A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans versus Omnivores
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Fecal Sample Collection
2.3. Enumeration of E. coli Strains and DNA Extraction
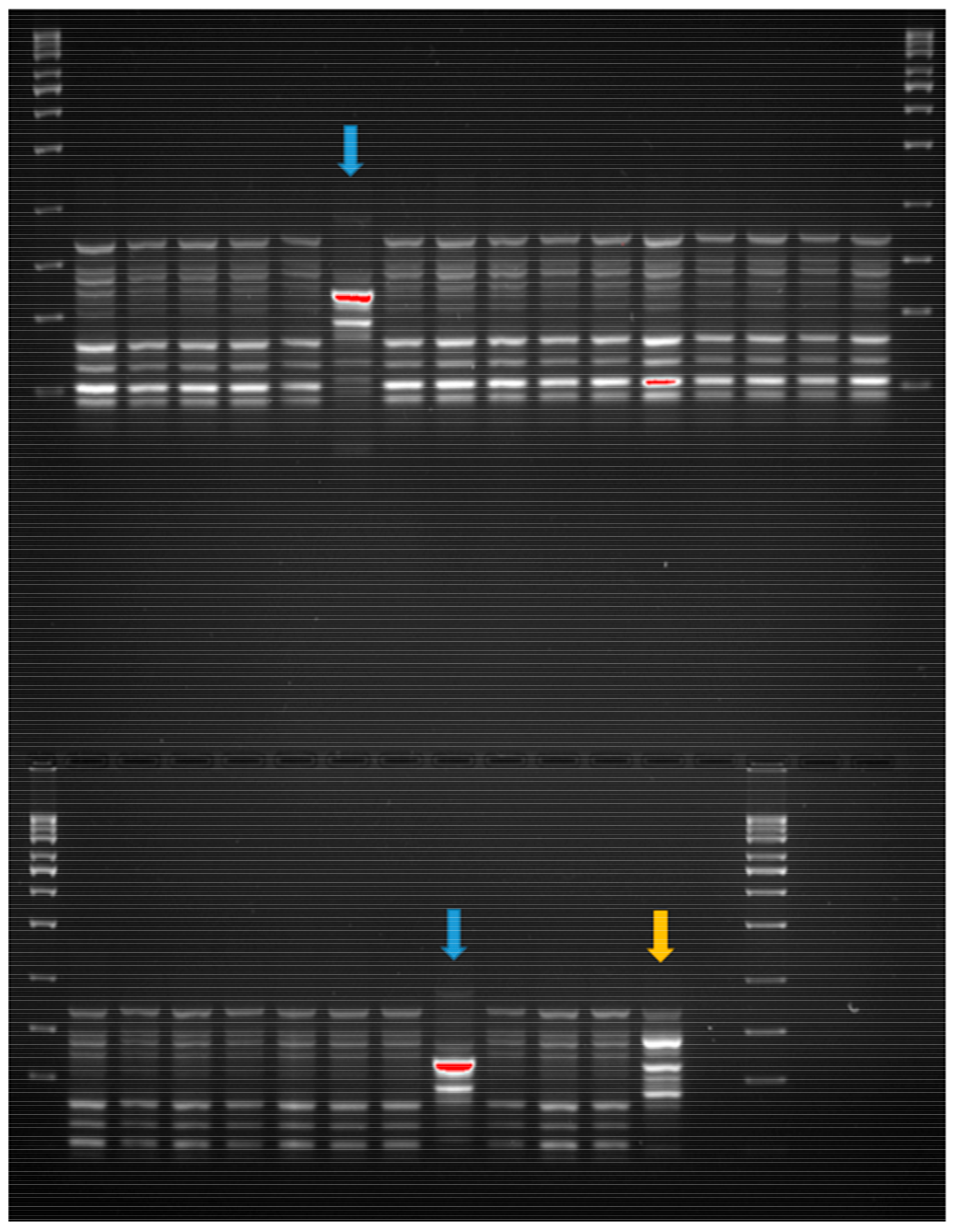
2.4. Clonality Analysis of E. coli Strains
2.5. Detection of AIEC Associated Virulence Genes
2.6. Diversity Profiling of Fecal Microbiota
2.7. Bioinformatics Analysis
2.8. Statistical Analysis
3. Results
3.1. Abundance and Diversity of E. coli
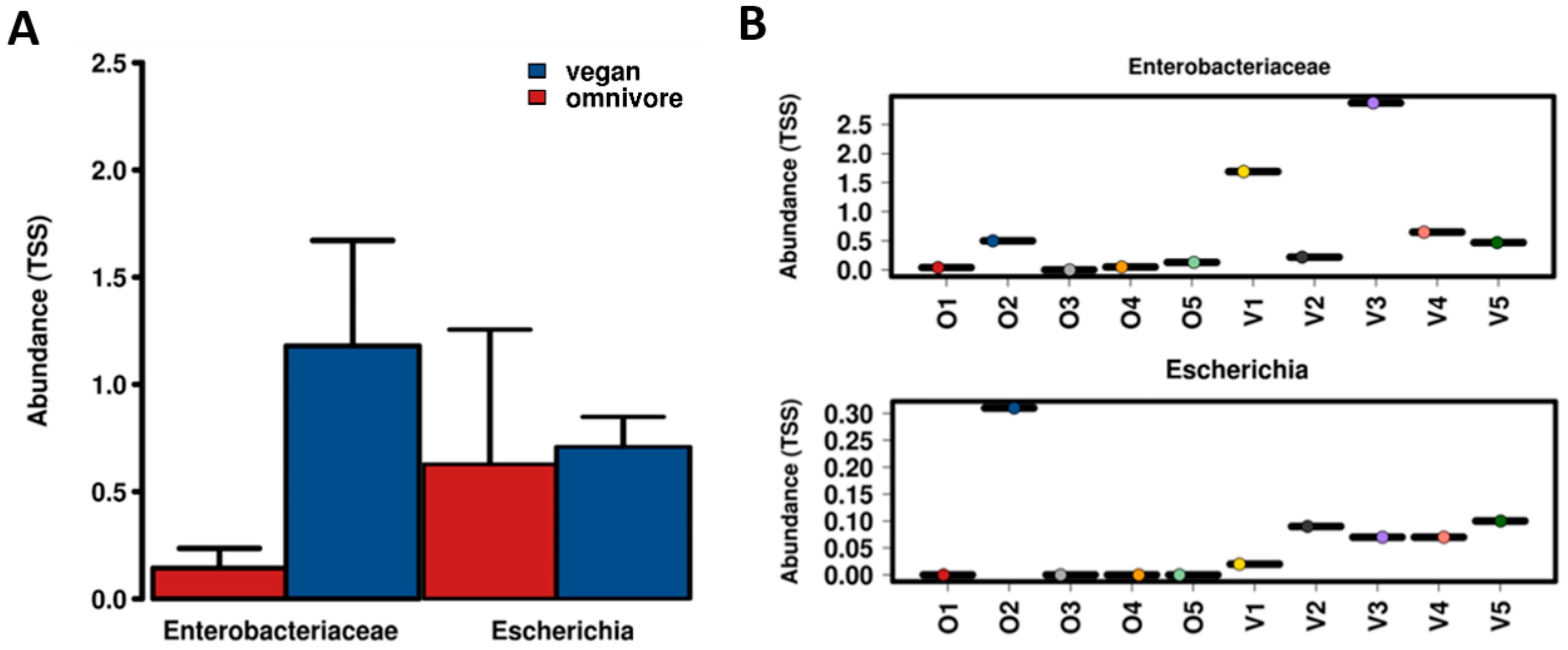
3.2. Prevalence of AIEC Associated VGs
3.3. Diversity of the Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Taxa | Type | Details | Omnivore Abundance | Vegan Abundance | Omnivore Occurrence | Vegan Occurrence |
|---|---|---|---|---|---|---|
| Victivallis | core | omnivore and vegan | 0.016 | 0.362 | 0.4 | 0.4 |
| Veillonella | unique | vegan | 0.008 | 0.046 | 0.2 | 1 |
| Uncultured_organism | unique | vegan | 0.038 | 0.622 | 0.2 | 1 |
| Uncultured_Firmicutes_bacterium | unique | vegan | 0 | 0.014 | 0 | 0.4 |
| Uncultured_bacterium_adhufec202 | unique | vegan | 0.038 | 0.082 | 0.2 | 0.4 |
| Uncultured_bacterium | core | omnivore and vegan | 0.504 | 2.15 | 0.6 | 1 |
| Uncultured | core | omnivore and vegan | 0.768 | 0.584 | 1 | 1 |
| Unclassified | core | omnivore and vegan | 5.086 | 12.826 | 1 | 1 |
| UBA1819 | unique | vegan | 0 | 0.022 | 0 | 0.4 |
| Tyzzerella_3 | unique | vegan | 0 | 0.052 | 0 | 0.8 |
| Tyzzerella | core | omnivore and vegan | 0.074 | 0.086 | 1 | 1 |
| Turicibacter | core | omnivore and vegan | 0.008 | 0.142 | 0.4 | 1 |
| Terrisporobacter | unique | vegan | 0.004 | 0.116 | 0.2 | 0.6 |
| Sutterella | core | omnivore and vegan | 3.33 | 2.566 | 1 | 1 |
| Subdoligranulum | core | omnivore and vegan | 1.344 | 0.552 | 1 | 1 |
| Streptococcus | core | omnivore and vegan | 0.036 | 0.074 | 0.8 | 0.8 |
| Slackia | unique | vegan | 0.018 | 0.124 | 0.2 | 0.8 |
| Ruminococcus_torques_group | core | omnivore and vegan | 0.64 | 0.154 | 1 | 1 |
| Ruminococcus_gauvreauii_group | core | omnivore and vegan | 0.298 | 0.156 | 1 | 1 |
| Ruminococcus_1 | core | omnivore and vegan | 0.484 | 0.952 | 0.8 | 1 |
| Ruminococcaceae_UCG014 | core | omnivore and vegan | 0.852 | 2.462 | 1 | 1 |
| Ruminococcaceae_UCG013 | core | omnivore and vegan | 0.11 | 0.326 | 0.8 | 1 |
| Ruminococcaceae_UCG010 | core | omnivore and vegan | 0.776 | 0.526 | 0.6 | 1 |
| Ruminococcaceae_UCG005 | core | omnivore and vegan | 0.302 | 0.524 | 0.6 | 1 |
| Ruminococcaceae_UCG004 | unique | vegan | 0.01 | 0.014 | 0.2 | 0.4 |
| Ruminococcaceae_UCG003 | core | omnivore and vegan | 0.49 | 0.446 | 0.8 | 1 |
| Ruminococcaceae_UCG002 | core | omnivore and vegan | 1.672 | 2.196 | 1 | 1 |
| Ruminococcaceae_NK4A214_group | core | omnivore and vegan | 0.242 | 0.72 | 0.6 | 1 |
| Ruminiclostridium_9 | core | omnivore and vegan | 0.184 | 0.08 | 1 | 0.8 |
| Ruminiclostridium_6 | core | omnivore and vegan | 0.098 | 0.536 | 0.6 | 0.6 |
| Ruminiclostridium_5 | core | omnivore and vegan | 0.044 | 0.076 | 0.6 | 0.8 |
| Ruminiclostridium | unique | vegan | 0.008 | 0.018 | 0.2 | 0.4 |
| Roseburia | core | omnivore and vegan | 1.308 | 1.666 | 1 | 1 |
| Romboutsia | core | omnivore and vegan | 0.404 | 0.324 | 0.8 | 1 |
| Rikenellaceae_RC9_gut_group | unique | vegan | 0.234 | 0.568 | 0.2 | 0.4 |
| Pseudomonas | core | omnivore and vegan | 0.01 | 0.018 | 0.4 | 0.4 |
| Prevotella_9 | core | omnivore and vegan | 10.928 | 9.87 | 0.6 | 0.8 |
| Prevotella_7 | core | omnivore and vegan | 1.11 | 0.38 | 0.6 | 0.4 |
| Prevotella_2 | core | omnivore and vegan | 6.08 | 0.68 | 0.6 | 0.4 |
| Phascolarctobacterium | core | omnivore and vegan | 1.312 | 0.734 | 0.8 | 0.8 |
| Peptococcus | unique | omnivore | 0.02 | 0.004 | 0.4 | 0.2 |
| Parasutterella | core | omnivore and vegan | 1.014 | 0.506 | 0.6 | 1 |
| Paraprevotella | core | omnivore and vegan | 0.29 | 0.388 | 0.8 | 0.8 |
| Parabacteroides | core | omnivore and vegan | 7.326 | 3.688 | 1 | 1 |
| Oxalobacter | core | omnivore and vegan | 0.026 | 0.062 | 0.4 | 1 |
| Oscillibacter | core | omnivore and vegan | 0.014 | 0.026 | 0.6 | 0.4 |
| Olsenella | unique | omnivore | 0.022 | 0 | 0.4 | 0 |
| Odoribacter | core | omnivore and vegan | 1.02 | 1.168 | 1 | 1 |
| Methanobrevibacter | unique | vegan | 0 | 0.02 | 0 | 0.6 |
| Metagenome | core | omnivore and vegan | 0.79 | 0.476 | 0.4 | 0.6 |
| Marvinbryantia | core | omnivore and vegan | 0.024 | 0.154 | 0.4 | 1 |
| Lactococcus | unique | vegan | 0 | 0.012 | 0 | 0.4 |
| Lactobacillus | unique | vegan | 0.188 | 0.184 | 0.2 | 0.4 |
| Lachnospiraceae_UCG010 | unique | omnivore | 0.012 | 0.006 | 0.6 | 0.2 |
| Lachnospiraceae_UCG008 | core | omnivore and vegan | 0.026 | 0.05 | 0.4 | 0.4 |
| Lachnospiraceae_UCG004 | core | omnivore and vegan | 0.152 | 0.12 | 1 | 0.8 |
| Lachnospiraceae_UCG001 | core | omnivore and vegan | 0.172 | 0.406 | 1 | 0.8 |
| Lachnospiraceae_NK4B4_group | unique | vegan | 0 | 0.014 | 0 | 0.4 |
| Lachnospiraceae_NK4A136_group | core | omnivore and vegan | 2.21 | 1.406 | 0.8 | 1 |
| Lachnospiraceae_ND3007_group | core | omnivore and vegan | 0.174 | 0.334 | 1 | 1 |
| Lachnospiraceae_FCS020_group | core | omnivore and vegan | 0.066 | 0.072 | 0.4 | 1 |
| Lachnospira | core | omnivore and vegan | 0.964 | 1.304 | 1 | 1 |
| Lachnoclostridium | core | omnivore and vegan | 0.592 | 0.2 | 1 | 1 |
| Klebsiella | unique | vegan | 0 | 0.352 | 0 | 0.6 |
| Holdemania | core | omnivore and vegan | 0.016 | 0.024 | 0.4 | 0.6 |
| Haemophilus | core | omnivore and vegan | 0.136 | 0.406 | 0.8 | 1 |
| Gut_metagenome | core | omnivore and vegan | 0.454 | 1.074 | 0.8 | 0.8 |
| Fusobacterium | unique | vegan | 0 | 0.024 | 0 | 0.4 |
| Fusicatenibacter | core | omnivore and vegan | 0.76 | 0.73 | 1 | 1 |
| Fournierella | unique | omnivore | 0.082 | 0.01 | 0.4 | 0.2 |
| Flavonifractor | core | omnivore and vegan | 0.072 | 0.034 | 0.4 | 0.6 |
| Family_XIII_AD3011_group | core | omnivore and vegan | 0.02 | 0.054 | 0.4 | 1 |
| Faecalibacterium | core | omnivore and vegan | 6.526 | 5.578 | 1 | 1 |
| Eubacterium_xylanophilum_group | core | omnivore and vegan | 0.452 | 0.346 | 0.6 | 1 |
| Eubacterium_ventriosum_group | core | omnivore and vegan | 0.128 | 0.278 | 0.8 | 1 |
| Eubacterium_ruminantium_group | core | omnivore and vegan | 0.25 | 0.334 | 0.4 | 0.4 |
| Eubacterium_nodatum_group | unique | omnivore | 0.006 | 0 | 0.4 | 0 |
| Eubacterium_hallii_group | core | omnivore and vegan | 0.29 | 0.676 | 1 | 1 |
| Eubacterium_eligens_group | core | omnivore and vegan | 0.652 | 0.552 | 1 | 1 |
| Eubacterium_coprostanoligenes_group | core | omnivore and vegan | 1.798 | 1.206 | 1 | 1 |
| EscherichiaShigella | unique | vegan | 0.062 | 0.07 | 0.2 | 1 |
| Erysipelotrichaceae_UCG004 | core | omnivore and vegan | 0.036 | 0.072 | 0.4 | 0.4 |
| Erysipelotrichaceae_UCG003 | core | omnivore and vegan | 0.848 | 0.692 | 1 | 1 |
| Erysipelatoclostridium | core | omnivore and vegan | 0.022 | 0.042 | 0.4 | 0.6 |
| Enterorhabdus | core | omnivore and vegan | 0.02 | 0.026 | 0.4 | 0.4 |
| Enterococcus | unique | vegan | 0 | 0.028 | 0 | 0.6 |
| Enterobacter | core | omnivore and vegan | 0.01 | 0.188 | 0.4 | 1 |
| Dorea | core | omnivore and vegan | 0.698 | 0.498 | 1 | 1 |
| Dialister | core | omnivore and vegan | 0.158 | 0.29 | 0.4 | 1 |
| Desulfovibrio | core | omnivore and vegan | 0.196 | 0.238 | 0.4 | 1 |
| Delftia | unique | omnivore | 0.05 | 0 | 0.8 | 0 |
| Coprococcus_3 | core | omnivore and vegan | 0.152 | 0.084 | 1 | 0.8 |
| Coprococcus_2 | core | omnivore and vegan | 0.778 | 1.59 | 0.8 | 1 |
| Coprococcus_1 | core | omnivore and vegan | 0.036 | 0.034 | 0.8 | 0.8 |
| Coprobacter | core | omnivore and vegan | 0.272 | 0.328 | 0.6 | 1 |
| Collinsella | core | omnivore and vegan | 0.69 | 0.482 | 1 | 1 |
| Clostridium_sensu_stricto_1 | core | omnivore and vegan | 0.376 | 0.188 | 0.6 | 1 |
| Cloacibacillus | unique | vegan | 0 | 0.012 | 0 | 0.4 |
| Christensenellaceae_R7_group | core | omnivore and vegan | 0.342 | 1.446 | 0.8 | 1 |
| Candidatus_Stoquefichus | unique | vegan | 0 | 0.006 | 0 | 0.4 |
| CAG56 | core | omnivore and vegan | 0.034 | 0.112 | 0.4 | 1 |
| CAG352 | unique | vegan | 0.008 | 0.054 | 0.2 | 0.4 |
| Butyricimonas | core | omnivore and vegan | 0.392 | 0.332 | 0.6 | 1 |
| Butyricicoccus | core | omnivore and vegan | 0.094 | 0.356 | 1 | 1 |
| Blautia | core | omnivore and vegan | 1.358 | 1.778 | 1 | 1 |
| Blastocystis_sp._subtype_3 | unique | vegan | 0.532 | 0.766 | 0.2 | 0.4 |
| Bilophila | core | omnivore and vegan | 0.204 | 0.092 | 1 | 1 |
| Bifidobacterium | core | omnivore and vegan | 0.614 | 1.182 | 1 | 1 |
| Barnesiella | core | omnivore and vegan | 0.276 | 0.768 | 0.8 | 1 |
| Bacteroides_pectinophilus_group | unique | vegan | 0.002 | 0.024 | 0.2 | 0.6 |
| Bacteroides | core | omnivore and vegan | 17.366 | 12.984 | 1 | 1 |
| Bacteroidales_bacterium_55_9 | unique | omnivore | 0.312 | 0.066 | 0.4 | 0.2 |
| Azospirillum_sp._47_25 | unique | vegan | 0 | 0.004 | 0 | 0.4 |
| Asteroleplasma | unique | vegan | 0.01 | 0.022 | 0.2 | 0.4 |
| Anaerostipes | core | omnivore and vegan | 1.068 | 1.132 | 1 | 1 |
| Anaerosporobacter | core | omnivore and vegan | 0.058 | 0.05 | 0.4 | 0.8 |
| Alloprevotella | core | omnivore and vegan | 0.952 | 0.082 | 0.4 | 0.4 |
| Alistipes | core | omnivore and vegan | 2.284 | 3.892 | 1 | 1 |
| Akkermansia | core | omnivore and vegan | 0.066 | 1.286 | 0.4 | 1 |
| Agathobacter | core | omnivore and vegan | 1.876 | 1.25 | 1 | 1 |
| Adlercreutzia | unique | vegan | 0.004 | 0.028 | 0.2 | 0.8 |
| Actinomyces | core | omnivore and vegan | 0.006 | 0.004 | 0.4 | 0.4 |
| Acidaminococcus | unique | vegan | 0.028 | 0.102 | 0.2 | 0.4 |
References
- Burisch, J.; Munkholm, P. The epidemiology of inflammatory bowel disease. Scand. J. Gastroenterol. 2015, 50, 942–951. [Google Scholar] [CrossRef]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Li, Y.-Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2013, 63, 761–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolhion, N.; Darfeuille-Michaud, A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Kohoutová, D.; Drahosova, M.; Cihak, M.; Moravkova, P.; Bures, J. Anti-Outer membrane protein C antibodies in colorectal neoplasia. Folia Microbiol. 2015, 61, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrede, J.-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahmouni, O.; Vignal, C.; Titécat, M.; Foligné, B.; Pariente, B.; Dubuquoy, L.; Desreumaux, P.; Neut, C. High carriage of adherent invasive E. coli in wildlife and healthy individuals. Gut Pathog. 2018, 10, 23. [Google Scholar] [CrossRef]
- Barnich, N.; Darfeuille-Michaud, A. Abnormal CEACAM6 expression in Crohn disease patients favors gut colonization and inflammation by Adherent-Invasive E. coli. Virulence 2010, 1, 281–282. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Albenberg, L.; Compher, C.; Baldassano, R.; Piccoli, D.; Lewis, J.D.; Wu, G.D. Diet in the Pathogenesis and Treatment of Inflammatory Bowel Diseases. Gastroenterology 2015, 148, 1087–1106. [Google Scholar] [CrossRef] [Green Version]
- Schreiner, P.; Yilmaz, B.; Franc, Y.; Rossel, J.-B.; Misselwitz, B.; Scharl, M.; Zeitz, J.; Frei, P.; Greuter, T.; Vavricka, S.; et al. P855 Vegetarian and gluten-free diet in patients with IBD—associated with a different microbiota compared with omnivore IBD patients. J. Crohn’s Colitis 2018, 12, S549. [Google Scholar] [CrossRef] [Green Version]
- Key, T.J.; Appleby, P.N.; Rosell, M.S. Health effects of vegetarian and vegan diets. Proc. Nutr. Soc. 2006, 65, 35–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary Intake and Risk of Developing Inflammatory Bowel Disease: A Systematic Review of the Literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, J.; Lange, B.; Frick, J.-S.; Sauer, H.; Zimmermann, K.; Schwiertz, A.; Rusch, K.; Klosterhalfen, S.; Enck, P. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur. J. Clin. Nutr. 2011, 66, 53–60. [Google Scholar] [CrossRef]
- Kim, M.-S.; Hwang, S.-S.; Park, E.-J.; Bae, J.W. Strict vegetarian diet improves the risk factors associated with metabolic diseases by modulating gut microbiota and reducing intestinal inflammation. Environ. Microbiol. Rep. 2013, 5, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Van Faassen, A.; Bol, J.; Van Dokkum, W.; A Pikaar, N.; Ockhuizen, T.; Hermus, R.J. Bile acids, neutral steroids, and bacteria in feces as affected by a mixed, a lacto-ovovegetarian, and a vegan diet. Am. J. Clin. Nutr. 1987, 46, 962–967. [Google Scholar] [CrossRef]
- Kabeerdoss, J.; Devi, R.S.; Mary, R.R.; Ramakrishna, B.S. Faecal microbiota composition in vegetarians: Comparison with omnivores in a cohort of young women in southern India. Br. J. Nutr. 2011, 108, 953–957. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.D.; Compher, C.; Chen, E.Z.; A Smith, S.; Shah, R.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef]
- Ferrocino, I.; Di Cagno, R.; De Angelis, M.; Turroni, S.; Vannini, L.; Bancalari, E.; Rantsiou, K.; Cardinali, G.; Neviani, E.; Cocolin, L. Fecal microbiota in healthy subjects following omnivore, vegetarian and vegan diets: Culturable populations and rRNA DGGE profiling. PLoS ONE 2015, 10, e0128669. [Google Scholar] [CrossRef] [Green Version]
- LoSasso, C.; Eckert, E.M.; Mastrorilli, E.; Villiger, J.; Mancin, M.; Patuzzi, I.; Di Cesare, A.; Cibin, V.; Barrucci, F.; Pernthaler, J.; et al. Assessing the influence of vegan, vegetarian and omnivore oriented westernized dietary styles on human gut microbiota: A cross sectional study. Front. Microbiol. 2018, 9, 317. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2013, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2012, 6, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2010, 108, 4554–4561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoetendal, E.; Rajilić-Stojanović, M.; De Vos, W.M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef]
- Walker, A.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010, 5, 220–230. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Casella, G.; Simell, O.; Krischer, J.; Wasserfall, C.; Schatz, D.; Atkinson, M.A.; Neu, J.; Triplett, E.W. Influence of fecal sample storage on bacterial community diversity. Open Microbiol. J. 2009, 3, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Griffiths, M.W. PCR differentiation of Escherichia coli from other gram-negative bacteria using primers derived from the nucleotide sequences flanking the gene encoding the universal stress protein. Lett. Appl. Microbiol. 1998, 27, 369–371. [Google Scholar] [CrossRef]
- Smith, S.I.; Fowora, M.A.; Otegbayo, J.A.; Abdulkareem, F.B.; Omonigbehin, E.A.; Adegboyega, A. Comparison of PCR with other diagnostic techniques for the detection of H. pylori infection in patients presenting with gastroduodenal symptoms in Nigeria. Int. J. Mol. Epidemiol. Genet. 2011, 2, 178–184. [Google Scholar]
- Ramos, N.L.; Saayman, M.L.; A Chapman, T.; Tucker, J.; Smith, H.V.; Faoagali, J.; Chin, J.C.; Brauner, A.; Katouli, M. Genetic relatedness and virulence gene profiles of Escherichia coli strains isolated from septicaemic and uroseptic patients. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 29, 15–23. [Google Scholar] [CrossRef]
- Hatje, E.; Neuman, C.; Stevenson, H.; Bowman, J.P.; Katouli, M. Population dynamics of vibrio and pseudomonas species isolated from farmed Tasmanian Atlantic salmon (Salmo salar L.): A seasonal study. Microb. Ecol. 2014, 68, 679–687. [Google Scholar] [CrossRef]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Lim, M.Y.; Song, E.-J.; Kim, S.H.; Lee, J.; Nam, Y.-D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst. Appl. Microbiol. 2018, 41, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Bharucha, T.; Breuer, J. Encephalitis diagnosis using metagenomics: Application of next generation sequencing for undiagnosed cases. J. Infect. 2018, 76, 225–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. Curr. Protoc. Microbiol. 2012, 27, 1E.5.1–1E.5.20. [Google Scholar]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.-J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinform. 2016, 33, 782–783. [Google Scholar] [CrossRef] [Green Version]
- Edberg, S.; Rice, E.; Karlin, R.; Allen, M. Escherichia coli: The best biological drinking water indicator for public health protection. Symp. Ser. Soc. Appl. Microbiol. 2000, 88, 106S–116S. [Google Scholar] [CrossRef]
- Owrangi, B.; Masters, N.; Vollmerhausen, T.; O’Dea, C.; Kuballa, A.; Katouli, M. Comparison between virulence characteristics of dominant and non-dominant Escherichia coli strains of the gut and their interaction with Caco-2 cells. Microb. Pathog. 2017, 105, 171–176. [Google Scholar] [CrossRef]
- Vollmerhausen, T.L.; Ramos, N.L.; Gundogdu, A.; Robinson, W.; Brauner, A.; Katouli, M. Population structure and uropathogenic virulence-associated genes of faecal Escherichia coli from healthy young and elderly adults. J. Med Microbiol. 2011, 60, 574–581. [Google Scholar] [CrossRef] [Green Version]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; De Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.G.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2010, 108, 4586–4591. [Google Scholar] [CrossRef] [Green Version]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Genet. 2010, 8, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Filippis, F.; Pellegrini, N.; Laghi, L.; Gobbetti, M.; Ercolini, D. Unusual sub-genus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome 2016, 4, 57. [Google Scholar] [CrossRef] [Green Version]
- Pulkrabek, M.; Rhee, Y.S.; Gibbs, P.; Hall, C. Flaxseed and Buckwheat-Supplemented Diets Altered Enterobacteriaceae diversity and prevalence in the cecum and feces of obese mice. J. Diet. Suppl. 2017, 14, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Rogerson, D. Vegan diets: Practical advice for athletes and exercisers. J. Int. Soc. Sports Nutr. 2017, 14, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, F. Vegetarian nutrition. Nutr. Bull. 2005, 30, 132–167. [Google Scholar] [CrossRef]



| Participant Diet | Participant No. | Female | Male | Age Range (Years) | Median Age (Years) | Months Being Vegan | Mean± SD Months Being Vegan |
|---|---|---|---|---|---|---|---|
| Vegan | 34 | 22 | 12 | 20–67 | 29 ± 1 | 1–108 | 36 ± 29 |
| Omnivore | 27 | 14 | 13 | 20–60 | 29 ± 1 | NA | NA |
| Gender | E. coli CFU/g of Faeces | ||
|---|---|---|---|
| Vegan | Omnivore | p-Value | |
| Female | 9.3 ± 4.4 × 105 | 7.9 ± 3.8 × 105 | 0.602 |
| Male | 9.7 ± 6.2 × 106 | 5.9 ± 3.1 × 105 | 0.079 |
| Combined Gender | 3.4 ± 2.2 × 106 | 9.6 ± 2.4 × 105 | 0.116 |
| E. coli Diversity | |||
| Female | 0.195 ± 0.06 | 0.199 ± 0.06 | 0.608 |
| Male | 0.201 ± 0.09 | 0.112 ± 0.06 | 0.519 |
| Combined Gender | 0.197 ± 0.05 | 0.157 ± 0.04 | 0.879 |
| No. of CTs | Average CTs pp | CT Range pp | Total STs | No. of STs | Average STs pp | ST Range pp | |
|---|---|---|---|---|---|---|---|
| Vegan | 59 | 1.7 | 1–5 | 48 | 30 | 0.9 | 0–8 |
| Omnivore | 40 | 1.5 | 1–4 | 18 | 0.7 | 0–6 |
| No. of CTs | No. of E. coli within CTs | Occurrence | ||||
|---|---|---|---|---|---|---|
| Virulence Genes | Vegan n = 59 | Omnivore n = 40 | Vegan n = 922 | Omnivore n = 738 | Vegan | Omnivore |
| htrA | 56 | 40 | 901 | 738 | 97% | 100% |
| lpfA | 25 | 18 | 500 | 316 | 54% | 43% |
| ompC | 59 | 40 | 922 | 738 | 100% | 100% |
| clbA | 7 | 6 | 77 | 113 | 8% | 15% |
| dsbA | 48 | 34 | 853 | 633 | 93% | 86% |
| afaC | 17 | 18 | 338 | 287 | 37% | 39% |
| afaC and lpfA | 11 | 14 | 241 | 213 | 26% | 29% |
| afaC and clbA | 1 | 0 | 8 | 0 | 1% | 0% |
| afaC and dsbA | 17 | 17 | 338 | 283 | 37% | 38% |
| dsbA and htrA | 48 | 34 | 853 | 633 | 93% | 86% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veca, R.; O’Dea, C.; Burke, J.; Hatje, E.; Kuballa, A.; Katouli, M. A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans versus Omnivores. Microorganisms 2020, 8, 1165. https://doi.org/10.3390/microorganisms8081165
Veca R, O’Dea C, Burke J, Hatje E, Kuballa A, Katouli M. A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans versus Omnivores. Microorganisms. 2020; 8(8):1165. https://doi.org/10.3390/microorganisms8081165
Chicago/Turabian StyleVeca, Rebecca, Christian O’Dea, Jarred Burke, Eva Hatje, Anna Kuballa, and Mohammad Katouli. 2020. "A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans versus Omnivores" Microorganisms 8, no. 8: 1165. https://doi.org/10.3390/microorganisms8081165
APA StyleVeca, R., O’Dea, C., Burke, J., Hatje, E., Kuballa, A., & Katouli, M. (2020). A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans versus Omnivores. Microorganisms, 8(8), 1165. https://doi.org/10.3390/microorganisms8081165






