Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Species and Culture Conditions
2.2. Amino Acid Auxotrophic Screening Assay
2.3. Pre-Culture and Fermentation Conditions
2.3.1. Amino Acid Selective Conditions
2.3.2. Co-Evolution of Yeast and Bacteria
2.4. Glucose, Fructose, and Malic Acid Measurement
2.5. Statistical Analysis
3. Results and Discussion
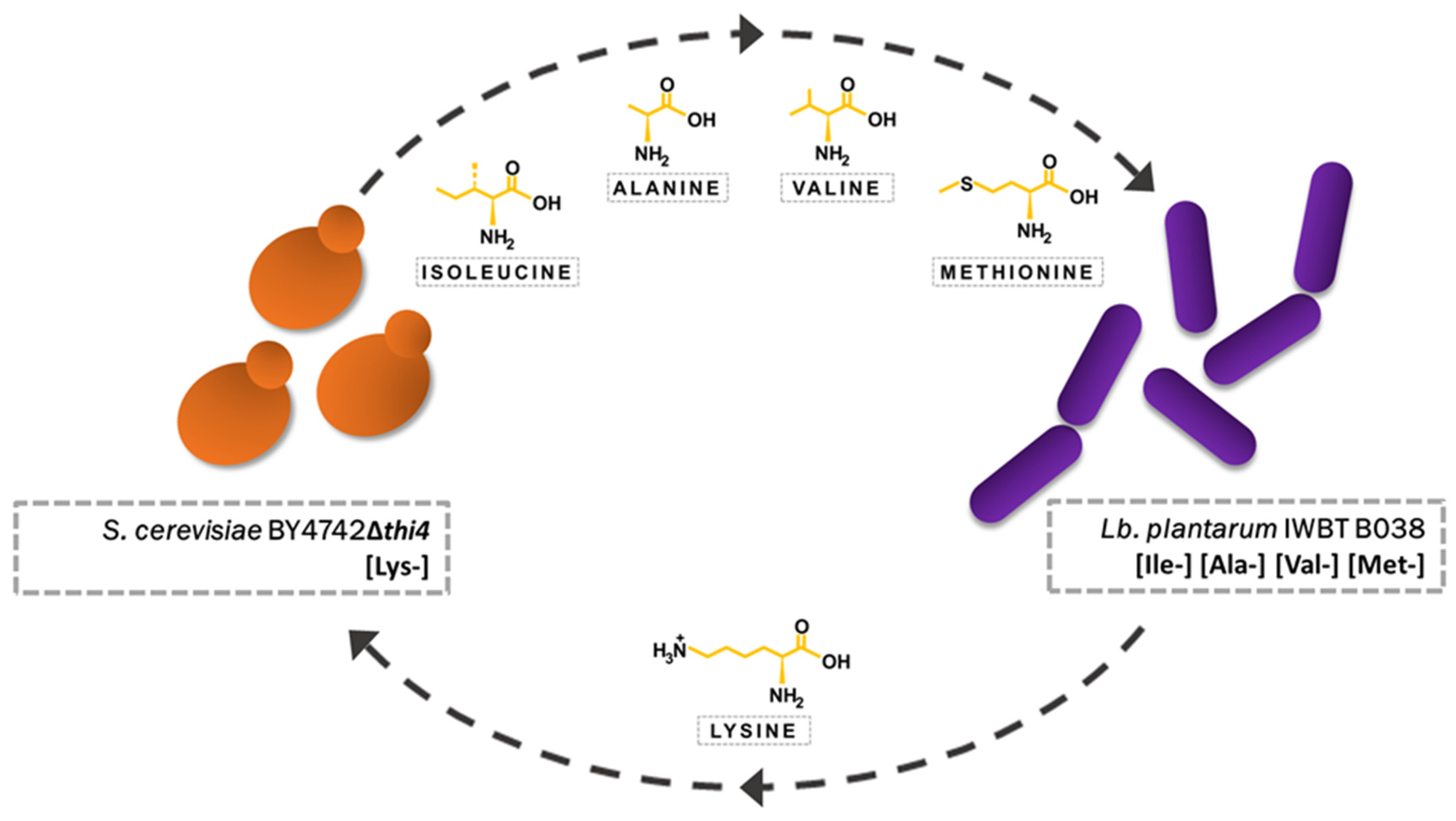
3.1. Initial Screening for Amino-Acid-Selective Conditions
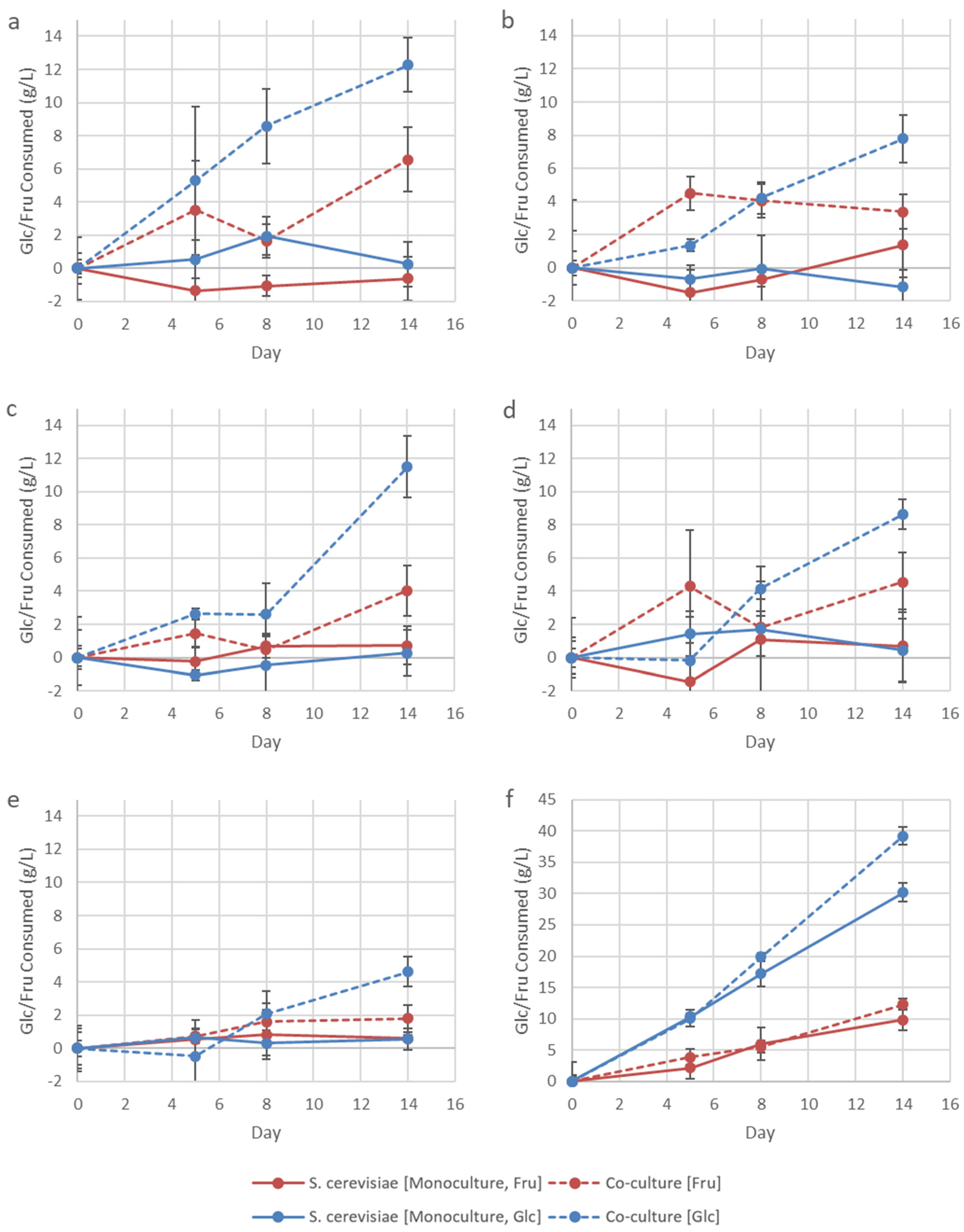
3.2. Addition of Lysine to Fermentations
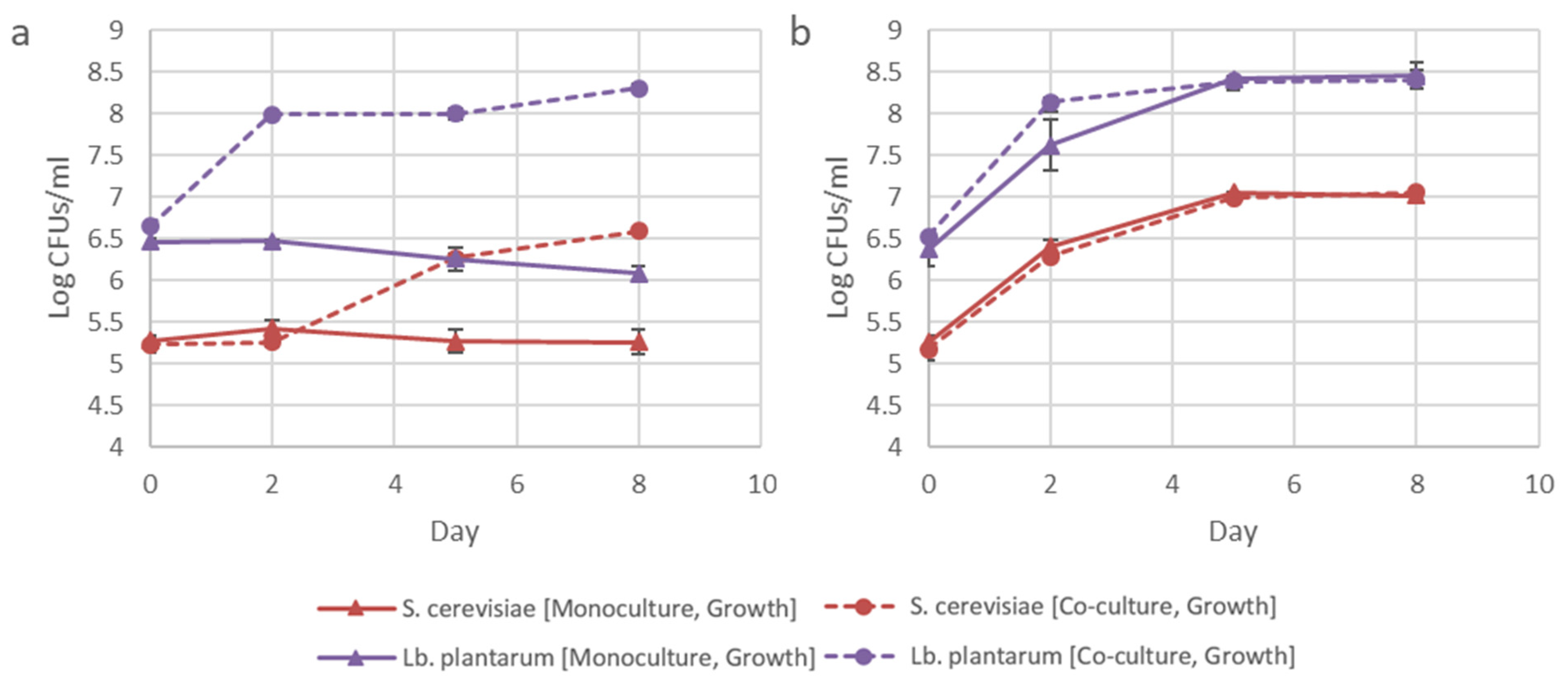
3.3. Influence of Inoculation Dosage
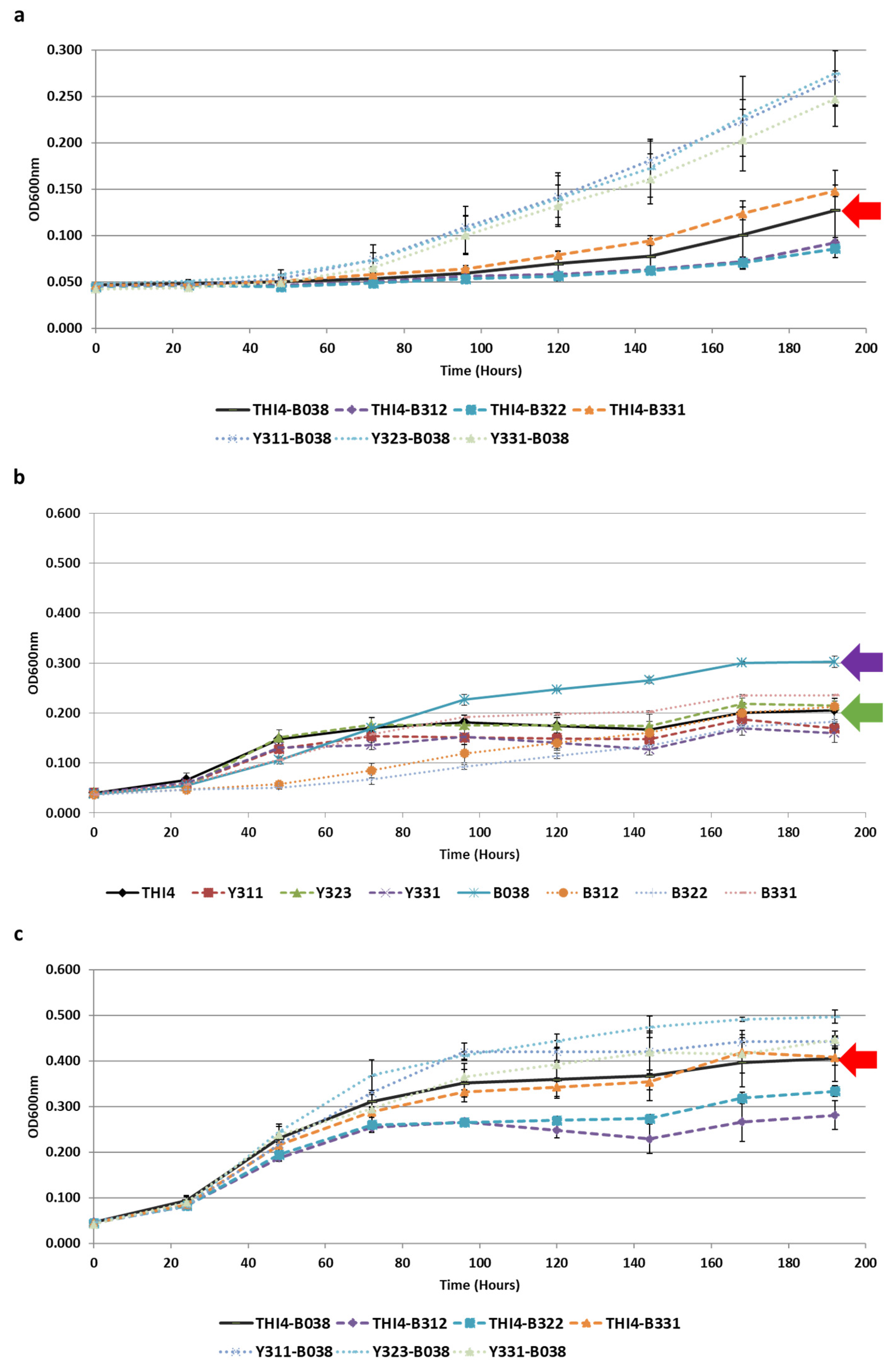
3.4. Co-Evolution of S. cerevisiae and Lb. plantarum
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bagheri, B.; Bauer, F.F.; Setati, M.E. The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. Front Microbiol. 2017, 8, 1988. [Google Scholar] [CrossRef]
- Liu, Y.; Rousseaux, S.; Tourdot-Maréchal, R.; Sadoudi, M.; Gougeon, R.; Schmitt-Kopplin, P.; Alexandre, H. Wine microbiome: A dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. 2017, 57, 856–873. [Google Scholar] [CrossRef]
- Lerm, E.; Engelbrecht, L.; du Toit, M. Malolactic fermentation: The ABC’s of MLF. S Afr. J. Enol. Vitic. 2010, 31, 186–212. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The next generation of malolactic fermentation starter cultures—an overview. Food Bioprocess Technol. 2011, 4, 876–906. [Google Scholar]
- Guzzon, R.; Villega, T.R.; Pedron, M.; Malacarne, M.; Nicolini, G.; Larcher, R. Simultaneous yeast-bacteria inoculum. A feasible solution for management of oenological fermentation in red must with low nitrogen content. Ann Microbiol. 2013, 63, 805–808. [Google Scholar] [CrossRef]
- Bartle, L.; Sumby, K.; Sundstrom, J.; Jiranek, V. The microbial challenge of winemaking: Yeast-bacteria compatibility. FEMS Yeast Res. 2019, 19, foz040. [Google Scholar] [CrossRef]
- Terrade, N.; Mira de Orduña, R. Determination of the essential nutrient requirements of wine-related bacteria from the genera Oenococcus and Lactobacillus. Int. J. Food Microbiol. 2009, 133, 8–13. [Google Scholar] [CrossRef]
- Bayrock, D.P.; Ingledew, W.M. Inhibition of yeast by lactic acid bacteria in continuous culture: Nutrient depletion and/or acid toxicity? J. Ind. Microbiol. Biotechnol. 2004, 31, 362–368. [Google Scholar] [CrossRef]
- Alexandre, H.; Costello, P.J.; Remize, F.; Guzzo, J.; Guilloux-Benatier, M. Saccharomyces cerevisiae—Oenococcus oeni interactions in wine: Current knowledge and perspectives. Int. J. Food Microbiol. 2004, 93, 141–154. [Google Scholar] [CrossRef]
- Malherbe, S.; Bauer, F.F.; du Toit, M. Understanding problem fermentations—A review. S. Afr. J. Enol. Vitic. 2007, 28, 169–186. [Google Scholar] [CrossRef]
- Nehme, N.; Mathieu, F.; Thaillandier, P. Quantitative study of the interactions between Saccharomyces cerevisiae and Oenococcus oeni strains. J. Ind. Microbiol. Biotechnol. 2008, 35, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Forcisi, S.; Harir, M.; Deleris-Bou, M.; Krieger-Weber, S.; Lucio, M.; Longin, C.; Degueurce, C.; Gougeon, R.D.; Schmitt-Kopplin, P.; et al. New molecular evidence of wine yeast-bacteria interaction unraveled by non-targeted exometabolomic profiling. Metabolomics 2016, 12, 1–16. [Google Scholar] [CrossRef]
- Tourdot-Maréchal, R.; Fortier, L.C.; Guzzo, J.; Lee, B.; Diviès, C. Acid sensitivity of neomycin-resistant mutants on Oenococcus oeni: A relationship between reduction of ATPase activity and lack of malolactic activity. FEMS Microbiol. Lett. 1999, 178, 319–326. [Google Scholar] [CrossRef]
- Capucho, I.; Romão, M.V.S. Effect of ethanol and fatty acids on malolactic activity of Leuconostoc oenos. Appl. Microbiol. Biotechnol. 1994, 42, 391–395. [Google Scholar] [CrossRef]
- Guilloux-Benatier, M.; Pageault, O.; Man, A.; Feuillat, M. Lysis of yeast cells by Oenococcus oeni enzymes. J. Ind. Microbiol. Biotechnol. 2000, 25, 193–197. [Google Scholar] [CrossRef]
- Yurdugül, S.; Bozoglu, F. Studies on an inhibitor produced by lactic acid bacteria of wines on the control of malolactic fermentation. Eur. Food Res. Technol. 2002, 215, 38–41. [Google Scholar] [CrossRef]
- Costello, P.J.; Henschke, P.A.; Markides, A.J. Standardised methodology for testing malolactic bacteria and wine yeast compatibility. Aust. J. Grape Wine Res. 2003, 9, 127–137. [Google Scholar] [CrossRef]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Implications of new research and technologies for malolactic fermentation in wine. Appl. Microbiol. Biotechnol. 2014, 98, 8111–8132. [Google Scholar] [CrossRef] [PubMed]
- Betteridge, A.L.; Sumby, K.M.; Sundstrom, J.F.; Grbin, P.R.; Jiranek, V. Application of directed evolution to develop ethanol tolerant Oenococcus oeni for more efficient malolactic fermentation. Appl. Microbiol. Biotechnol. 2017, 102, 921–932. [Google Scholar] [CrossRef]
- Henschke, P.A.; Jiranek, V. Yeasts—metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Reading, UK, 1993; pp. 77–164. [Google Scholar]
- Bely, M.; Sablayrolles, J.M.; Barre, P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Ponomarova, O.; Gabrielli, N.; Sévin, D.C.; Mülleder, M.; Zirngibl, K.; Bulyha, K.; Andrejev, S.; Kafkia, E.; Typas, A.; Sauer, U.; et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst 2017, 5, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Swamy, K.B.S.; Leu, J.Y.; McDonald, M.J.; Galafassi, S.; Compagno, C.; Piškur, J. Coevolution with bacteria drives the evolution of aerobic fermentation in Lachancea kluyveri. PLoS ONE 2017, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Mendes, F.; Sieuwerts, S.; de Hulster, E.; Almering, M.J.H.; Luttik, M.A.H.; Pronk, J.T.; Smid, E.J.; Bron, P.A.; Daran-Lapujade, P. Transcriptome-based characterization of interactions between Saccharomyces cerevisiae and Lactobacillus delbrueckii subsp. bulgaricus in lactose-grown chemostat cocultures. Appl. Environ. Microbiol. 2013, 79, 5949–5961. [Google Scholar] [CrossRef] [PubMed]
- Hom, E.F.Y.; Murray, A.W. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 2014, 345, 94–98. [Google Scholar] [CrossRef]
- Oosthuizen, J.R.; Naidoo, R.K.; Rossouw, D.; Bauer, F.F. Evolution of mutualistic behaviour between Chlorella sorokiniana and Saccharomyces cerevisiae within a synthetic environment. J. Ind. Microbiol. Biot. 2020, 47, 357–372. [Google Scholar] [CrossRef]
- Spano, G.; Massa, S. Environmental stress response in wine lactic acid bacteria: Beyond Bacillus subtilis. Crit. Rev. Microbiol. 2006, 32, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Van Bokhorst-van de Veen, H.; Abee, T.; Tempelaars, M.; Bron, P.A.; Kleerebezem, M.; Marco, M.L. Short- and long-term adaptation to ethanol stress and its cross-protective consequences in Lactobacillus plantarum. Appl. Environ. Microbiol. 2011, 77, 5247–5256. [Google Scholar] [CrossRef]
- Zhou, N.; Bottagisi, S.; Katz, M.; Schacherer, J.; Friedrich, A.; Gojkovic, Z.; Swamy, K.B.S.; Knecht, W.; Compagno, C.; Piškur, J. Yeast-bacteria competition induced new metabolic traits through large-scale genomic rearrangements in Lachancea kluyveri. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Morrison-Whittle, P.; Lee, S.A.; Fedrizzi, B.; Goddard, M.R. Co-evolution as tool for diversifying flavor and aroma profiles of wines. Front Microbiol. 2018, 9, 910. [Google Scholar] [CrossRef] [PubMed]

| Compound Group | Compound | Amount per Liter |
|---|---|---|
| Carbon sources | Glucose | 100 g |
| Fructose | 100 g | |
| Acids | Potassium L-Tartrate Monobasic | 2.5 g |
| L-Malic acid | 4 g | |
| Citric acid | 0.2 g | |
| Salts | Potassium phosphate dibasic (K2HPO4) | 1.14 g |
| Magnesium sulphate heptahydrate (MgSO4.7H2O) | 1.23 g | |
| Calcium chloride dihydrate (CaCl2.2H2O) | 0.44 g | |
| Nitrogen sources | Ammonium sulphate | 0.3 g |
| Amino acids (prepared as 100X stock solution in 20 g/L NaHCO3 buffer solution) | ||
| - Tyrosine | 0.014 g | |
| - Tryptophane | 0.137 g | |
| - Isoleucine | 0.025 g | |
| - Aspartic acid | 0.034 g | |
| - Glutamic acid | 0.092 g | |
| - Arginine | 0.286 g | |
| - Leucine | 0.037 g | |
| - Threonine | 0.058 g | |
| - Glycine | 0.014 g | |
| - Glutamine | 0.386 g | |
| - Alanine | 0.111 g | |
| - Valine | 0.034 g | |
| - Methionine | 0.024 g | |
| - Phenylalanine | 0.029 g | |
| - Serine | 0.060 g | |
| - Histidine | 0.025 g | |
| - Lysine | 0.013 g | |
| - Cysteine | 0.010 g | |
| - Proline | 0.468 g | |
| Trace elements (prepared as 100X stock solution) | Manganese(II) chloride tetrahydrate (MnCl2.4H2O) | 200 µg |
| Zinc(II) chloride (ZnCl) | 135 µg | |
| Iron(II) chloride (FeCl2) | 30 µg | |
| Copper(II) chloride (CuCl2) | 15 µg | |
| Boric acid (H3BO3) | 5 µg | |
| Cobalt(II) nitrate hexahydrate (Co(NO3)2.6H2O) | 30 µg | |
| Sodium molybdate dihydrate (NaMoO4.2H2O) | 25 µg | |
| Potassium iodate (KIO3) | 10 µg | |
| Vitamins (prepared as 100X stock solution) | Myo-inositol | 100 mg |
| Pyridoxine hydrochloride | 2 mg | |
| Nicotinic acid | 2 mg | |
| Calcium pantothenate | 1 mg | |
| Thiamin hydrochloride | 0.5 mg | |
| Para-aminobenzoic acid (PABA)-K | 0.2 mg | |
| Riboflavin | 0.2 mg | |
| Biotin | 0.125 mg | |
| Folic acid | 0.2 mg | |
| Anaerobic factors (prepared as 10X stock solution in hot 96% EtOH) | Ergosterol | 10 mg |
| Tween 80 | 0.5 mL |
| Treatment Name | Amino Acids Omitted | Yeast Assimilable Nitrogen (YAN) Concentration (mg/L) |
|---|---|---|
| Control | None | 322.48 |
| Lys-Ile | Lysine | 317.32 |
| Isoleucine | ||
| Lys-Ala | Lysine | 302.54 |
| Alanine | ||
| Lys-Val | Lysine | 315.93 |
| Valine | ||
| Lys-Met | Lysine | 317.74 |
| Methionine | ||
| Lys-Ile-Ala-Val-Met | Lysine | 293.55 |
| Isoleucine | ||
| Alanine | ||
| Valine | ||
| Methionine |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
du Toit, S.C.; Rossouw, D.; du Toit, M.; Bauer, F.F. Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum. Microorganisms 2020, 8, 1109. https://doi.org/10.3390/microorganisms8081109
du Toit SC, Rossouw D, du Toit M, Bauer FF. Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum. Microorganisms. 2020; 8(8):1109. https://doi.org/10.3390/microorganisms8081109
Chicago/Turabian Styledu Toit, S. Christine, Debra Rossouw, Maret du Toit, and Florian F. Bauer. 2020. "Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum" Microorganisms 8, no. 8: 1109. https://doi.org/10.3390/microorganisms8081109
APA Styledu Toit, S. C., Rossouw, D., du Toit, M., & Bauer, F. F. (2020). Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum. Microorganisms, 8(8), 1109. https://doi.org/10.3390/microorganisms8081109






