Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design
2.2. Sample Collection and Analysis
2.3. Antimicrobial Susceptibility Testing
2.4. DNA Extraction
2.5. Whole Genome Sequencing
2.6. Draft Assembly of Whole Genome Sequences (WGS), in Silico Multi-Locus Sequence Typing (MLST) and Phylogeny Analysis
2.7. Testing for Resistance Genes
2.8. Statistical Analysis
3. Results
3.1. Study Population
3.2. Risk Factors of ESBL-Enterobacteriacae Carriage
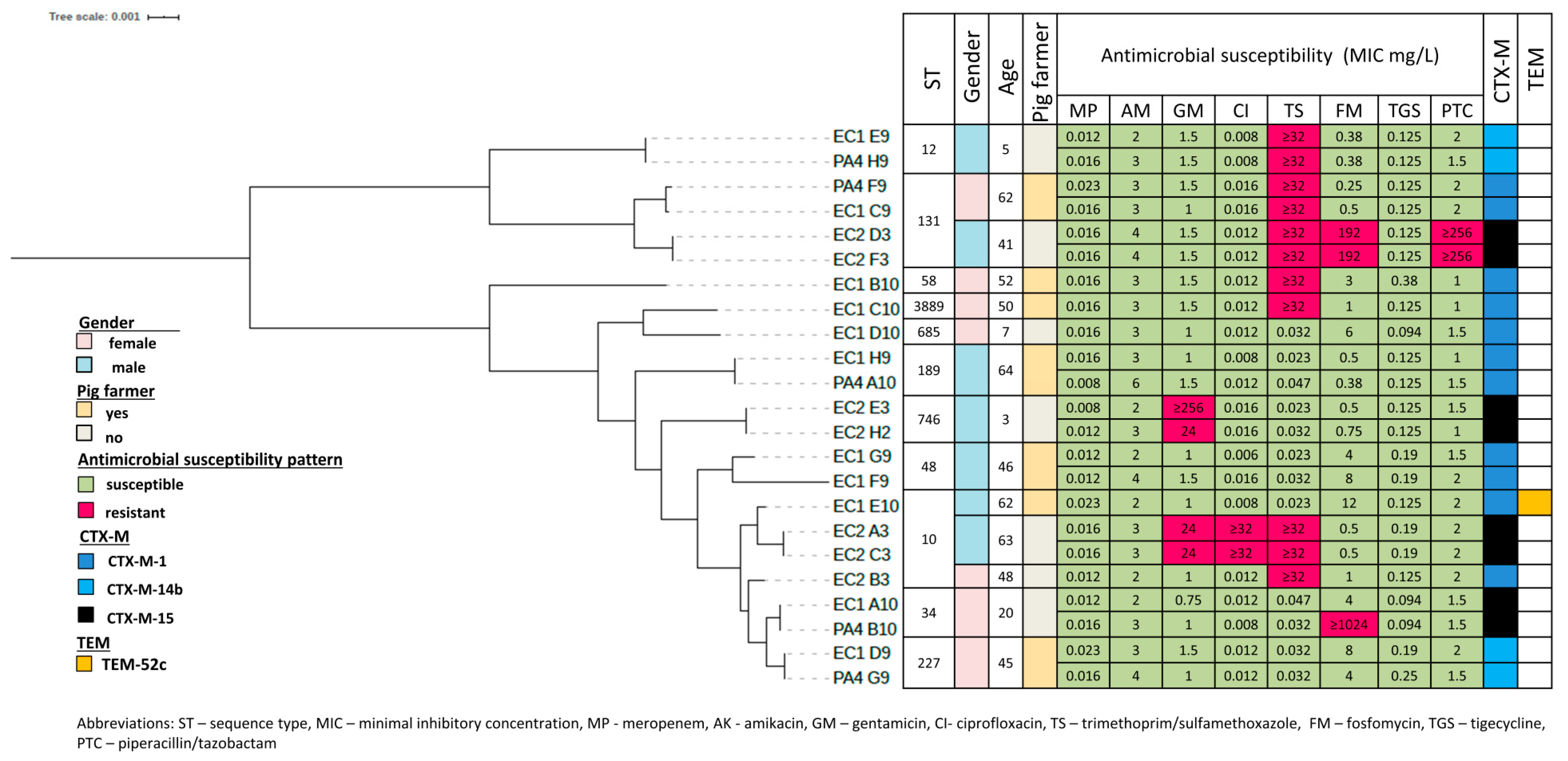
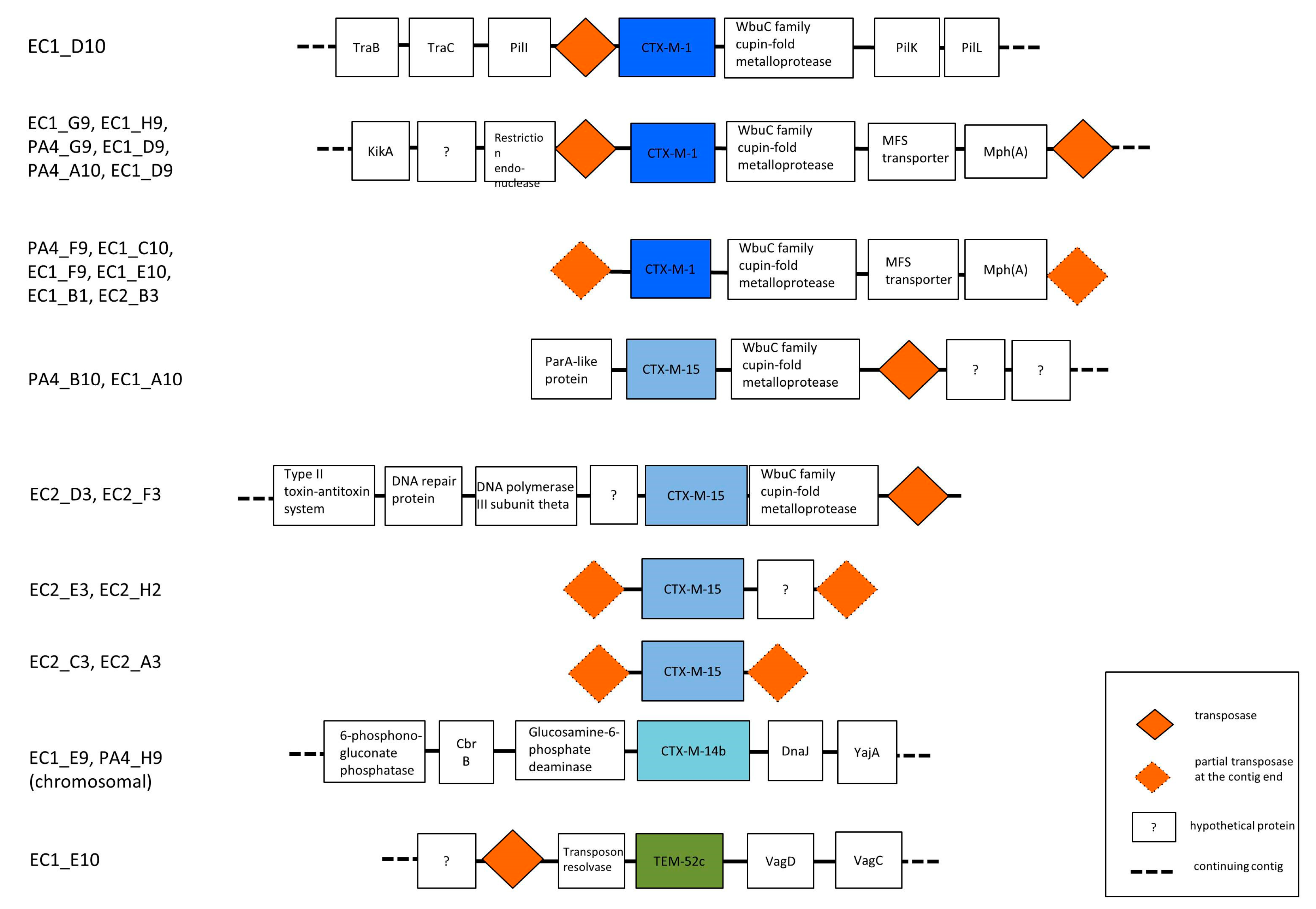
3.3. Phylogenetic Grouping, Antimicrobial Susceptibility and ESBL Encoding Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum beta-lactamases (ESBLs) in the developed world. J. Travel Med. 2017, 24, S44–S51. [Google Scholar] [CrossRef] [PubMed]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal Colonization with Extended-spectrum Beta-lactamase-Producing Enterobacteriaceae and Risk Factors Among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. 2016, 63, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Ny, S.; Kozlov, R.; Dumpis, U.; Edquist, P.; Grondahl-Yli-Hannuksela, K.; Kling, A.M.; Lis, D.O.; Lubbert, C.; Pomorska-Wesolowska, M.; Palagin, I.; et al. Large variation in ESBL-producing Escherichia coli carriers in six European countries including Russia. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2347–2354. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, X.; Zhang, J.; Tao, X.; Deng, Z.; Hu, Y.; Li, M.; Yang, X.; Wang, M.; Yang, Z. High Prevalence of CTX-M Beta-Lactamases in Enterobacteriaceae from Healthy Individuals in Guangzhou, China. Microb. Drug Resist. 2015, 21, 398–403. [Google Scholar] [CrossRef]
- Rodrigues, C.; Machado, E.; Fernandes, S.; Peixe, L.; Novais, A. An update on faecal carriage of ESBL-producing Enterobacteriaceae by Portuguese healthy humans: Detection of the H30 subclone of B2-ST131 Escherichia coli producing CTX-M-27. J. Antimicrob. Chemother. 2016, 71, 1120–1122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M beta-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control Homepage. Surveillance of Antimicrobial Resistance in Europe. 2017. Available online: http://ecdc.europa.eu/en/antimicrobial-resistance/surveillance-and-disease-data (accessed on 1 December 2019).
- Lubbert, C.; Straube, L.; Stein, C.; Makarewicz, O.; Schubert, S.; Mossner, J.; Pletz, M.W.; Rodloff, A.C. Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae in international travelers returning to Germany. Int. J. Med. Microbiol. 2015, 305, 148–156. [Google Scholar] [CrossRef]
- Ny, S.; Lofmark, S.; Borjesson, S.; Englund, S.; Ringman, M.; Bergstrom, J.; Naucler, P.; Giske, C.G.; Byfors, S. Community carriage of ESBL-producing Escherichia coli is associated with strains of low pathogenicity: A Swedish nationwide study. J. Antimicrob. Chemother. 2017, 72, 582–588. [Google Scholar] [CrossRef]
- Woerther, P.L.; Andremont, A.; Kantele, A. Travel-acquired ESBL-producing Enterobacteriaceae: Impact of colonization at individual and community level. J. Travel Med. 2017, 24, S29–S34. [Google Scholar] [CrossRef]
- Madec, J.Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum beta-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- De Been, M.; Lanza, V.F.; De Toro, M.; Scharringa, J.; Dohmen, W.; Du, Y.; Hu, J.; Lei, Y.; Li, N.; Tooming-Klunderud, A.; et al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 2014, 10, e1004776. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, B.; Paterson, D.L.; Mollinger, J.L.; Rogers, B.A. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 2015, 60, 439–452. [Google Scholar] [CrossRef]
- Doi, Y.; Park, Y.S.; Rivera, J.I.; Adams-Haduch, J.; Hingwe, A.; Sordillo, E.M.; Lewis, J.S.; Howard, W.J.; Johnson, L.E.; Polsky, B.; et al. Community-Associated Extended-Spectrum β-Lactamase–Producing Escherichia coli Infection in the United States. Clin. Infect. Dis. 2013, 56, 641–648. [Google Scholar] [CrossRef]
- Sepp, E.; Andreson, R.; Balode, A.; Bilozor, A.; Brauer, A.; Egorova, S.; Huik, K.; Ivanova, M.; Kaftyreva, L.; Koljalg, S.; et al. Phenotypic and Molecular Epidemiology of ESBL-, AmpC-, and Carbapenemase-Producing Escherichia coli in Northern and Eastern Europe. Front. Microbiol. 2019, 10, 2465. [Google Scholar] [CrossRef]
- Bonnet, R.; Recule, C.; Baraduc, R.; Chanal, C.; Sirot, D.; De Champs, C.; Sirot, J. Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 2003, 52, 29–35. [Google Scholar] [CrossRef]
- European Society of Clinical Microbiology and Infectious Diseases Homepage. Clinical Breakpoints and Dosing. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 25 May 2019).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.; Wertheim-van Dillen, P. Rapid purification of ribosomal RNAs from neutral agarose gels. Nucleic Acids Res. 1990, 18, 2195. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Marttinen, P.; Hanage, W.P.; Croucher, N.J.; Connor, T.R.; Harris, S.R.; Bentley, S.D.; Corander, J. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012, 40, e6. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.J.; Zhang, Z.; Miller, W.; Lipman, J.D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- StataCorp. Stata Statistical Software: Release 14; StataCorp LP: College Station, TX, USA, 2015. [Google Scholar]
- European Society of Clinical Microbiology and Infectious Diseases Homepage. Guideline for the Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Available online: http://www.eucast.org/resistance_mechanisms/ (accessed on 22 May 2019).
- Niumsup, P.R.; Tansawai, U.; Na-Udom, A.; Jantapalaboon, D.; Assawatheptawee, K.; Kiddee, A.; Romgaew, T.; Lamlertthon, S.; Walsh, T.R. Prevalence and risk factors for intestinal carriage of CTX-M-type ESBLs in Enterobacteriaceae from a Thai community. Eur. J. Clin. Microbiol. Infect. Dis. 2017. [Google Scholar] [CrossRef]
- Zhong, Y.M.; Liu, W.E.; Liang, X.H.; Li, Y.M.; Jian, Z.J.; Hawkey, P.M. Emergence and spread of O16-ST131 and O25b-ST131 clones among faecal CTX-M-producing Escherichia coli in healthy individuals in Hunan Province, China. J. Antimicrob. Chemother. 2015, 70, 2223–2227. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Lester, C.H.; Jakobsen, L.; Porsbo, L.J. Faecal carriage of extended-spectrum beta-lactamase-producing and AmpC beta-lactamase-producing bacteria among Danish army recruits. Clin. Microbiol. Infect. 2011, 17, 566–568. [Google Scholar] [CrossRef]
- Ulstad, C.R.; Solheim, M.; Berg, S.; Lindbaek, M.; Dahle, U.R.; Wester, A.L. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob. Resist. Infect. Control 2016, 5, 57. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control Homepage. Available online: https://www.ecdc.europa.eu/en/antimicrobial-consumption/database/rates-country (accessed on 20 April 2020).
- Dahms, C.; Hubner, N.O.; Kossow, A.; Mellmann, A.; Dittmann, K.; Kramer, A. Occurrence of ESBL-Producing Escherichia coli in Livestock and Farm Workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE 2015, 10, e0143326. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, W.; Van Gompel, L.; Schmitt, H.; Liakopoulos, A.; Heres, L.; Urlings, B.A.; Mevius, D.; Bonten, M.J.M.; Heederik, D.J.J. ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol. Infect. 2017, 145, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Hille, K.; Ruddat, I.; Mellmann, A.; Kock, R.; Kreienbrock, L. Simultaneous occurrence of MRSA and ESBL-producing Enterobacteriaceae on pig farms and in nasal and stool samples from farmers. Vet. Microbiol. 2017, 200, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Schmithausen, R.M.; Schulze-Geisthoevel, S.V.; Stemmer, F.; El-Jade, M.; Reif, M.; Hack, S.; Meilaender, A.; Montabauer, G.; Fimmers, R.; Parcina, M.; et al. Analysis of Transmission of MRSA and ESBL-E among Pigs and Farm Personnel. PLoS ONE 2015, 10, e0138173. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, Z.; Li, Q.; Liu, L.; Guo, S.; Yang, L.; Ye, C.; Chang, W.; Zhai, J. Characterization of Extended-Spectrum beta-Lactamase-Producing Escherichia coli Isolates from Pigs and Farm Workers. J. Food Prot. 2016, 79, 1630–1634. [Google Scholar] [CrossRef]
- Aasmae, B.; Hakkinen, L.; Kaart, T.; Kalmus, P. Antimicrobial resistance of Escherichia coli and Enterococcus spp. isolated from Estonian cattle and swine from 2010 to 2015. Acta Vet. Scand. 2019, 61, 5. [Google Scholar] [CrossRef]
- Dohmen, W.; Schmitt, H.; Bonten, M.; Heederik, D. Air exposure as a possible route for ESBL in pig farmers. Environ. Res. 2017, 155, 359–364. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Larsen, J.; Andersen, V.D.; Lester, C.H.; Skytte, T.S.S.; Hansen, F.; Olsen, S.S.; Mordhorst, H.; Skov, R.L.; Aarestrup, F.M.; et al. Characterization of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 2014, 69, 2650–2657. [Google Scholar] [CrossRef]
- Cortes-Cortes, G.; Lozano-Zarain, P.; Torres, C.; Alonso, C.A.; Rios-Torres, A.M.; Castaneda, M.; Lopez-Pliego, L.; Navarro, A.; Del Carmen Rocha-Gracia, R. Extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans in Mexico, including subclone ST131-B2-O25:H4-H30-Rx. J. Glob. Antimicrob. Resist. 2017, 9, 130–134. [Google Scholar] [CrossRef]
- Valenza, G.; Nickel, S.; Pfeifer, Y.; Eller, C.; Krupa, E.; Lehner-Reindl, V.; Holler, C. Extended-spectrum-beta-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob. Agents Chemother. 2014, 58, 1228–1230. [Google Scholar] [CrossRef]
- Lei, T.; Tian, W.; He, L.; Huang, X.-H.; Sun, Y.-X.; Deng, Y.-T.; Sun, Y.; Lv, D.-H.; Wu, C.-M.; Huang, L.-Z.; et al. Antimicrobial resistance in Escherichia coli isolates from food animals, animal food products and companion animals in China. Vet. Microbiol. 2010, 146, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Naaber, P.; Balode, A.; Egorova, S.; Huik, K.; Ivanova, M.; Kaftyreva, L.; Kõljalg, S.; Kõressaar, T.; Lillo, J.; Löhr, I.; et al. The epidemiology of CTX-M group genes among clinical Escherichia coli strains in Estonia, Latvia, Lithuania, Norway and Russia. In Proceedings of the 115th General Meeting of American Society for Microbiology, New Orleans, LA, USA, 30 May–2 June 2015. [Google Scholar]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef] [PubMed]
| ESBL-Enterobacteriaceae Carriage | Univariate Analysis | |||
|---|---|---|---|---|
| Negative (n = 193) | Positive (n = 14) | OR (95%CI) | p-Value | |
| Male sex (%) | 56 (29) | 7 (50) | 2.4 (0.8–7.3) | 0.109 |
| Age: median (range) | 40 (0–82) | 47 (3–64) | NA | |
| Occupation (%) Other/not working a Veterinarian Pig farmer | 141 (73.1) 30 (15.5) 22 (11.4) | 5 (35.7) 2 (14) 7 (50) | 1 1.9 (0.3–10.1) 9.0 (2.6–30.8) | 0.463 <0.001 * |
| Source of food b | ||||
| Milk products (%) Purchased from market/shop Own produce/local farmer Meat (%) Purchased from market/shop Own produce/local farmer Fruit and vegetables Purchased from market/shop Own produce/local farmer | 154 (82.4) 33 (17.7) 156 (80.8) 34 (17.9) 93 (48.2) 98 (51.3) | 10 (71.4) 4 (28.6) 11 (78.6) 3 (21.4) 2 (14.3) 12 (85.8) | 1 1.9 (0.5–6.3) 1 1.2 (0.3–4.7) 1 5.7 (1.2–26.1) | 0.316 0.741 0.025 * |
| Source of the drinking water (%) Central water supply Well Bottled water Various sources | 128 (67.0) 55 (28.8) 3 (1.6) 5 (2.6) | 6 (42.8) 6 (42.9) 2 (14.3) 0 (0) | 1 2.3 (0.7–7.5) 14.2 (2.0–101.7) NA | 0.159 0.008 * |
| Smoking (%) | 21 (10.9) | 4 (28.6) | 3.3 (0.9–11.3) | 0.063 |
| Owning domestic animals (%) Dog Cat Farm animals c | 150 (77.7) 126 (65.3) 92 (47.7) 21 (10.9) | 10 (71.4) 8 (57.1) 8 (57.1) 2 (14.3) | 0.7 (0.2–2.4) 0.7 (0.2–2.1) 1.5 (0.5–4.3) 1.3 (0.3–6.5) | 0.589 0.539 0.495 0.697 |
| Travel abroad in past 12 months (%) Europe Outside Europe | 110 (57) 105 (54.4) 17 (8.8) | 3 (21.4) 3 (21.4) 2 (14.3) | 0.2 (0.1–0.8) 0.2 (0.1–0.8) 1.7 (0.4–8.4) | 0.018 * 0.023 * 0.498 |
| Eating outside home (%) d Few times (<3 times) a year/never Frequently (≥3 times a month) | 90 (46.9) 102 (53.1) | 11 (78.6) 3 (21.4) | 1 0.2 (0.1–0.9) | 0.033 * |
| Antibacterial treatment in previous year (%) | 53 (27.5) | 2 (14.3) | 2.3 (0.5–10.5) | 0.293 |
| Hospitalization in previous year (%) | 21 (11.1) | 0 (0) | NA | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Telling, K.; Brauer, A.; Laht, M.; Kalmus, P.; Toompere, K.; Kisand, V.; Maimets, M.; Remm, M.; Tenson, T.; Lutsar, I. Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia. Microorganisms 2020, 8, 1130. https://doi.org/10.3390/microorganisms8081130
Telling K, Brauer A, Laht M, Kalmus P, Toompere K, Kisand V, Maimets M, Remm M, Tenson T, Lutsar I. Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia. Microorganisms. 2020; 8(8):1130. https://doi.org/10.3390/microorganisms8081130
Chicago/Turabian StyleTelling, Kaidi, Age Brauer, Mailis Laht, Piret Kalmus, Karolin Toompere, Veljo Kisand, Matti Maimets, Maido Remm, Tanel Tenson, and Irja Lutsar. 2020. "Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia" Microorganisms 8, no. 8: 1130. https://doi.org/10.3390/microorganisms8081130
APA StyleTelling, K., Brauer, A., Laht, M., Kalmus, P., Toompere, K., Kisand, V., Maimets, M., Remm, M., Tenson, T., & Lutsar, I. (2020). Characteristics of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae and Contact to Animals in Estonia. Microorganisms, 8(8), 1130. https://doi.org/10.3390/microorganisms8081130






