Should Networks Supplant Tree Building?
Abstract
1. Introduction
2. HGT is the Hobgoblin of Bifurcation or Vertical Divergence
3. Bifurcation as an Evolutionary Pattern
4. Bacterial and Archaeal Divergence: Nothing Special?
5. Tree Thinking, Concatenation and Bacteria
“The goal of the total evidence approach to phylogenetic research is based in the idea of increasing explanatory power over background knowledge through test and corroboration, rather than to bolster support for nodes in a tree. In this context, the testing of phylogenetic data is a falsificationist endeavor that includes [italics added] the possibility of not rejecting the null hypothesis that there is no tree-like structure in molecular phylogenetic data.”
6. Three Simple Falsificationist Hypotheses that Test for the Existence of the Tree of Life
7. Obscured Pattern or Obscured Process
8. Why a Tree of Life Infected with HGT Still Bifurcates
9. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baptese, E.; Van Iersel, L.; Janke, A.; Kelchner, S.; Kelk, S.; McInerney, J.O.; Morrison, D.A.; Nakhleh, L.; Steel, M.; Stougie, L.; et al. Networks: Expanding evolutionary thinking. Trends Genet. 2013, 29, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Papale, F.; Saget, J.; Bapteste, É. Networks consolidate the core concepts of evolution by natural selection. Trends Microbiol. 2020, 28, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.K.; Habib, M.; Bapteste, É. Phylosystemics: Merging phylogenomics, systems biology, and ecology to study evolution. Trends Microbiol. 2020, 28, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.K.; Lannes, R.; Pathmanathan, J.S.; Méheust, R.; Karkar, S.; Colson, P.; Corel, E.; Lopez, P.; Bapteste, É. The methodology behind network thinking: Graphs to analyze microbial complexity and evolution. In Evolutionary Genomics; Anisimova, M., Ed.; Humana: New York, NY, USA, 2019; pp. 271–308. [Google Scholar]
- Booth, A.; Mariscal, C.; Doolittle, W.F. The modern synthesis in the light of microbial genomics. Annu. Rev. Microbiol. 2016, 70, 279–297. [Google Scholar] [CrossRef]
- Bapteste, E.; Huneman, P. Towards a dynamic interaction network of life to unify and expand the evolutionary theory. BMC Biol. 2018, 16, 56. [Google Scholar] [CrossRef]
- Corel, E.; Lopez, P.; Méheust, R.; Bapteste, E. Network-thinking: Graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016, 24, 224–237. [Google Scholar] [CrossRef]
- Morrison, D.A. Is the tree of life the best metaphor, model, or heuristic for phylogenetics? Syst. Biol. 2014, 63, 628–638. [Google Scholar] [CrossRef]
- Doolittle, W.F.; Brunet, T. What is the tree of life? PLoS Genet. 2016, 12, e1005912. [Google Scholar] [CrossRef][Green Version]
- Puigbò, P.; Wolf, Y.I.; Koonin, E.V. Genome-wide comparative analysis of phylogenetic trees: The prokaryotic forest of life. Methods Mol. Biol. 2012, 856, 53–79. [Google Scholar]
- Koonin, E.V. Horizontal gene transfer: Essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Doolittle, W.F. Eradicating typological thinking in prokaryotic systematics and evolution. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F. Early evolution without a tree of life. Biol. Direct. 2011, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.; Huelsenbeck, J.; Swofford, D. Hobgoblin of phylogenetics? Nature 1994, 369, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F.; Bapteste, E. Pattern pluralism and the tree of life hypothesis. Proc. Natl. Acad. Sci. USA 2007, 104, 2043–2049. [Google Scholar] [CrossRef]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2128. [Google Scholar] [CrossRef]
- Bapteste, E.; Boucher, Y. Epistemological impacts of horizontal gene transfer on classification in microbiology. In Horizontal Gene Transfer; Gogarten, M.B., Olendzenski, L., Gogarten, J.P., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 55–72. [Google Scholar]
- Boucher, Y.; Bapteste, E. Revisiting the concept of lineage in prokaryotes: A phylogenetic perspective. Bioessays 2009, 31, 526–536. [Google Scholar] [CrossRef]
- Creevey, C.J.; Fitspatrick, D.A.; Philip, G.K.; Kinsella, R.J.; O’Connell, M.J.; Pentony, M.M.; Travers, S.A.; Wilkinson, M.; McInerney, J.O. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc. Biol. Sci. 2004, 271, 2551. [Google Scholar] [CrossRef]
- Bapteste, E.; Boucher, Y.; Leigh, J.; Doolittle, W.F. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 2004, 12, 406–411. [Google Scholar] [CrossRef]
- Koonin, E.V. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009, 37, 1011–1034. [Google Scholar] [CrossRef]
- Kohn, D. Darwin’s keystone: The principle of divergence. In The Cambridge Companion to the “Origin of Species”; Ruse, M., Richards, R.J., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 242–278. [Google Scholar]
- Mayr, E. Systematics and the Origin of Species, from the Viewpoint of a Zoologist; Harvard University Press: Cambridge, MA, USA, 1942. [Google Scholar]
- Huelsenbeck, J.P.; Rannala, B. Phylogenetic methods come of age: Testing hypotheses in an evolutionary context. Science 1997, 276, 227–232. [Google Scholar] [CrossRef]
- Planet, P.J. Tree disagreement: Measuring and testing incongruence in phylogenies. J. Biomed. Inform. 2006, 39, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Lienau, E.K.; DeSalle, R.; Allard, M.; Brown, E.W.; Swofford, D.; Rosenfeld, J.A.; Sarkar, I.N.; Planet, P.J. The mega-matrix tree of life: Using genome-scale horizontal gene transfer and sequence evolution data as information about the vertical history of life. Cladistics 2010, 27, 417–427. [Google Scholar] [CrossRef]
- Doolittle, W.F.; Zhaxybayeva, O. On the origin of prokaryotic species. Genome Res. 2009, 19, 744. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T. Universal species concept: Pipe dream or a step toward unifying biology? J. Ind. Microbiol. Biotechnol. 2009, 36, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.; Ochman, H. Biological species are universal across Life’s domains. Genome Biol. Evol. 2017, 9, 491–501. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Zaneveld, J.R.; Nemergut, D.R.; Knight, R. Are all horizontal gene transfers created equal? Prospects for mechanism-based studies of HGT patterns. Microbiology 2008, 154, 1–15. [Google Scholar] [CrossRef]
- Davison, J. Genetic exchange between bacteria in the environment. Plasmid 1999, 42, 73–91. [Google Scholar] [CrossRef]
- Beiko, R.G.; Harlow, T.J.; Ragan, M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 14332–14337. [Google Scholar] [CrossRef]
- Wertz, J.E.; Goldstone, C.; Gordon, D.; Riley, M.A. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J. Evol. Biol. 2003, 16, 1236–1248. [Google Scholar] [CrossRef]
- Lerat, E.; Daubin, V.; Ochman, H.; Moran, N.A. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005, 3, e130. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.A.; Lizotte-Waniewski, M. Population genomics and the bacterial species concept. In Horizontal Gene Transfer; Gogarten, M.B., Olendzenski, L., Gogarten, J.P., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 367–377. [Google Scholar]
- Glasner, J.D.; Perna, N.T. Comparative genomics of E. coli. Microbiol. Today 2004, 31, 125. [Google Scholar]
- Mau, B.; Glasner, J.D.; Darling, A.E.; Perna, N.T. Genome-wide detection and analysis of homologous recombination among sequenced strains of Escherichia coli. Genome Biol. 2006, 7, R44. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.V.; Fertil, B.; Giron, A.; Deschavanne, P.J. A genomic schism in birds revealed by phylogenetic analysis of DNA strings. Syst. Biol. 2002, 51, 599–613. [Google Scholar] [CrossRef]
- Waterfield, N.R.; Daborn, P.J.; Dowling, A.J.; Yang, G.W.; Hares, M.; Ffrench-Constant, R.H. The insecticidal toxin makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol. Lett. 2003, 229, 265–270. [Google Scholar] [CrossRef]
- Coleman, M.L.; Sullivan, M.B.; Martiny, A.C.; Steglich, C.; Barry, K.; Delong, E.F.; Chisolm, S.W. Genomic islands and the ecology and evolution of Prochlorococcus. Science 2006, 311, 1768–1770. [Google Scholar] [CrossRef]
- Juhas, M.; Crook, D.W.; Dimopoulou, I.D.; Lunter, G.; Harding, R.M.; Ferguson, D.J.P.; Hood, D.W. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 2007, 189, 761–771. [Google Scholar] [CrossRef]
- Brown, J.R.; Volker, C. Phylogeny of gamma proteobacteria: Resolution of one branch of the universal tree? Bioessays 2004, 26, 463–468. [Google Scholar] [CrossRef]
- Woodward, M.J.; Sojka, M.; Sprigings, K.A.; Humphrey, T.J. The role of SEF14 and SEF17 fimbriae in the adherence of Salmonella enterica serotype Enteritidis to inanimate surfaces. J. Med. Microbiol. 2000, 49, 481–487. [Google Scholar] [CrossRef][Green Version]
- Godoy, D.; Randle, G.; Simpson, A.J.; Aanensen, D.M.; Pitt, T.L.; Kinoshita, R.; Spratt, B.G. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003, 41, 2068–2079. [Google Scholar] [CrossRef]
- Thompson, F.L.; Gevers, D.; Thompson, C.C.; Dawyndt, P.; Naser, S.; Hoste, B.; Munn, C.B.; Swings, J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 2005, 71, 5107–5115. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, R.J.; Grogan, D.W.; Taylor, J.W. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobusislandicus. Mol. Biol. Evol. 2005, 22, 2354–2361. [Google Scholar] [CrossRef] [PubMed]
- Lerat, E.; Daubin, V.; Moran, N.A. From gene trees to organismal phylogeny in prokaryotes: The case of the gamma-Proteobacteria. PLoS Biol. 2003, 1, e19. [Google Scholar] [CrossRef]
- Hanage, W.P.; Kaijalainen, T.; Herva, E.; Saukkoriipi, A.; Syrjanen, R.; Spratt, B.G. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 2005, 187, 6223–6230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Popoff, M.Y.; Kersters, K.; Kiredjian, M.; Miras, I.; Coynault, C. Taxonomic position of Agrobacterium strains of hospital origin. Ann. Microbiol. (Paris) 1984, 135A, 427–442. [Google Scholar]
- Mougel, C.; Thioulouse, J.; Perriere, G.; Nesme, X. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 2002, 52, 573–586. [Google Scholar] [CrossRef]
- Portier, P.; Saux, M.F.; Mougel, C.; Lerondelle, C.; Chapulliot, D.; Thioulouse, J.; Nesme, X. Identification of genomic species in Agrobacterium biovar 1 by AFLP genomic markers. Appl. Environ. Microbiol. 2006, 72, 7123–7131. [Google Scholar] [CrossRef][Green Version]
- Majewski, J. Sexual isolation in bacteria. FEMS Microbiol. Lett. 2001, 199, 161–169. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 3160–3165. [Google Scholar] [CrossRef]
- Mysara, M.; Vandamme, P.; Props, R.; Kerckhof, F.; Leys, N.; Boon, N.; Raes, J.; Monsieurs, P. Reconciliation between operational taxonomic units and species boundaries. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Venter, S.N.; Palmer, M.; Beukes, C.W.; Chan, W.Y.; Shin, G.; van Zyl, E.; Seale, T.; Coutinho, T.A.; Steenkamp, E.T. Practically delineating bacterial species with genealogical concordance. Antonie Van Leeuwenhoek 2017, 110, 1311–1325. [Google Scholar] [CrossRef] [PubMed]
- Haber, M.H. Species in the Age of Discordance. Philos. Theory Pract. Biol. 2019, 11. [Google Scholar] [CrossRef]
- Lean, C.H. Biodiversity realism: Preserving the tree of life. Biol. Philos. 2017, 32, 1083–1103. [Google Scholar] [CrossRef]
- Suárez, J. Bacterial species pluralism in the light of medicine and endosymbiosis. THEORIA. Rev. Teoría Hist. Fundam. Cienc. 2016, 31, 91–105. [Google Scholar]
- Lienau, E.K.; DeSalle, R. Evidence, content and corroboration and the Tree of Life. Acta Biotheor. 2007, 57, 187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maddison, W. Reconstructing character evolution on polytomous cladograms. Cladistics 1989, 5, 365–377. [Google Scholar] [CrossRef]
- Bapteste, E.; Boucher, Y. Lateral gene transfer challenges principles of microbial systematics. Trends Microbiol. 2008, 16, 200–207. [Google Scholar] [CrossRef]
- Velasco, J.D.; Sober, E. Testing for treeness: Lateral gene transfer, phylogenetic inference, and model selection. Biol. Philos. 2010, 25, 675–687. [Google Scholar] [CrossRef]
- Degnan, J.H.; Rosenberg, N.A. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006, 2, 762–768. [Google Scholar] [CrossRef]
- Degnan, J.H.; Salter, L.A. Gene tree distributions under the coalescent process. Evolution 2005, 59, 24–37. [Google Scholar] [CrossRef]
- Kubatko, L.S.; Degnan, J.H. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 2007, 56, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 14, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Lienau, E.K.; DeSalle, R.; Rosenfeld, J.; Planet, P.J. Reciprocal illumination in the gene content ToL. Syst. Biol. 2006, 55, 441. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.; Tannier, E.; Gouy, M.; Daubin, V. Lateral gene transfer as a support for the tree of life. Proc. Natl. Acad. Sci. USA 2012, 109, 4962–4967. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Rokas, A.; Williams, B.L.; King, N.; Carroll, S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003, 425, 798–804. [Google Scholar] [CrossRef]
- Yu, Y.; Degnan, J.H.; Nakhleh, L. The probability of a gene tree topology within a phylogenetic network with applications to hybridization detection. PLoS Genet. 2012, 8, e1002660. [Google Scholar] [CrossRef]
- Gatesy, J. How many genes should a systematist sample? Conflicting insights from a phylogenomic matrix characterized by replicated incongruence. Syst. Biol. 2007, 56, 355–363. [Google Scholar] [CrossRef]
- Gatesy, J.; Baker, R.H. Hidden likelihood support in genomic data: Can forty-five wrongs make a right? Syst. Biol. 2005, 54, 483–492. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Payne, A.; DeSalle, R. Random roots and lineage sorting. Mol. Phylogenetics Evol. 2012, 64, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.; Donoghue, M.J.; Sober, E. Against consensus. Syst. Zool. 1991, 40, 486–493. [Google Scholar] [CrossRef]
- Gatesy, J.; O’Grady, P.; Baker, R.H. Corroboration among data sets in simultaneous analysis: Hidden support for phylogenetic relationships among higher level artiodactyl taxa. Cladistics 1999, 15, 271–313. [Google Scholar] [CrossRef]
- Sánchez-Pacheco, S.J.; Kong, S.; Pulido-Santacruz, P.; Murphy, R.W.; Kubatko, L. Median-joining network analysis of SARS-CoV-2 genomes is neither phylogenetic nor evolutionary. Proc. Natl. Acad. Sci. USA 2020, 117, 12518–12519. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Sánchez-Pacheco, S.J.; Murphy, R.W. On the use of median-joining networks in evolutionary biology. Cladistics 2016, 32, 691–699. [Google Scholar] [CrossRef]
- Gogarten, J.; Townsend, J. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005, 3, 679–687. [Google Scholar] [CrossRef]
- DeSalle, R. The Twin Phylogenomic Challenges. In Darwin Evolution and Life; NIBR Symposium: Inchon, Korea, 2009; pp. 23–30. [Google Scholar]
- Zamani-Dahaj, S.A.; Okasha, M.; Kosakowski, J.; Higgs, P.G. Estimating the frequency of horizontal gene transfer using phylogenetic models of gene gain and loss. Mol. Biol. Evol. 2016, 33, 1843–1857. [Google Scholar] [CrossRef]
- Davín, A.A.; Tannier, E.; Williams, T.A.; Boussau, B.; Daubin, V.; Szöllősi, G.J. Gene transfers can date the tree of life. Nat. Ecol. Evol. 2018, 2, 904–909. [Google Scholar] [CrossRef]
- Daubin, V.; Szöllősi, G.J. Horizontal gene transfer and the history of life. Cold Spring Harb. Perspect. Biol. 2016, 8, a018036. [Google Scholar] [CrossRef]
- Dagan, T.; Martin, W. The tree of one percent. Genome Biol. 2006, 7, 118. [Google Scholar] [CrossRef][Green Version]
- Di Bonaventura, M.P.; Lee, E.K.; DeSalle, R.; Planet, P.J. A whole-genome phylogeny of the family Pasteurellaceae. Mol. Phylogenetics Evol. 2010, 54, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.; Sneath, P. A numerical phenotypic taxonomic study of the genus Neisseria. Microbiology 1994, 140, 2867–2891. [Google Scholar] [CrossRef] [PubMed]
- Darwin, C. The Origin of Species by Means of Natural Selection, or the Preservation of Favored Races in the Struggle for Life; Murray, J., Ed.; W. Clowes and Sons: London, UK, 1859. [Google Scholar]
- Shute, L.A.; Gutteridge, C.S.; Norris, J.R.; Berkeley, R.C. Curie-point pyrolysis mass spectrometry applied to characterization and identification of selected Bacillus species. J. Gen. Microbiol. 1984, 130, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.; Stevens, M. A numerical taxonomic study of Actinobacillus, Pasteurella, and Yersinia. J. Gen. Microbiol. 1985, 131, 2711–2738. [Google Scholar]
- Mauchline, W.; Keevil, C. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl. Environ. Microbiol. 1991, 57, 3345–3349. [Google Scholar] [CrossRef]
- Kirschner, C.; Maquelin, K.; Pina, P.; Thi, N.N.; Choo-Smith, L.; Sockalingum, G.; Sandt, C.; Ami, D.; Orsini, F.; Doglia, S.; et al. Classification and identification of enterococci: A comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 2001, 39, 1763–1770. [Google Scholar] [CrossRef]
- Cohan, F. Sexual isolation and speciation in bacteria. Genetica 2002, 116, 359–370. [Google Scholar] [CrossRef]
- Lan, R.; Reeves, P.R. Intraspecific variation in bacterial genomes: The need for a species genome concept. Trends Microbiol. 2000, 8, 396–401. [Google Scholar] [CrossRef]
- Thompson, J.; Pacocha, S.; Pharino, C.; Klepac-Ceraj, V.; Hunt, D.; Benoit, J.; Sarma-Rupavtarm, R.; Distel, D.; Polz, M. Genotypic diversity within a natural coastal bacterioplankton population. Science 2005, 307, 1311–1313. [Google Scholar] [CrossRef]
- Godoy, A.; Ribeiro, M.; Benvengo, Y.; Vitiello, L.; Miranda, C.M.; Mendonca, S.; Pedrazzoli, J.J. Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates. BMC Gastroenterol. 2003, 3, 20. [Google Scholar] [CrossRef]
- Thompson, C.C.; Amaral, R.G.; Campeão, M.; Edwards, R.A.; Polz, M.F.; Dutilh, B.E.; Ussery, D.W.; Sawabe, T.; Swings, J.; Thompson, F.L. Microbial taxonomy in the post-genomic era: Rebuilding from scratch? Arch. Microbiol. 2015, 197, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Malaterre, C. Going small: The challenges of microbial diversity. In The Routledge Handbook of Philosophy of Biodiversity; Garson, J., Plutynski, A., Sarkar, S., Eds.; Routledge: New York, NY, USA, 2016; pp. 153–166. [Google Scholar]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.-M. The Prokaryotic Species Concept and Challenges. In The Pangenome; Tettelin, H., Medini, D., Eds.; Springer: Cham, Switzerland, 2020; pp. 21–49. [Google Scholar]
- Louca, S.; Mazel, F.; Doebeli, M.; Parfrey, L.W. A census-based estimate of Earth’s bacterial and archaeal diversity. PLoS Biol. 2019, 17, e3000106. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Shih, P.M.; Pennell, M.W.; Fischer, W.W.; Parfrey, L.W.; Doebeli, M. Bacterial diversification through geological time. Nat. Ecol. Evol. 2018, 2, 1458–1467. [Google Scholar] [CrossRef]
- Palmer, M.; Venter, S.N.; Coetzee, M.P.A.; Steenkamp, E.T. Prokaryotic species are sui generis evolutionary units. Syst. Appl. Microbiol. 2019, 42, 145–158. [Google Scholar] [CrossRef]
- Hayashi Sant’Anna, F.; Bach, E.; Porto, R.Z.; Guella, F.; Sant’Anna, E.H.; Passaglia, L.M.P. Genomic metrics made easy: What to do and where to go in the new era of bacterial taxonomy. Crit. Rev. Microbiol. 2019, 45, 182–200. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
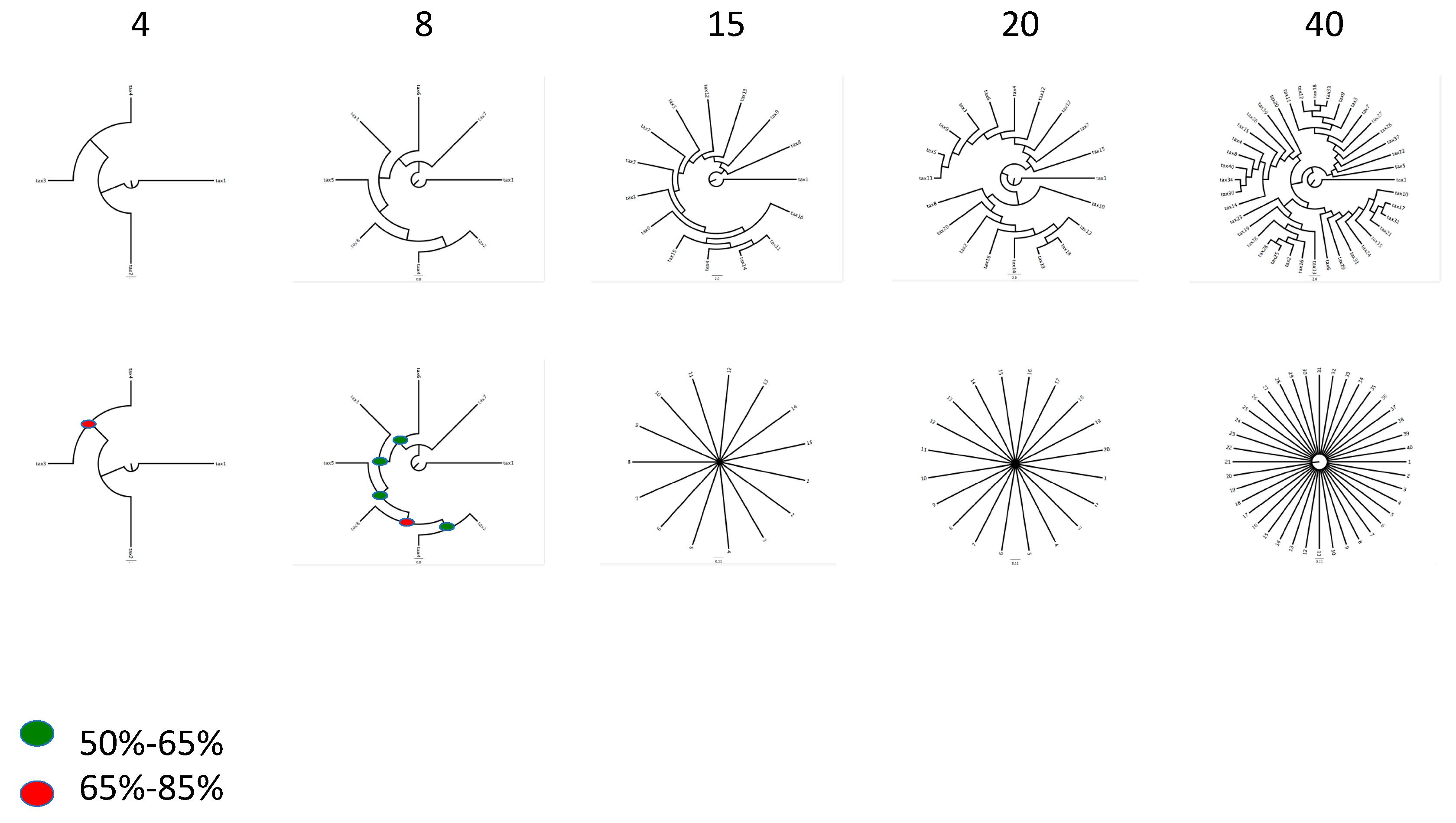
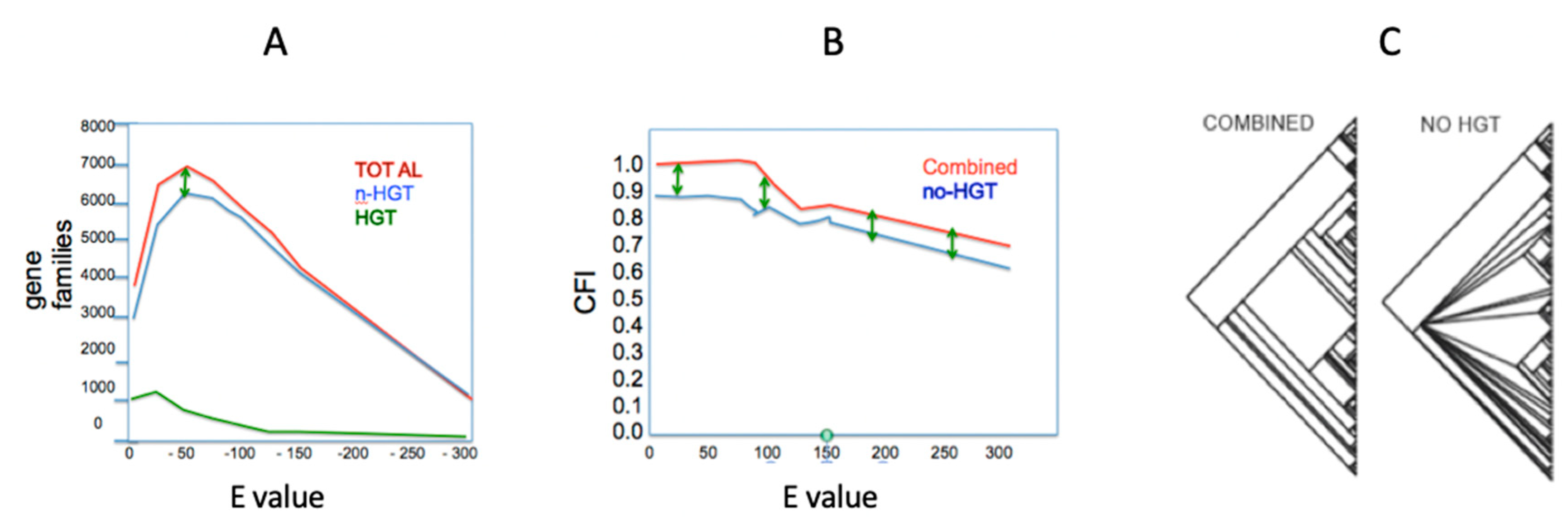
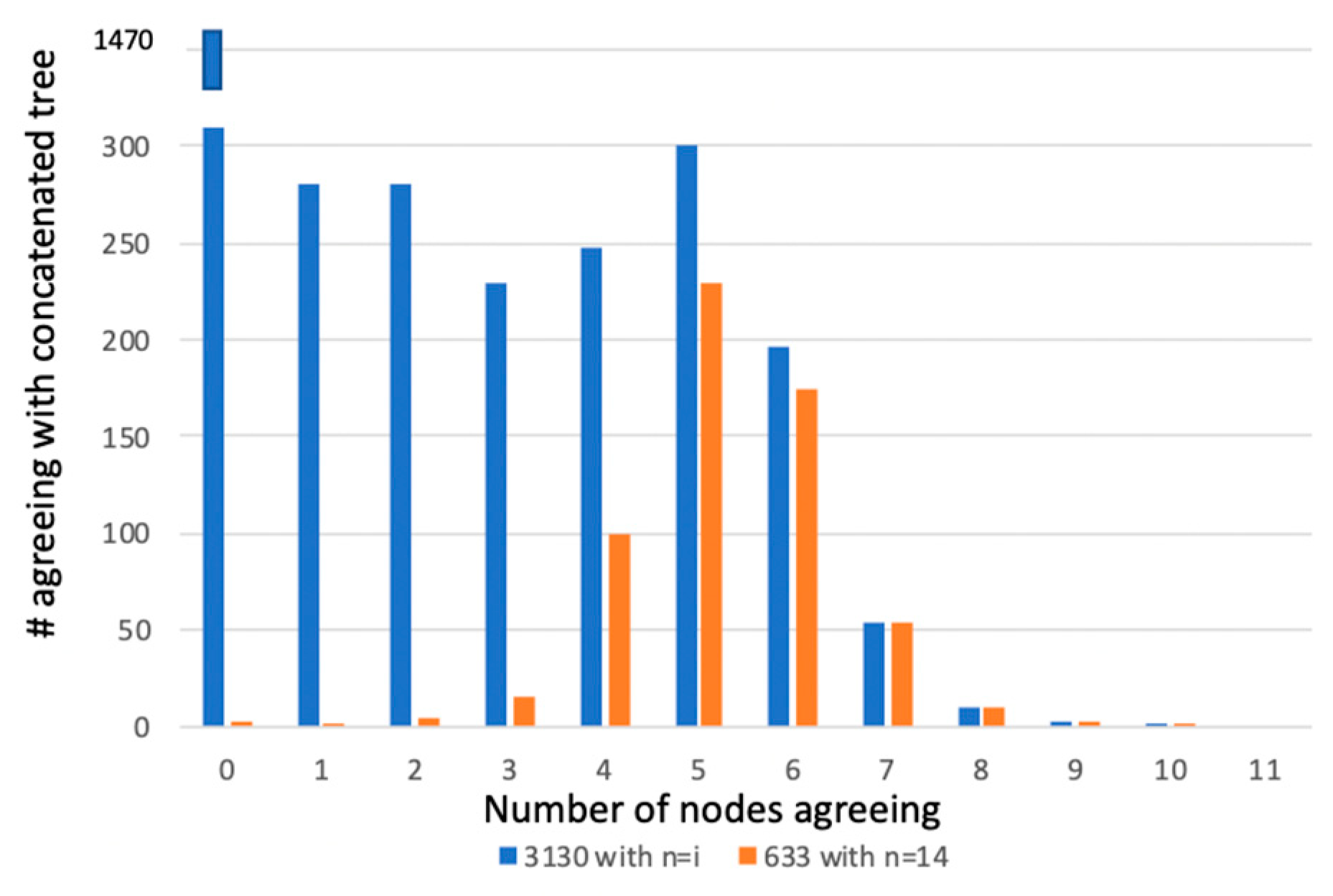
| Hypothesis 1 | A Massively Concatenated Matrix of Genome-Based Information Results in a Generally Unresolved Phylogenetic Tree. |
| If this hypothesis can be rejected then, | |
| Hypothesis 2 | The Tree Generated from a Massively Concatenated Matrix is not Robust. |
| If this hypothesis can be rejected then, | |
| Hypothesis 3 | The Robust Tree Generated from a Massively Concatenated Matrix does not Make Biological Sense (i.e., is in Conflict with Accepted Taxonomic Knowledge). |
| If this hypothesis can be rejected then, | No vertical tree of life exists. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeSalle, R.; Riley, M. Should Networks Supplant Tree Building? Microorganisms 2020, 8, 1179. https://doi.org/10.3390/microorganisms8081179
DeSalle R, Riley M. Should Networks Supplant Tree Building? Microorganisms. 2020; 8(8):1179. https://doi.org/10.3390/microorganisms8081179
Chicago/Turabian StyleDeSalle, Rob, and Margaret Riley. 2020. "Should Networks Supplant Tree Building?" Microorganisms 8, no. 8: 1179. https://doi.org/10.3390/microorganisms8081179
APA StyleDeSalle, R., & Riley, M. (2020). Should Networks Supplant Tree Building? Microorganisms, 8(8), 1179. https://doi.org/10.3390/microorganisms8081179





