Marine Plastics from Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Identification of Bacterial Strains
2.3. Genomic DNA Extraction and Illumina Sequencing
2.4. Genome Sequence Assembly and Screening for ARGs
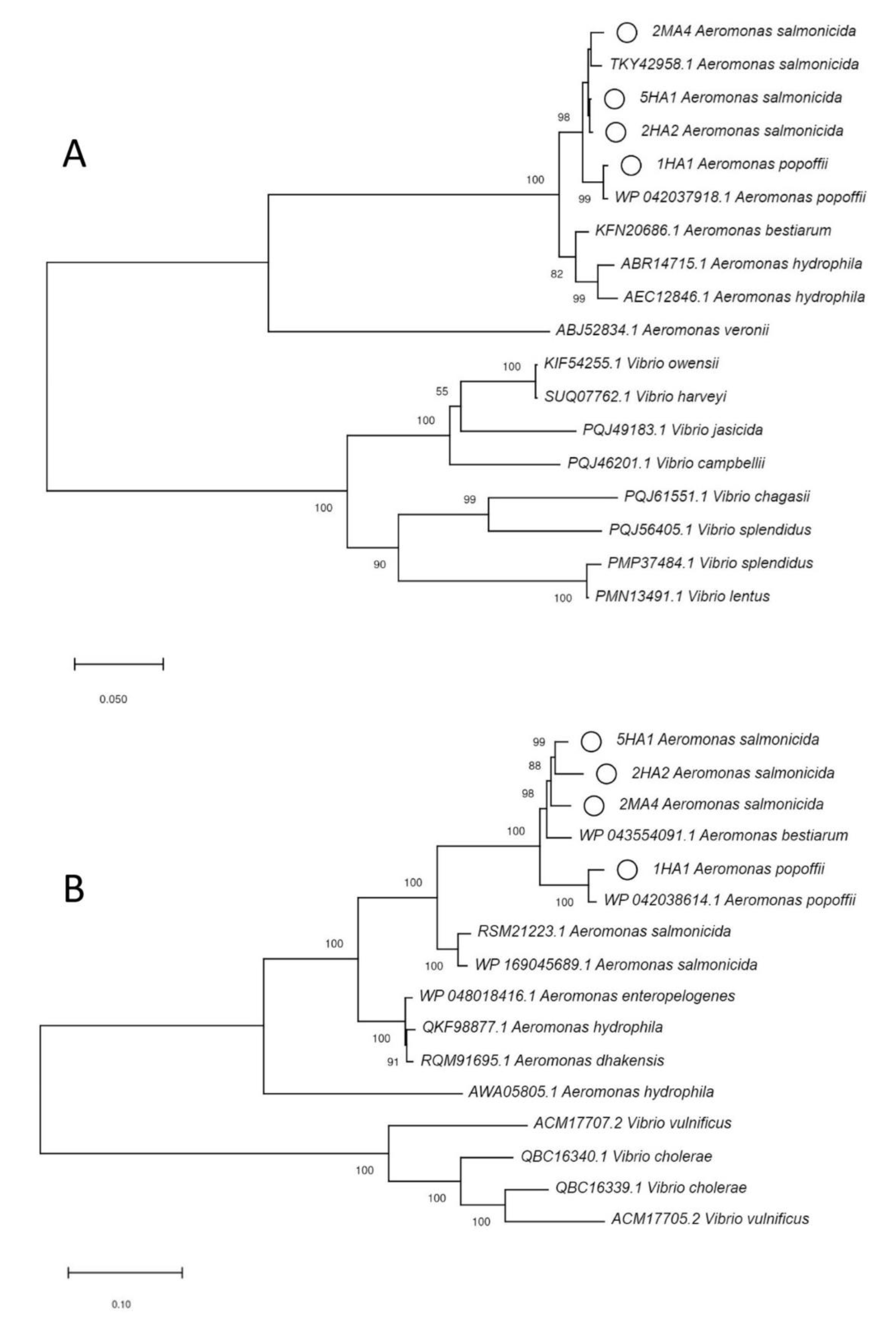
2.5. Phylogenetic Analysis
2.6. Antibiotic Susceptibility Testing
2.7. Identifying Plastic Polymer Type
3. Results
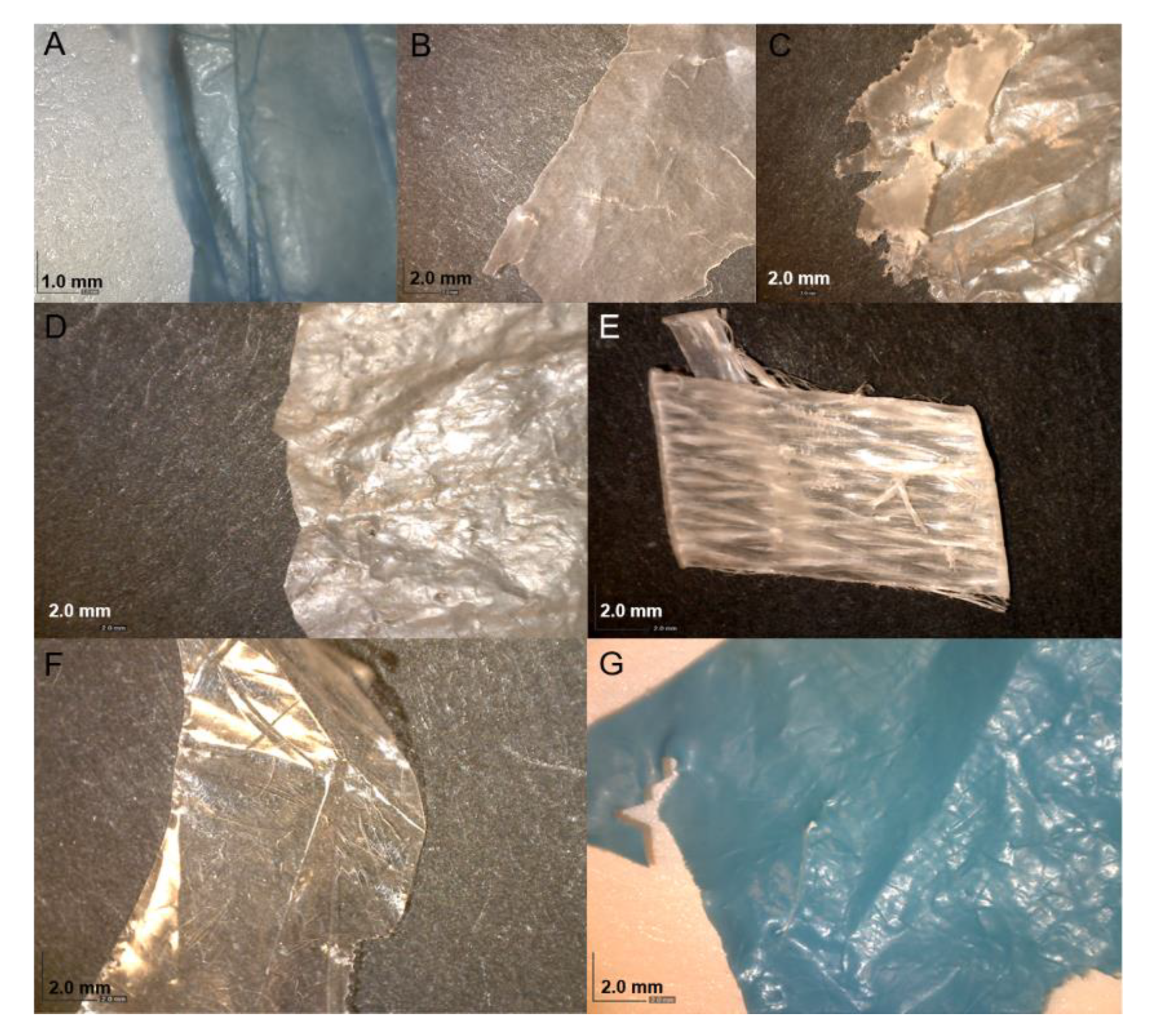
3.1. Identification of the Plastic Polymers and Bacterial Strains
3.2. Antibiotic Resistance and ARGs
3.3. Virulence Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data
References
- Shah, A.A.; Hasan, F.; Hameed, A.; Ahmed, S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008, 26, 246–265. [Google Scholar] [CrossRef]
- Derraik, J.G. The pollution of the marine environment by plastic debris: A review. Mar. Pollut. Bull. 2002, 44, 842–852. [Google Scholar] [CrossRef]
- Wilber, R.J. Plastic in the North Atlantic. Oceanus 1987, 30, 61–68. [Google Scholar]
- Pruter, A.T. Sources, quantities and distribution of persistent plastics in the marine environment. Mar. Pollut. Bull. 1987, 18, 305–310. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, G.; Song, W.; Ye, C.; Lin, H.; Li, Z.; Liu, W. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 2019, 123, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Blair, R.M.; Waldron, S.; Phoenix, V.; Gauchotte-Lindsay, C. Micro-and nanoplastic pollution of freshwater and wastewater treatment systems. Springer Sci. Rev. 2017, 5, 19–30. [Google Scholar] [CrossRef]
- Koelmans, A.A.; Besseling, E.; Shim, W.J. Nanoplastics in the Aquatic Environment. Critical Review. In Marine Anthropogenic Litter; Springer: Cham, Switzerland, 2015; pp. 325–340. [Google Scholar]
- Avio, C.G.; Gorbi, S.; Regoli, F. Plastics and microplastics in the oceans: From emerging pollutants to emerged threat. Mar. Environ. Res. 2017, 128, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.J. Synthetic polymers in the marine environment: A rapidly increasing, long-term threat. Environ. Res. 2008, 108, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.R. Environmental implications of plastic debris in marine settings entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos. Trans. R. Soc. B 2009, 364, 2013–2025. [Google Scholar] [CrossRef]
- Jepsen, E.M.; de Bruyn, P.J.N. Pinniped entanglement in oceanic plastic pollution: A global review. Mar. Pollut. Bull. 2019, 145, 295–305. [Google Scholar] [CrossRef]
- Kühn, S.; Bravo Rebolledo, E.L.; van Franeker, J.A. Deleterious Effects of Litter on Marine Life. In Marine Anthropogenic Litter; Bergmann, M., Gutow, L., Klages, M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 75–116. [Google Scholar] [CrossRef]
- Kögel, T.; Bjorøy, Ø.; Toto, B.; Bienfait, A.M.; Sanden, M. Micro- and nanoplastic toxicity on aquatic life: Determining factors. Sci. Total Environ. 2020, 709, 136050. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Tse, H.F.; Fok, L. Plastic waste in the marine environment: A review of sources, occurrence and effects. Sci. Total Environ. 2016, 566–567, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Zarfl, C.; Matthies, M. Are marine plastic particles transport vectors for organic pollutants to the Arctic? Mar. Pollut. Bull. 2010, 60, 1810–1814. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Parsons, S.; Clift, S.; Stolte, A.; McManus, M.C. Collected marine litter—A growing waste challenge. Mar. Pollut. Bull. 2018, 128, 162–174. [Google Scholar] [CrossRef]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.A.; Fong, A.A.; Thomas, K.A.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. MSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Loeder, M.G.J.; Gerdts, G.; Osborn, A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014, 90, 478–492. [Google Scholar] [CrossRef]
- Eriksen, M.; Lebreton, L.C.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic pollution in the world’s oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef]
- Arias-Andres, M.; Klümper, U.; Rojas-Jimenez, K.; Grossart, H.-P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018, 237, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.; Kubowicz, S.; Beegle-Krause, C.; Skancke, J.; Nordam, T.; Landsem, E.; Throne-Holst, M.; Jahren, S. Microplastic in Global and Norwegian Marine Environments: Distributions, Degradation Mechanisms and Transport. Miljødirektoratet M-918; SINTEF: Trondheim, Norway, 2017; pp. 1–147. [Google Scholar]
- Jensen, S.; Lindbaek, M.; Gradmann, C.; Lie, A.K. Reflections about antibiotic resistance history in Norway. Erindr. Antibiot. Hist. Nor. 2010, 130, 2494. [Google Scholar] [CrossRef]
- Amalina, N.Z.; Santha, S.; Zulperi, D.; Amal, M.N.A.; Yusof, M.T.; Zamri-Saad, M.; Ina-Salwany, M.Y. Prevalence, antimicrobial susceptibility and plasmid profiling of Vibrio spp. isolated from cultured groupers in Peninsular Malaysia. BMC Microbiol. 2019, 19, 251. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Granier, S.A.; Larvor, E.; Jouy, E.; Cineux, M.; Wilhelm, A.; Gassilloud, B.; Le Bouquin, S.; Kempf, I.; Chauvin, C. Aeromonas diversity and antimicrobial susceptibility in freshwater—An attempt to set generic epidemiological cut-off values. Front. Microbiol. 2017, 8, 503. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Ciufo, S.; Li, W. Prokaryotic Genome Annotation Pipeline. In The NCBI Handbook [Internet], 2nd ed.; Bethesda, US: Center for Biotechnology Information: Rockville, MD, USA, 2013; pp. 1–12. [Google Scholar]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Primpke, S.; Wirth, M.; Lorenz, C.; Gerdts, G. Reference database design for the automated analysis of microplastic samples based on Fourier transform infrared (FTIR) spectroscopy. Anal. Bioanal. Chem. 2018, 410, 5131–5141. [Google Scholar] [CrossRef]
- Viršek, M.K.; Lovšin, M.N.; Koren, Š.; Kržan, A.; Peterlin, M. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar. Pollut. Bull. 2017, 125, 301–309. [Google Scholar] [CrossRef]
- Parker, J.L.; Shaw, J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011, 62, 109–118. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012, 2012, 1–13. [Google Scholar] [CrossRef]
- Reith, M.E.; Singh, R.K.; Curtis, B.; Boyd, J.M.; Bouevitch, A.; Kimball, J.; Munholland, J.; Murphy, C.; Sarty, D.; Williams, J. The genome of Aeromonas salmonicida subsp. salmonicida A449: Insights into the evolution of a fish pathogen. BMC Genom. 2008, 9, 427. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish; Springer: London, UK, 1999. [Google Scholar]
- Wiles, T.J.; Mulvey, M.A. The RTX pore-forming toxin α-hemolysin of uropathogenic Escherichia coli: Progress and perspectives. Future Microbiol. 2013, 8, 73–84. [Google Scholar] [CrossRef]
- Suarez, G.; Khajanchi, B.K.; Sierra, J.C.; Erova, T.E.; Sha, J.; Chopra, A.K. Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 2012, 506, 369–376. [Google Scholar] [CrossRef]
- Bücker, R.; Krug, S.M.; Rosenthal, R.; Günzel, D.; Fromm, A.; Zeitz, M.; Chakraborty, T.; Fromm, M.; Epple, H.-J.; Schulzke, J.-D. Aerolysin from Aeromonas hydrophila Perturbs Tight Junction Integrity and Cell Lesion Repair in Intestinal Epithelial HT-29/B6 Cells. J. Infect. Dis. 2011, 204, 1283–1292. [Google Scholar] [CrossRef]
- Rasmussen-Ivey, C.R.; Figueras, M.J.; McGarey, D.; Liles, M.R. Virulence Factors of Aeromonas hydrophila: In the Wake of Reclassification. Front. Microbiol. 2016, 7, 1337. [Google Scholar] [CrossRef] [PubMed]
- Wedekind, J.E.; Trame, C.B.; Dorywalska, M.; Koehl, P.; Raschke, T.M.; McKee, M.; FitzGerald, D.; Collier, R.J.; McKay, D.B. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 2001, 314, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Howard, S.P. ExeA binds to peptidoglycan and forms a multimer for assembly of the type II secretion apparatus in Aeromonas hydrophila. Mol. Microbiol. 2010, 76, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Schoenhofen, I.C.; Stratilo, C.; Howard, S.P. An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: Effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 1998, 29, 1237–1247. [Google Scholar] [CrossRef]
- Suarez, G.; Sierra, J.C.; Sha, J.; Wang, S.; Erova, T.E.; Fadl, A.A.; Foltz, S.M.; Horneman, A.J.; Chopra, A.K. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 2008, 44, 344–361. [Google Scholar] [CrossRef]
- Sha, J.; Rosenzweig, J.A.; Kozlova, E.V.; Wang, S.; Erova, T.E.; Kirtley, M.L.; van Lier, C.J.; Chopra, A.K. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology 2013, 159, 1120. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Mazel, D.; Soussy, C.-J.; Nordmann, P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 2007, 51, 2650–2651. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Rodriguez-Martinez, J.-M.; Mammeri, H.; Liard, A.; Nordmann, P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005, 49, 3523–3525. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob. Agents Chemother. 2011, 55, 4405–4407. [Google Scholar] [CrossRef]
- Ebmeyer, S.; Kristiansson, E.; Larsson, D.G.J. The mobile FOX AmpC beta-lactamases originated in Aeromonas allosaccharophila. Int. J. Antimicrob. Agents 2019, 54, 798–802. [Google Scholar] [CrossRef]
- Ebmeyer, S.; Kristiansson, E.; Larsson, D.J. CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three aeromonas species. J. Antimicrob. Chemother. 2019, 74, 1202–1206. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Zhang, H. Adsorption of antibiotics on microplastics. Environ. Pollut. 2018, 237, 460–467. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Blaak, H.; de Jong, M.C.; Graat, E.A.; Vandenbroucke-Grauls, C.M.; de Roda Husman, A.M. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Caruso, G. Microplastics as vectors of contaminants. Mar. Pollut. Bull. 2019, 146, 921–924. [Google Scholar] [CrossRef]
- Jutkina, J.; Marathe, N.P.; Flach, C.F.; Larsson, D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Tucci, V.; Isenberg, H.D. Hospital cluster epidemic with Morganella morganii. J. Clin. Microbiol. 1981, 14, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Mastroianni, A.; Coronado, O.; Chiodo, F. Morganella morganii meningitis in a patient with AIDS. J. Infect. 1994, 29, 356–357. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Seifert, H.; Towner, K.J. Acinetobacter infection—An emerging threat to human health. IUBMB Life 2011, 63, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.T.; Bollet, C.; Tercian, S.; Drancourt, M.; Raoult, D. Aeromonas popoffii urinary tract infection. J. Clin. Microbiol. 2004, 42, 5427–5428. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar]
- Silva, M.M.; Maldonado, G.C.; Castro, R.O.; de Sá Felizardo, J.; Cardoso, R.P.; dos Anjos, R.M.; de Araújo, F.V. Dispersal of potentially pathogenic bacteria by plastic debris in Guanabara Bay, RJ, Brazil. Mar. Pollut. Bull. 2019, 141, 561–568. [Google Scholar] [CrossRef]
- Stévant, P.; Rebours, C.; Chapman, A. Seaweed aquaculture in Norway: Recent industrial developments and future perspectives. Aquac. Int. 2017, 25, 1373–1390. [Google Scholar] [CrossRef]
- Fisheries, F. The State of World Fisheries and Aquaculture 2010; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. [Google Scholar]
- Sentralbyrå, S. Akvakultur, 2018, Årlig Endelige Tall. Available online: https://www.ssb.no/jord-skog-jakt-og-fiskeri/statistikker/fiskeoppdrett/aar (accessed on 5 May 2020).
- FAO. Globefish highlights—A quarterly update on world seafood markets—April 2019 issues, with Jan.–Dec. 2018 statistics. Globefish Highlights 2019, 2, 1–68. [Google Scholar]
| Isolate | Name | MIC (µg/mL) | Isolation Source | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT | TC | CI | AM | MP | SM | TR | GM | IP | CL | |||
| 2MA1 | Morganella morganii | 0.023 | 256 | 0.008 | >256 | 0.032 | 4 | 0.125 | 0.75 | 0.50 | 24 | Polyethylene/Ethylene vinyl acetate copolymer |
| 5HA1 | Aeromonas salmonicida | 0.023 | 0.25 | 0.003 | >256 | 0.032 | 4 | 0.032 | 0.75 | 0.25 | 0.38 | Polyethylene |
| 2HA2 | Aeromonas salmonicida | 0.047 | 0.50 | 0.002 | >256 | 0.023 | 8 | 0.25 | 1.5 | 0.25 | 1.0 | Polyethylene/Ethylene vinyl acetate copolymer |
| 2MA4 | Aeromonas salmonicida | 0.094 | 0.38 | 0.003 | >256 | 0.032 | 8 | 0.50 | 1.5 | 0.38 | 0.75 | Polyethylene/Ethylene vinyl acetate copolymer |
| 1HA1 | Aeromonas popoffii | 0.064 | 0.19 | <0.002 | >256 | 0.004 | 6 | 0.125 | 0.75 | 0.047 | 0.38 | Polyethylene |
| 2HC4 | Acinetobacter beijerinckii | 12 | 1.5 | 0.094 | >256 | 0.50 | 2 | 6 | 1.0 | 0.19 | 24 | Polyethylene/Ethylene vinyl acetate copolymer |
| Isolate | Name | Gene | Closest Blast Hit | Percent Identity (Amino Acid) |
|---|---|---|---|---|
| 2MA1 | Morganella morganii | blaDHA | DHA family class C β-lactamase | 100.00% |
| tet(D) | tetracycline efflux MFS transporter Tet(D) | 100.00% | ||
| aac(3) | aminoglycoside 3-N-acetyltransferase | 100.00% | ||
| catB | antibiotic acetyltransferase | 100.00% | ||
| 5HA1 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 98.43% |
| blaOXA | class D β-lactamase | 100.00% | ||
| cphA | CphA family subclass B2 metallo-β-lactamase | 97.64% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 98.64% | ||
| 2HA2 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 97.90% |
| cphA | CphA family subclass B2 metallo- β-lactamase | 99.61% | ||
| blaOXA | class D β-lactamase | 100.00% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 98.64% | ||
| 2MA4 | Aeromonas salmonicida | ampC | FOX/MOX family class C β-lactamase | 96.33% |
| blaOXA | class D β-lactamase | 99.24% | ||
| cphA | CphA family subclass B2 metallo-β-lactamase | 96.85% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 99.09% | ||
| 1HA1 | Aeromonas popoffii | blaOXA | class D β-lactamase | 99.62% |
| ampC | FOX/MOX family class C β-lactamase | 98.95% | ||
| qnrA | Qnr family pentapeptide repeat protein | 100.00% | ||
| catB | antibiotic acetyltransferase | 99.10% | ||
| 2HC4 | Acinetobacter beijerinckii | ampC | class C β-lactamase | 96.46% |
| bla | class A β-lactamase | 96.11% | ||
| aac(6′)-I | AAC(6′)-Ighjkrstuvwx family aminoglycoside N-acetyltransferase | 97.93% | ||
| catB | antibiotic acetyltransferase | 96.24% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radisic, V.; Nimje, P.S.; Bienfait, A.M.; Marathe, N.P. Marine Plastics from Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes. Microorganisms 2020, 8, 1200. https://doi.org/10.3390/microorganisms8081200
Radisic V, Nimje PS, Bienfait AM, Marathe NP. Marine Plastics from Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes. Microorganisms. 2020; 8(8):1200. https://doi.org/10.3390/microorganisms8081200
Chicago/Turabian StyleRadisic, Vera, Priyank S. Nimje, André Marcel Bienfait, and Nachiket P. Marathe. 2020. "Marine Plastics from Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes" Microorganisms 8, no. 8: 1200. https://doi.org/10.3390/microorganisms8081200
APA StyleRadisic, V., Nimje, P. S., Bienfait, A. M., & Marathe, N. P. (2020). Marine Plastics from Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes. Microorganisms, 8(8), 1200. https://doi.org/10.3390/microorganisms8081200






