance genes mediated by class I integrons. Environ. Sci. Technol. Lett. 2014, 1, 266–270. [Google Scholar] [CrossRef]
- Liao, J.; Huang, H.; Chen, Y. CO2 promotes the conjugative transfer of multiresistance genes by facilitating cellular contact and plasmid transfer. Environ. Int. 2019, 129, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1689. [Google Scholar] [CrossRef] [PubMed]
- Buckner, M.M.C.; Ciusa, M.L.; Piddock, L.J.V. Strategies to combat antimicrobial resistance: Anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 2018, 42, 781–804. [Google Scholar] [CrossRef] [Green Version]
- Jalasvuori, M.; Friman, V.P.; Nieminen, A.; Bamford, J.K.; Buckling, A. Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol. Lett. 2011, 7, 902–905. [Google Scholar] [CrossRef] [Green Version]
- Cabezón, E.; De la Cruz, F.; Arechaga, I. Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 2017, 8, 2329. [Google Scholar] [CrossRef] [Green Version]
- Kamruzzaman, M.; Shoma, S.; Thomas, C.M.; Partridge, S.R.; Iredell, J.R. Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS ONE 2017, 12, e0172913. [Google Scholar] [CrossRef]
- Garcillán-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [Green Version]
- Lujan, S.A.; Guogas, L.M.; Ragonese, H.; Matson, S.W.; Redinbo, M.R. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc. Natl. Acad. Sci. USA 2007, 104, 12282–12287. [Google Scholar] [CrossRef] [Green Version]
- Redzej, A.; Ukleja, M.; Connery, S.; Trokter, M.; Felisberto-Rodrigues, C.; Cryar, A.; Thalassinos, K.; Hayward, R.D.; Orlova, E.V.; Waksman, G. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. 2017, 36, 3080–3095. [Google Scholar] [CrossRef] [Green Version]
- Paschos, A.; den Hartigh, A.; Smith, M.A.; Atluri, V.L.; Sivanesan, D.; Tsolis, R.M.; Baron, C. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun. 2011, 79, 1033–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keyser, P.; Elofsson, M.; Rosell, S.; Wolf-Watz, H. Virulence blockers as alternatives to antibiotics: Type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 2008, 264, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Coinçon, M.; Paschos, A.; Jolicoeur, B.; Lavallée, P.; Sygusch, J.; Baron, C. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem. Biol. 2012, 19, 1041–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casu, B.; Smart, J.; Hancock, M.A.; Smith, M.; Sygusch, J.; Baron, C. Structural analysis and inhibition of TraE from the pKM101 type IV secretion system. J. Biol. Chem. 2016, 291, 23817–23829. [Google Scholar] [CrossRef] [Green Version]
- Casu, B.; Arya, T.; Bessette, B.; Baron, C. Fragment-based screening identifies novel targets for inhibitors of conjugative transfer of antimicrobial resistance by plasmid pKM101. Sci. Rep. 2017, 7, 14907. [Google Scholar] [CrossRef] [Green Version]
- Ripoll-Rozada, J.; García-Cazorla, Y.; Getino, M.; Machón, C.; Sanabria-Ríos, D.; de la Cruz, F.; Cabezón, E.; Arechaga, I. Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol. Microbiol. 2016, 100, 912–921. [Google Scholar] [CrossRef]
- Getino, M.; Sanabria-Ríos, D.J.; Fernández-López, R.; Campos-Gómez, J.; Sánchez-López, J.M.; Fernández, A.; Carballeira, N.M.; de la Cruz, F. Synthetic fatty acids prevent plasmid-mediated horizontal gene transfer. mBio 2015, 6, e01032-15. [Google Scholar] [CrossRef] [Green Version]
- Getino, M.; Fernández-López, R.; Palencia-Gándara, C.; Campos-Gómez, J.; Sánchez-López, J.M.; Martínez, M.; Fernández, A.; de la Cruz, F. Tanzawaic Acids, a chemically novel set of bacterial conjugation inhibitors. PLoS ONE 2016, 11, e0148098. [Google Scholar] [CrossRef]
- Anthony, K.G.; Sherburne, C.; Sherburne, R.; Frost, L.S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 1994, 13, 939–953. [Google Scholar] [CrossRef]
- Shaffer, C.L.; Good, J.A.; Kumar, S.; Krishnan, K.S.; Gaddy, J.A.; Loh, J.T.; Chappell, J.; Almqvist, F.; Cover, T.L.; Hadjifrangiskou, M. Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio 2016, 7, e00221-16. [Google Scholar] [CrossRef] [Green Version]
- Salvadori, G.; Junges, R.; Morrison, D.A.; Petersen, F.C. Competence in Streptococcus pneumoniae and close commensal relatives: Mechanisms and implications. Front. Cell. Infect. Microbiol. 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engholm, D.H.; Kilian, M.; Goodsell, D.S.; Andersen, E.S.; Kjærgaard, R.S. A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 2017, 41, 854–879. [Google Scholar] [CrossRef] [PubMed]
- Domenech, A.; Brochado, A.R.; Sender, V.; Hentrich, K.; Henriques-Normark, B.; Typas, A.; Veening, J.W. Proton motive force disruptors block bacterial competence and horizontal gene transfer. Cell Host Microbe 2020, 27, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Buckner, M.M.C.; Ciusa, M.L.; Meek, R.W.; Moorey, A.R.; McCallum, G.E.; Prentice, E.L.; Reid, J.P.; Alderwick, L.J.; Di Maio, A.; Piddock, L.J.V. HIV drugs inhibit transfer of plasmids carrying extended-spectrum β-lactamase and carbapenemase genes. mBio 2020, 11, e03355-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Jia, Y.; Yang, K.; Li, R.; Xiao, X.; Wang, Z. Anti-HIV agent azidothymidine decreases Tet(X)-mediated bacterial resistance to tigecycline in Escherichia coli. Commun. Biol. 2020, 3, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Qi, H.; Zhu, M.; Gong, S.; Huang, Z.; Zhang, Y.; Chen, X.; Jiao, X. MoS(2) decorated nanocomposite: Fe2O3@MoS2 inhibits the conjugative transfer of antibiotic resistance genes. Ecotoxicol. Environ. Saf. 2019, 186, 109781. [Google Scholar] [CrossRef]
- Kudo, H.; Usui, M.; Nagafuji, W.; Oka, K.; Takahashi, M.; Yamaguchi, H.; Tamura, Y. Inhibition effect of flavophospholipol on conjugative transfer of the extended-spectrum β-lactamase and vanA genes. J. Antibiot. 2019, 72, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Kwapong, A.A.; Stapleton, P.; Gibbons, S. Inhibiting plasmid mobility: The effect of isothiocyanates on bacterial conjugation. Int. J. Antimicrob. Agents 2019, 53, 629–636. [Google Scholar] [CrossRef]
- Rosch, J.W.; Tuomanen, E.I. Caging and COM-bating antibiotic resistance. Cell Host Microbe 2020, 27, 489–490. [Google Scholar] [CrossRef]
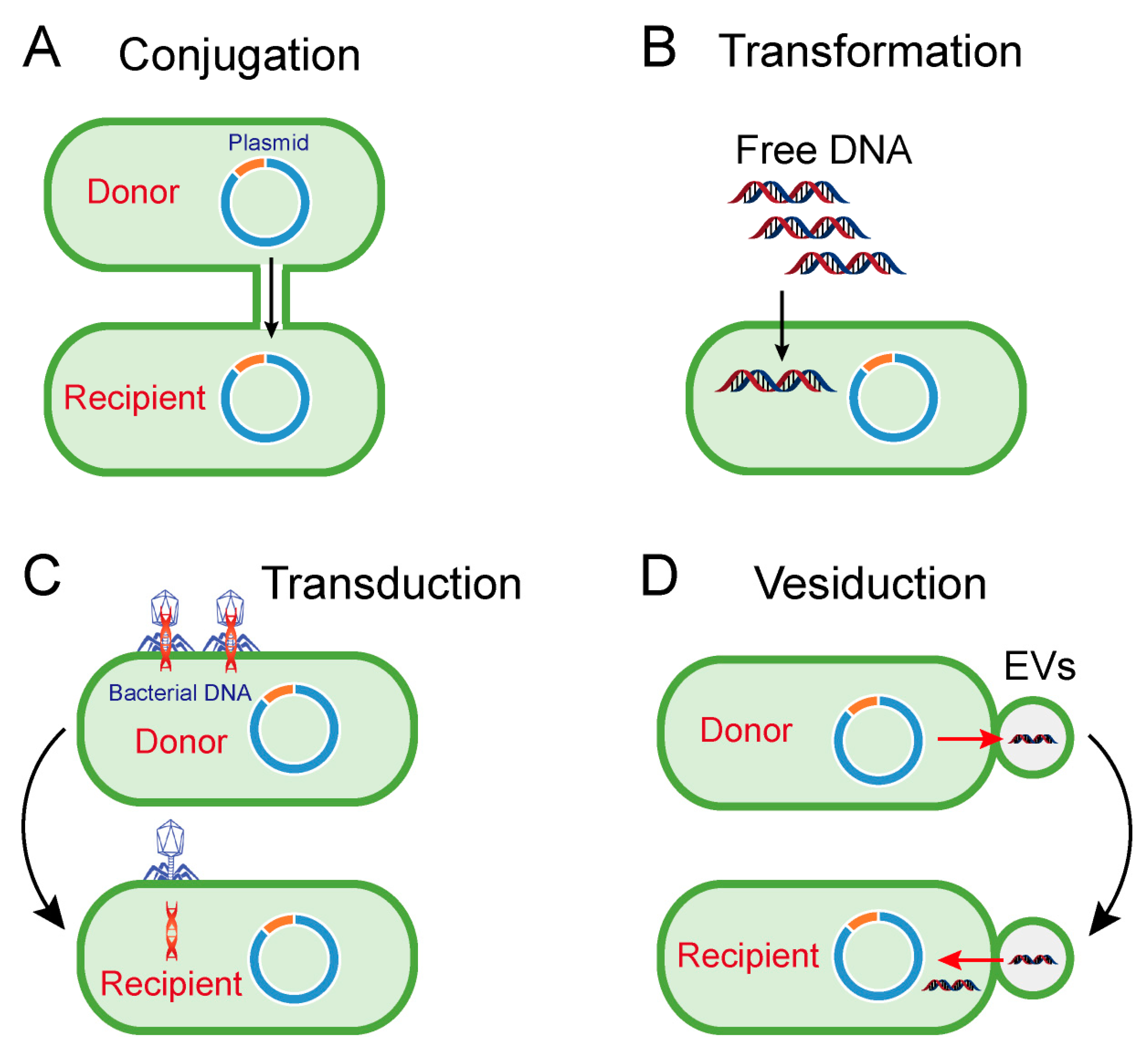
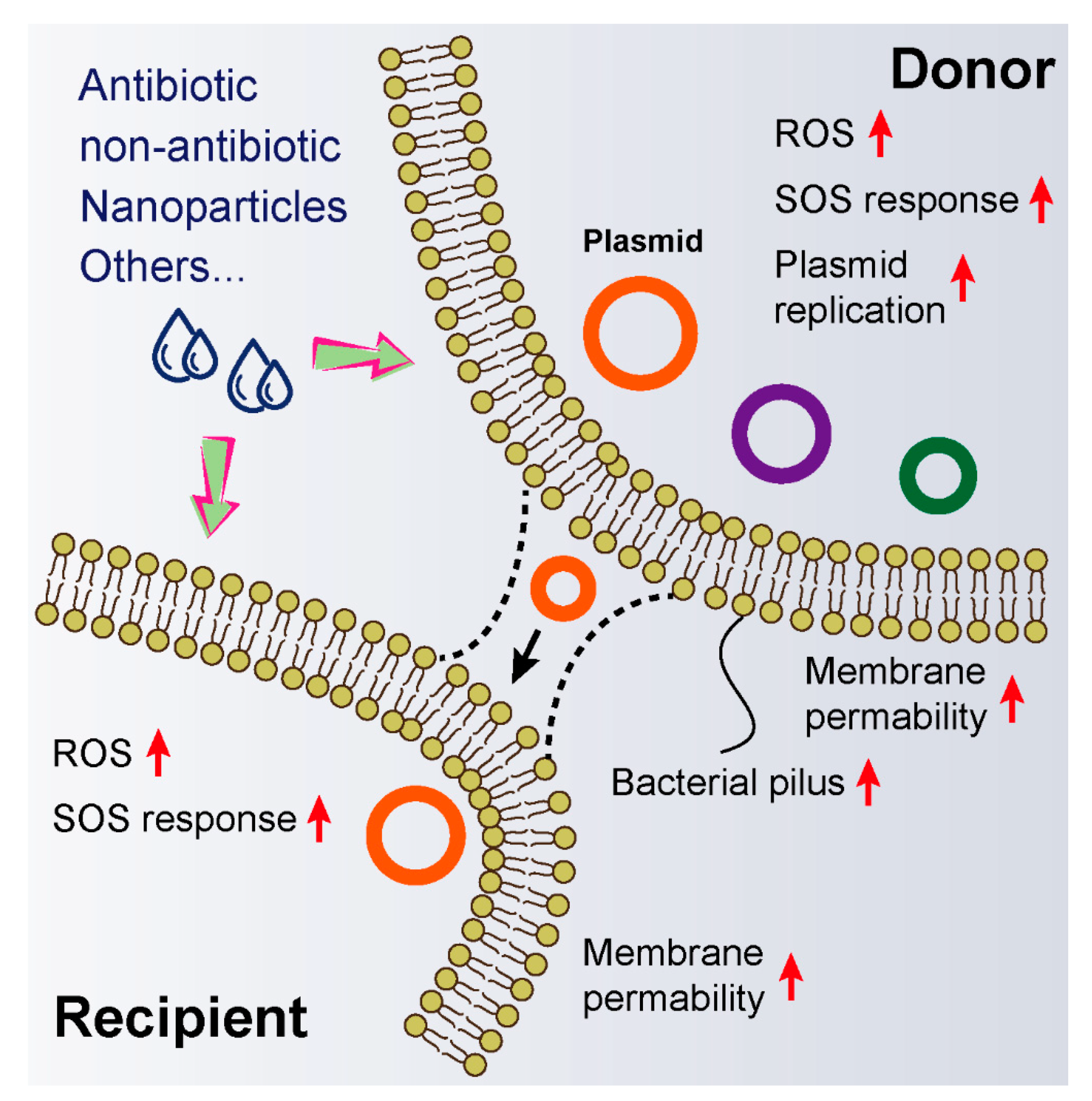
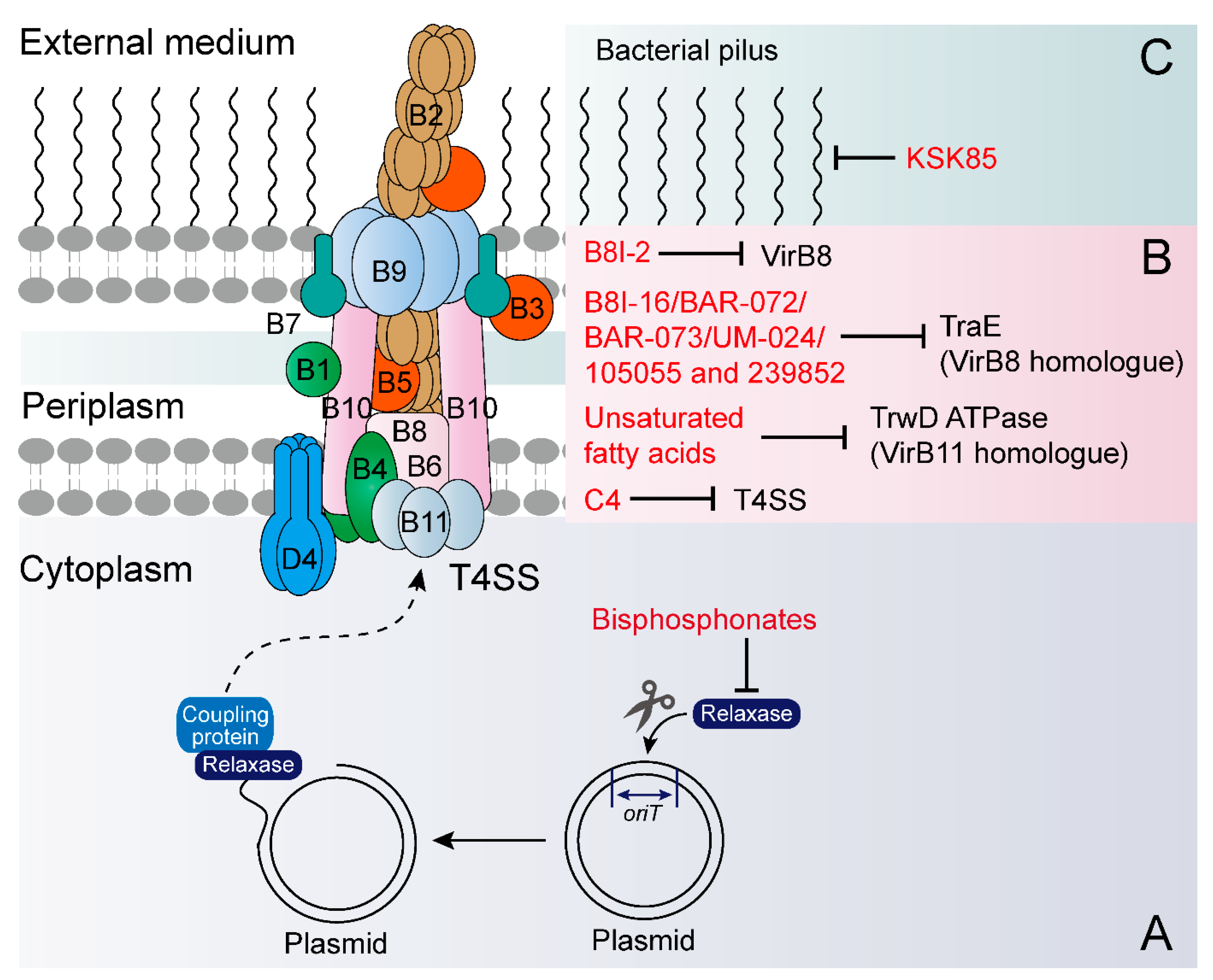
| Compounds | Year | Species | Plasmid/ARGs | Mechanisms |
|---|---|---|---|---|
| Silver ions/nanoparticles | 2020 | E. coli → P. putida | RP4 | ROS generation, membrane damage and the SOS response. |
| Triclosan | 2020 | Plasmid → E. coli | pUC19 | Triggers ROS over-production, damages cell membrane barrier, mediates the pilus capture of plasmid and the translocation of plasmid via cell membrane channels. |
| Preservatives | 2020 | E. coli → E. coli | pCM194-Cm | Stimulates radical-induced RpoS regulon and SOS response, increases cell membrane permeability, and regulates conjugative transfer-related genes. |
| Copper nanoparticles/ions | 2019 | E. coli → P. putida | RP4 | Over-production of ROS. |
| Carbamazepine | 2019 | E. coli → E. coli; E. coli → P. putida; | -- | Increases ROS and the SOS response and enhances cell membrane permeability and pilus generation. |
| CO2 | 2019 | E. coli → E. coli; E. coli → S. typhimurium; | RP4 | Reduces intercellular repulsion and increases PMF. |
| Triclosan | 2018 | E. coli → E. coli; E. coli → P. putida; | RP4 | Promotes ROS generation and damages bacterial membrane, and causes increased expression of the SOS response regulatory genes. |
| Disinfectants (free chlorine, chloramine, and hydrogen peroxide) | 2017 | E. coli → E. coli; E. coli → S. typhimurium; | -- | Intracellular ROS formation, SOS response, increases cell membrane permeability, and alters expressions of conjugation relevant genes. |
| Polycyclic aromatic hydrocarbons (PAHs) | 2017 | intI1-positive bacterium Aerococcus sp. → intI1-negative bacterium Pseudoalteromonas sp. | intI1, sulI and aadA2 genes | -- |
| Gentamicin | 2017 | E. coli → P. aeruginosa | RP4 | Inhibits quorum sensing. |
| Ionic liquid ((BMIm)(PF6)) | 2015 | E. coil K12 → Salmonella spp.; E. coil K12 → microbacterium spp. | RP4 | Enhances cell membrane permeability. |
| Ionic liquid ((BMIm)(PF6)) | 2014 | intI-positive bacteria to other bacterial strains | sulI and intI genes | Increases cell membrane permeability. |
| Nanoalumina | 2012 | E. coli → Salmonella; E. coli → E. coli; G+ → G−; Enterococci → Enterococci | RP4 | Damages bacterial membranes, enhances the expression of conjugative genes and represses global regulatory factor genes. |
| Compounds | Year | Species | Plasmid | ARGs | Mechanisms |
|---|---|---|---|---|---|
| COM-blockers (triclosan, hydrochloride and pimozide) | 2020 | S. pneumoniae → E. coli | -- | -- | Inhibits PMF, thereby preventing the export of a quorum-sensing CSP |
| Azidothymidine | 2020 | E. coli → E. coli | Tet(X)-producing plasmids | tet(X3/X4) | -- |
| Anti-HIV drugs (abacavir and azidothymidine) | 2020 | E. coli → E. coli K. pneumoniae → K. pneumoniae | ESBL-producing plasmid (pCT) and carbapenemase-producing plasmid (pKpQIL) | Extended spectrum β-lactamase and carbapenemase genes | -- |
| Fe2O3@MoS2 | 2019 | E. coli → E. coli; E. coli → E. faecalis; E. faecalis → E. faecalis | RP4-7 | -- | Promotes the expression of global regulatory gene (trbA) and inhibits the expression of conjugative transfer genes |
| Isothiocyanates | 2019 | E. coli → E. coli | pKM101 (IncN), TP114 (IncI2), pUB307 (IncP) and R7K (IncW) | -- | -- |
| Flavophospholipol | 2019 | E. coli → E. coli; E. faecalis → E. faecalis | Self-transmissible plasmids carrying the βlactamase genes or vanA genes | Extended-spectrum β-lactamase and vanA genes | -- |
| Molecules (105055 and 239852) | 2017 | E. coli → E. coli | pKM101 | -- | Targets the TraE protein |
| Unsaturated fatty acids | 2016 | E. coli → E. coli | IncW, IncH, IncF IncI, IncL/M and IncX | -- | Targets type IV traffic ATPase TrwD |
| Tanzawaic acids | 2016 | E. coli → E. coli | IncW and IncFII | -- | -- |
| B8I-16, BAR-072, BAR-073 and UM-024 | 2016 | E. coli → E. coli | pKM101 | -- | Targets the TraE protein |
| Peptidomimetic | 2016 | E. coli → E. coli | IncF plasmid | -- | Disrupts type IV secretion system |
| Synthetic fatty acids | 2015 | Escherichia, Salmonella, Pseudomonas and Acinetobacter spp | IncF, IncW, IncH, IncI, IncL/M, and IncX plasmids | -- | -- |
| Bisphosphonates | 2007 | E. coli → E. coli | F plasmid | -- | Inhibits the conjugative DNA relaxase |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Tong, Z.; Shi, J.; Jia, Y.; Yang, K.; Wang, Z. Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes. Microorganisms 2020, 8, 1211. https://doi.org/10.3390/microorganisms8081211
Liu Y, Tong Z, Shi J, Jia Y, Yang K, Wang Z. Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes. Microorganisms. 2020; 8(8):1211. https://doi.org/10.3390/microorganisms8081211
Chicago/Turabian StyleLiu, Yuan, Ziwen Tong, Jingru Shi, Yuqian Jia, Kangni Yang, and Zhiqiang Wang. 2020. "Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes" Microorganisms 8, no. 8: 1211. https://doi.org/10.3390/microorganisms8081211
APA StyleLiu, Y., Tong, Z., Shi, J., Jia, Y., Yang, K., & Wang, Z. (2020). Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes. Microorganisms, 8(8), 1211. https://doi.org/10.3390/microorganisms8081211