Microbial Diversity Associated with Gwell, a Traditional French Mesophilic Fermented Milk Inoculated with a Natural Starter
Abstract
1. Introduction
2. Materials and Methods
2.1. Gwell Sampling and Data Collection
2.2. Sensory Evaluation of Gwell
2.2.1. Two-Steps Sensory Analysis
2.2.2. Statistical Analysis
2.3. Microbial Enumeration
2.4. Bacterial Isolation and Phenotypic Sorting
2.5. Identification and Phenotypic Characterization of Strains
2.5.1. API 50CH—Biochemical Characterization
2.5.2. Identification of the Biovar diacetylactis
2.5.3. DNA Extraction for 16S rDNA Sequencing and Species-Specific PCR
2.5.4. 16S rDNA Sequencing
2.5.5. Species-Specific PCR
2.5.6. Pulse-Field Gel Electrophoresis (PFGE)
2.5.7. Acidifying Capacity of Strains
2.6. Metabarcoding
2.6.1. DNA Extraction
2.6.2. Sequencing on Illumina MiSeq
2.6.3. Bioinformatics Analysis
2.7. A Participatory Research Framework
3. Results
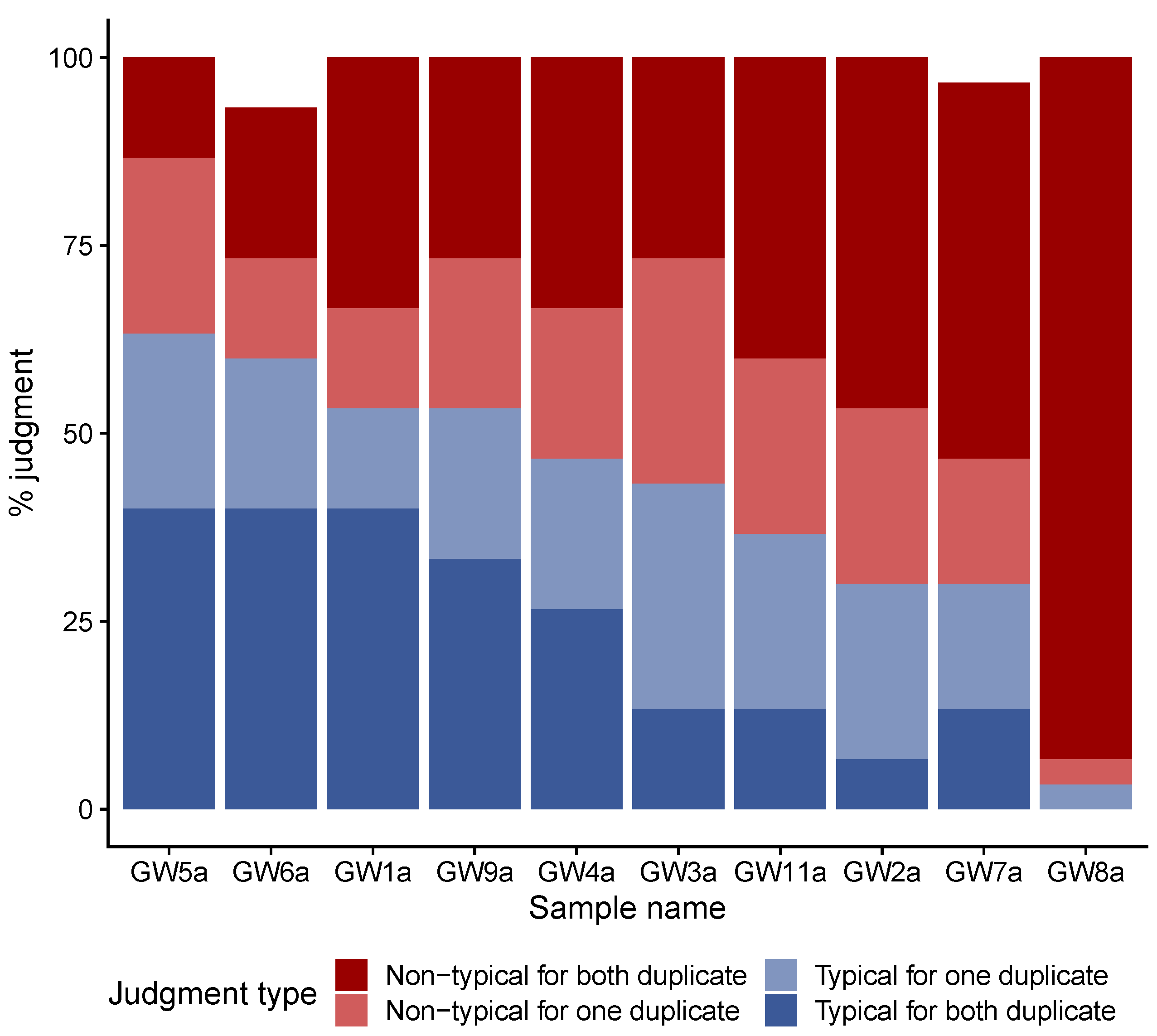
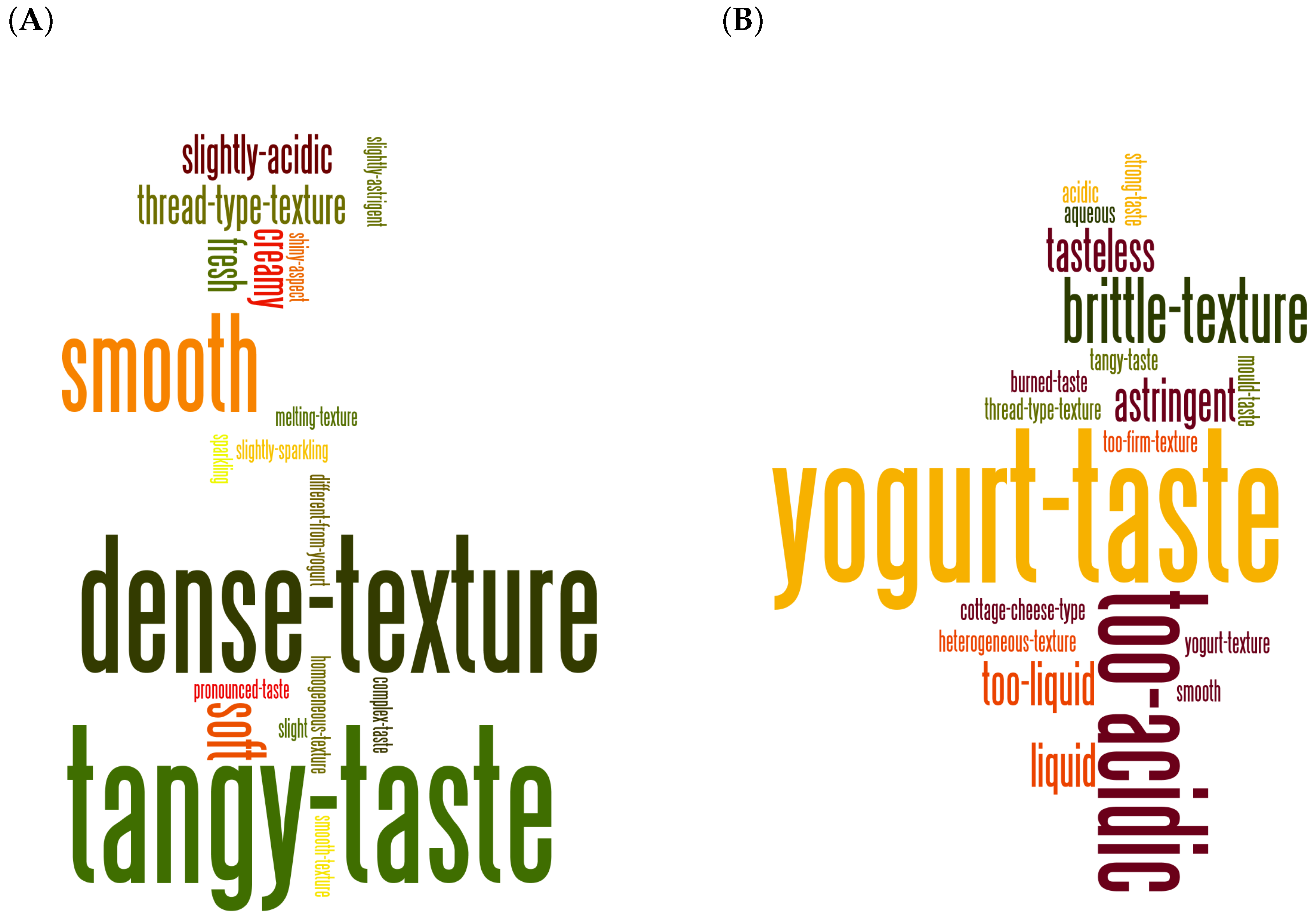
3.1. Consensual Descriptors of Gwell Samples Considered to Be Typical
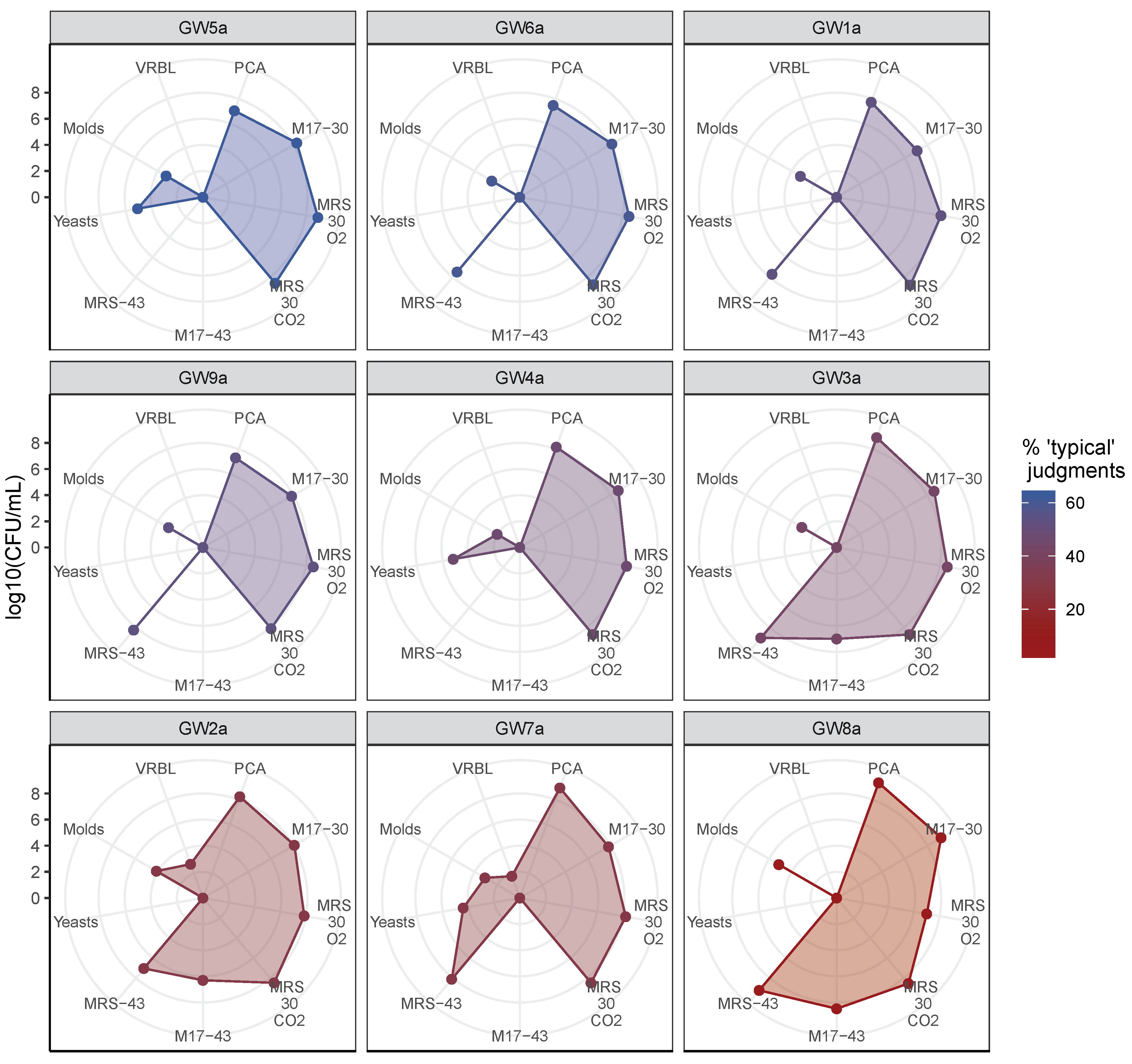
3.2. Microbial Enumeration of Gwell
3.3. Selection of Five Gwell Samples for the Culture-Dependent Characterization of Lab Diversity
3.4. Characterization of LAB Diversity
3.4.1. Selection and Identification of Isolates Representative of LAB Diversity
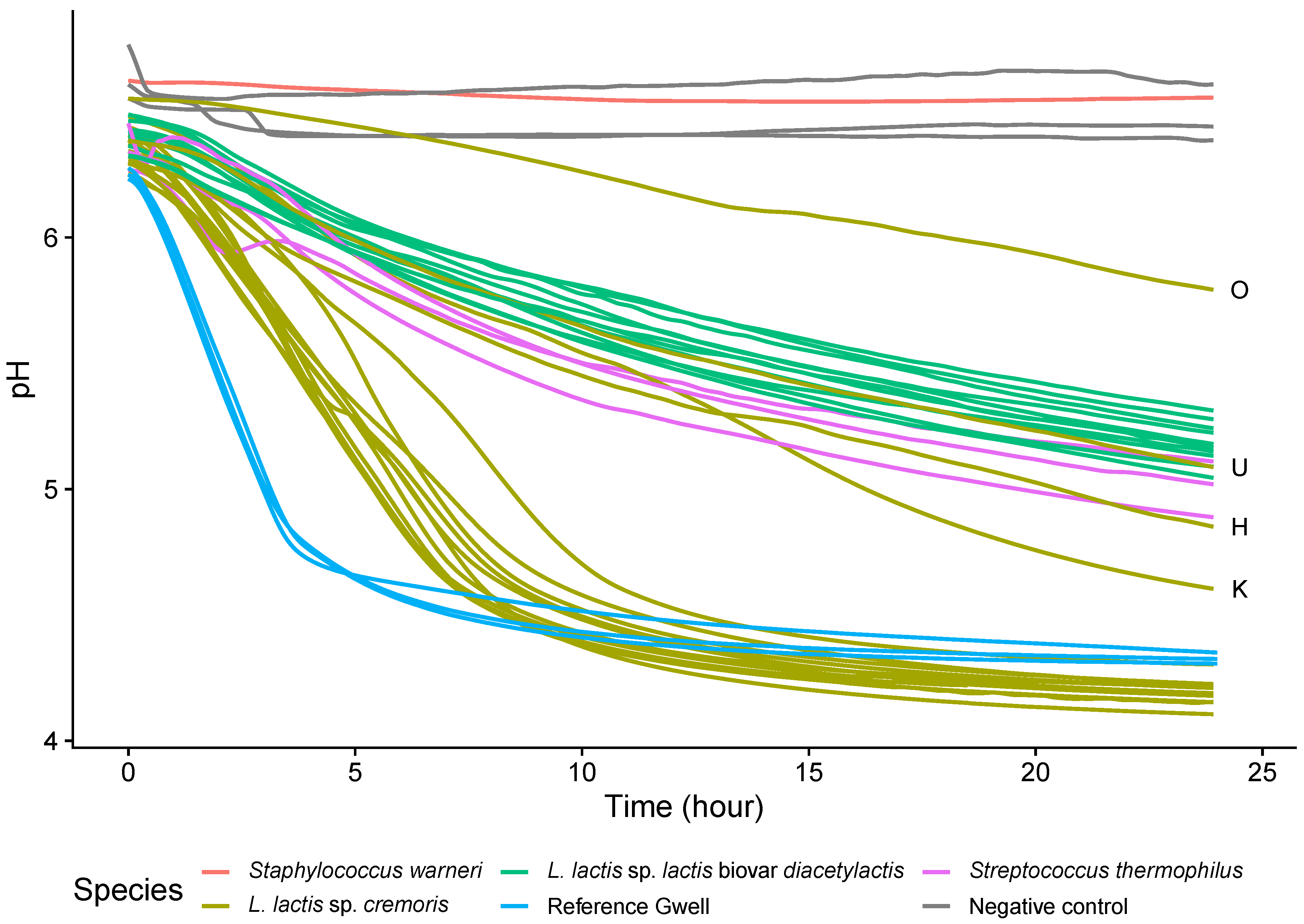
3.4.2. Acidifying Capacity of Lactococcus lactis Strains
3.5. Culture-Independent Characterization of Gwell Microbial Communities
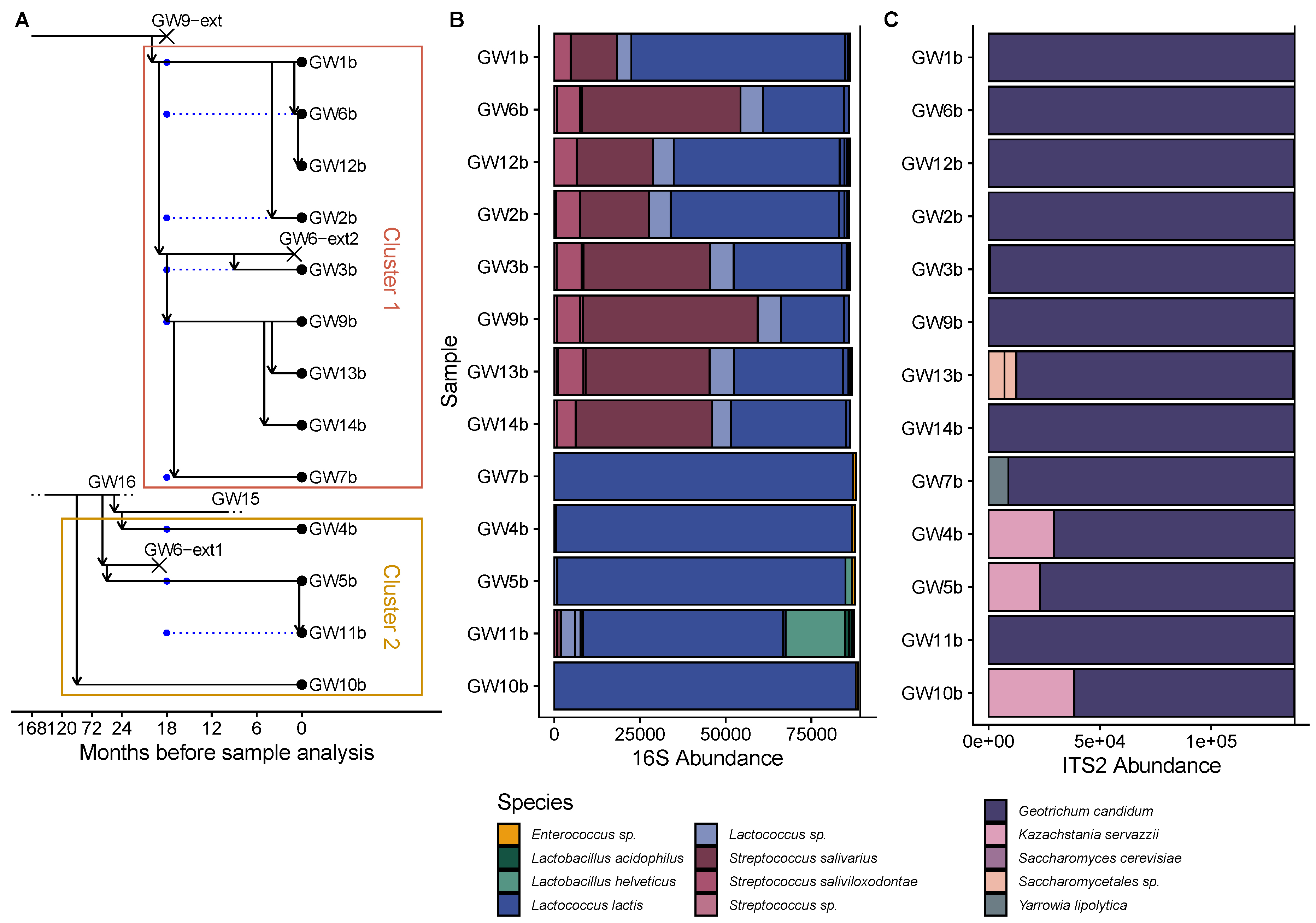
3.6. The History of Gwell Exchanges between Producers Explains Their Microbial Composition
4. Discussion
4.1. Microbiological Characterization of Gwell
4.2. Microbial Composition of Typical and Non-Typical Gwell
4.3. The Evolution of Gwell over Time and Exchanges between Producers
4.4. Gwell Loss
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Holzapfel, W. Use of Starter Cultures in Fermentation on a Household Scale. Food Control 1997, 8, 241–258. [Google Scholar] [CrossRef]
- Holzapfel, W.H. Appropriate Starter Culture Technologies for Small-Scale Fermentation in Developing Countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef]
- Tamang, J.; Kailasapathy, K. Fermented Foods and Beverages of the World; CRCNET Books; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Spitaels, F.; Wieme, A.D.; Janssens, M.; Aerts, M.; Daniel, H.M.; Van Landschoot, A.; De Vuyst, L.; Vandamme, P. The Microbial Diversity of Traditional Spontaneously Fermented Lambic Beer. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Tormo, H.; Ali Haimoud Lekhal, D.; Roques, C. Phenotypic and Genotypic Characterization of Lactic Acid Bacteria Isolated from Raw Goat Milk and Effect of Farming Practices on the Dominant Species of Lactic Acid Bacteria. Int. J. Food Microbiol. 2015, 210, 9–15. [Google Scholar] [CrossRef]
- Sternes, P.R.; Lee, D.; Kutyna, D.R.; Borneman, A.R. A Combined Meta-Barcoding and Shotgun Metagenomic Analysis of Spontaneous Wine Fermentation. Gigascience 2017, 6, gix040. [Google Scholar] [CrossRef]
- Demarigny, Y.; Gerber, P. Usefulness of Natural Starters in Food Industry: The Example of Cheeses and Bread. FNS 2014, 5, 1679–1691. [Google Scholar] [CrossRef]
- Gibbons, J.G.; Rinker, D.C. The Genomics of Microbial Domestication in the Fermented Food Environment. Curr. Opin. Genet. Dev. 2015, 35, 1–8. [Google Scholar] [CrossRef]
- Leborgne, A. (UBPN, Maison de l’agriculture, GIE Elevages de Bretagne). Personal communication, Étude Géographique et Socio-Historique Du Gros Lait. 1998.
- Morinière, C. Quand la valorisation alimentaire encourage la conservation d’une race: Le cas de la race brettonne pie-noir. Ethnozootechnie 2017, 103, 13–18. [Google Scholar]
- Wirawati, C.U.; Sudarwanto, M.B.; Lukman, D.W.; Wientarsih, I.; Srihanto, E.A. Diversity of Lactic Acid Bacteria in Dadih Produced by Either Back-Slopping or Spontaneous Fermentation from Two Different Regions of West Sumatra, Indonesia. Vet. World 2019, 12, 823–829. [Google Scholar] [CrossRef]
- Akuzawa, R.; Miura, T.; Surono, I. Fermented Milks | Asian Fermented Milks. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 507–511. [Google Scholar] [CrossRef]
- Kahala, M.; Mäki, M.; Lehtovaara, A.; Tapanainen, J.M.; Katiska, R.; Juuruskorpi, M.; Juhola, J.; Joutsjoki, V. Characterization of Starter Lactic Acid Bacteria from the Finnish Fermented Milk Product Viili. J. Appl. Microbiol. 2008, 105, 1929–1938. [Google Scholar] [CrossRef]
- Bakry, A.M. Mini-Review on Functional Characteristics of Viili and Manufacturing Process. J. Food Biotechnol. Res. 2018, 2, 7. [Google Scholar]
- Harun-ur-Rashid, M.; Togo, K.; Ueda, M.; Miyamoto, T. Identification and Characterization of Dominant Lactic Acid Bacteria Isolated from Traditional Fermented Milk Dahi in Bangladesh. World J. Microbiol. Biotechnol. 2007, 23, 125–133. [Google Scholar] [CrossRef]
- Venema, K.; Surono, I. Microbiota Composition of Dadih—A Traditional Fermented Buffalo Milk of West Sumatra. Lett. Appl. Microbiol. 2019, 68, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Tormo, H. (Université de Toulouse, Ecole d’Ingénieurs de Purpan, Toulouse, France). Personal communication, 2019.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: A Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses; R Package Version 1.0.6; 2019. [Google Scholar]
- Nickels, C.; Leesment, H. Methode Zur Differenzierung und Quantitativen Bestimmung von Säureweckerbakterien. Milchwissenschaft 1964, 19, 37–378. [Google Scholar]
- Pu, Z.Y.; Dobos, M.; Limsowtin, G.K.Y.; Powell, I.B. Integrated Polymerase Chain Reaction-Based Procedures for the Detection and Identification of Species and Subspecies of the Gram-Positive Bacterial Genus Lactococcus. J. Appl. Microbiol. 2002, 93, 353–361. [Google Scholar] [CrossRef]
- Forsman, P.; Tilsaia-Timisjärvi, A.; Alatossava, T. Identification of Staphylococcal and Streptococcal Causes of Bovine Mastitis Using 16S-23S rRNA Spacer Regions. Microbiology 1997, 143, 3491–3500. [Google Scholar] [CrossRef]
- Lortal, S.; Rouault, A.; Guezenec, S.; Gautier, M. Lactobacillus Helveticus: Strain Typing and Genome Size Estimation by Pulsed Field Gel Electrophoresis. Curr. Microbiol. 1997, 34, 180–185. [Google Scholar] [CrossRef]
- Liu, Z.; Lozupone, C.; Hamady, M.; Bushman, F.D.; Knight, R. Short Pyrosequencing Reads Suffice for Accurate Microbial Community Analysis. Nucleic Acids Res. 2007, 35, e120. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F.; et al. Shotgun Metagenomes and Multiple Primer Pair-Barcode Combinations of Amplicons Reveal Biases in Metabarcoding Analyses of Fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. In Disease Gene Identification: Methods and Protocols; DiStefano, J.K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 223–232. [Google Scholar] [CrossRef]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- ADRIA Quimper (UBPN, Maison de l’agriculture, GIE Elevages de Bretagne). Personal communication, Caractérisation et Maîtrise de La Fabrication d’un Produit Régional, “le Gros Lait”. 1995.
- Gänzle, M.; Ripari, V. Composition and Function of Sourdough Microbiota: From Ecological Theory to Bread Quality. Int. J. Food Microbiol. 2016, 239, 19–25. [Google Scholar] [CrossRef]
- Doslash, R.; Osvik, V.; Sperstad, S.; Breines, E.; Hareide, E.; Godfroid, J.; Zhou, Z.; Ren, P.; Geoghegan, C.; Holzapfel, W.; et al. Bacterial Diversity of aMasi, a South African Fermented Milk Product, Determined by Clone Library and Denaturing Gradient Gel Electrophoresis Analysis. AJMR 2013, 7, 4146–4158. [Google Scholar] [CrossRef]
- Shangpliang, H.N.J.; Sharma, S.; Rai, R.; Tamang, J.P. Some Technological Properties of Lactic Acid Bacteria Isolated from Dahi and Datshi, Naturally Fermented Milk Products of Bhutan. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Passerini, D.; Laroute, V.; Coddeville, M.; Le Bourgeois, P.; Loubière, P.; Ritzenthaler, P.; Cocaign-Bousquet, M.; Daveran-Mingot, M.L. New Insights into Lactococcus Lactis Diacetyl- and Acetoin-Producing Strains Isolated from Diverse Origins. Int. J. Food Microbiol. 2013, 160, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Aryana, K.J.; Olson, D.W. A 100-Year Review: Yogurt and Other Cultured Dairy Products. J. Dairy Sci. 2017, 100, 9987–10013. [Google Scholar] [CrossRef]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Fundamentals of Cheese Science, 2nd ed.; Springer: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Vaningelgem, F.; Zamfir, M.; Adriany, T.; Vuyst, L.D. Fermentation Conditions Affecting the Bacterial Growth and Exopolysaccharide Production by Streptococcus Thermophilus ST 111 in Milk-Based Medium. J. Appl. Microbiol. 2004, 97, 1257–1273. [Google Scholar] [CrossRef]
- Smid, E.; Kleerebezem, M. Production of Aroma Compounds in Lactic Fermentations. Annu. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef]
- Lhomme, E.; Lattanzi, A.; Dousset, X.; Minervini, F.; De Angelis, M.; Lacaze, G.; Onno, B.; Gobbetti, M. Lactic Acid Bacterium and Yeast Microbiotas of Sixteen French Traditional Sourdoughs. Int. J. Food Microbiol. 2015, 215, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the Lactic Acid Bacterium and Yeast Microbiota in the Switch from Firm- to Liquid-Sourdough Fermentation. Appl. Environ. Microbiol. 2014, 80, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Jood, I.; Hoff, J.W.; Setati, M.E. Evaluating Fermentation Characteristics of Kazachstania Spp. and Their Potential Influence on Wine Quality. World J. Microbiol. Biotechnol. 2017, 33, 129. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Mei, J.; Guo, Q.; Wu, Y.; Li, Y. Microbial Diversity of a Camembert-Type Cheese Using Freeze-Dried Tibetan Kefir Coculture as Starter Culture by Culture-Dependent and Culture-Independent Methods. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Garneau, J.E.; Moineau, S. Bacteriophages of Lactic Acid Bacteria and Their Impact on Milk Fermentations. Microb. Cell Fact. 2011, 10, S20. [Google Scholar] [CrossRef] [PubMed]
- Erkus, O.; de Jager, V.C.; Spus, M.; van Alen-Boerrigter, I.J.; van Rijswijck, I.M.; Hazelwood, L.; Janssen, P.W.; van Hijum, S.A.; Kleerebezem, M.; Smid, E.J. Multifactorial Diversity Sustains Microbial Community Stability. ISME J. 2013, 7, 2126–2136. [Google Scholar] [CrossRef]
- Herve-Jimenez, L.; Guillouard, I.; Guedon, E.; Boudebbouze, S.; Hols, P.; Monnet, V.; Maguin, E.; Rul, F. Postgenomic Analysis of Streptococcus Thermophilus Cocultivated in Milk with Lactobacillus Delbrueckii Subsp. Bulgaricus: Involvement of Nitrogen, Purine, and Iron Metabolism. Appl. Environ. Microbiol. 2009, 75, 2062–2073. [Google Scholar] [CrossRef] [PubMed]

| First Gwell Sample Collection (May, 2017) | Second Gwell Sample collection (November, 2018) | ||||||
|---|---|---|---|---|---|---|---|
| Producer | Gwell Sample Code | Sensory Analysis | Microbial Enumeration | Bacterial Isolation | Gwell Sample Code | Metabarcoding | |
| Step1 | Step2 | ||||||
| 1 | GW1a | x | x | x | x | GW1b | x |
| 2 | GW2a | x | - | x | - | GW2b | x |
| 3 | GW3a | x | x | x | - | GW3b | x |
| 4 | GW4a | x | x | x | x | GW4b | x |
| 5 | GW5a | x | x | x | x | GW5b | x |
| 6 | GW6a | x | x | x | x | GW6b | x |
| 7 | GW7a | x | x | x | - | GW7b | x |
| 8 | Gw8a | x | x | x | - | GW8b | - |
| 9 | GW9a | x | x | x | x | GW9b | x |
| 10 | GW10b | x | |||||
| 11 | GW11a | x | - | - | - | GW11b | x |
| 12 | GW12b | x | |||||
| 13 | GW13b | x | |||||
| 14 | GW14b | x | |||||
| Name of the Medium | Composition or Reference of the Medium | Targeted Microorganisms | Temperature of Incubation | Condition of Incubation (Aerobiosis or Anaerobiosis) |
|---|---|---|---|---|
| M17 | Biokar Diagnostic | Mesophilic and thermophilic Lactococcocal and streptococcal strains | 30 C or 43 C | |
| MRS | Biokar Diagnostic | Mesophilic and thermophilic lactobacilli strains | 30 C or 43 C | & |
| VRBL | Difco Laboratories, detroit, MI, USA | Coliforms | 30C | |
| KF | Biokar Diagnostic | Enterococcus | 30 C | |
| PCA | Biokar Diagnostic | Mesophilic and aerobic total flora | 30 C | |
| OGA | Biokar Diagnostic | Yeast and filamentous fungi | 25 C |
| Gwell Sample | Clone ID | API Strips 50CH Profile | Species-Specific PCR Identification | KCA Positive Strains | PFGE Profil Type | Validated Identification |
|---|---|---|---|---|---|---|
| GW1 | B | - | - | - | - | Staphylococcus warneri (Identified by 16S sequencing) |
| Q | 2 | L. lactis sp. cremoris | No | P13 | Lactococcus lactis sp. cremoris | |
| C | 2 | L. lactis sp. cremoris | No | P13 | Lactococcus lactis sp. cremoris | |
| R | 13 | L. lactis sp. lactis | Yes | P10 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| D | 4 | L. lactis sp. cremoris | No | P15 | Lactococcus lactis sp. cremoris | |
| A | 12 | L. lactis sp. lactis | Yes | P6 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| GW1 | - | S. thermophilus | - | - | Streptococcus thermophilus | |
| GW4 | F | 9 | L. lactis sp. lactis | Yes | P2 | Lactococcus lactis sp. lactis biovar diacetylactis |
| S | 11 | L. lactis sp. lactis | Yes | P3 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| E | 5 | L. lactis sp. cremoris | No | P14 | Lactococcus lactis sp. cremoris | |
| GW5 | H | 6 | L. lactis sp. cremoris | No | P11 | Lactococcus lactis sp. cremoris |
| I | 4 | L. lactis sp. cremoris | No | P1 | Lactococcus lactis sp. cremoris | |
| U | 6 | L. lactis sp. cremoris | No | P12 | Lactococcus lactis sp. cremoris | |
| Jp | 9 | L. lactis sp. lactis | Yes | P4 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| Jg | 9 | L. lactis sp. lactis | Yes | P5 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| V | 11 | L. lactis sp. lactis | Yes | P4 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| G | 4 | L. lactis sp. cremoris | No | P1 | Lactococcus lactis sp. cremoris | |
| GW6 | L | 8 | L. lactis sp. lactis | Yes | P7 | Lactococcus lactis sp. lactis biovar diacetylactis |
| M | 7 | L. lactis sp. cremoris | No | P13 | Lactococcus lactis sp. cremoris | |
| K | 3 | L. lactis sp. cremoris | No | P13 | Lactococcus lactis sp. cremoris | |
| GW6 | - | S. thermophilus | - | - | Streptococcus thermophilus | |
| GW9 | W | 10 | L. lactis sp. lactis | Yes | P9 | Lactococcus lactis sp. lactis biovar diacetylactis |
| P | 3 | L. lactis sp. cremoris | No | P13 | Lactococcus lactis sp. cremoris | |
| N | 9 | L. lactis sp. lactis | Yes | P8 | Lactococcus lactis sp. lactis biovar diacetylactis | |
| O | 1 | L. lactis sp. cremoris | No | P15 | Lactococcus lactis sp. cremoris | |
| GW9 | - | S. thermophilus | - | - | Streptococcus thermophilus |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Gastrow, L.; Madec, M.-N.; Chuat, V.; Lubac, S.; Morinière, C.; Lé, S.; Santoni, S.; Sicard, D.; Valence, F. Microbial Diversity Associated with Gwell, a Traditional French Mesophilic Fermented Milk Inoculated with a Natural Starter. Microorganisms 2020, 8, 982. https://doi.org/10.3390/microorganisms8070982
von Gastrow L, Madec M-N, Chuat V, Lubac S, Morinière C, Lé S, Santoni S, Sicard D, Valence F. Microbial Diversity Associated with Gwell, a Traditional French Mesophilic Fermented Milk Inoculated with a Natural Starter. Microorganisms. 2020; 8(7):982. https://doi.org/10.3390/microorganisms8070982
Chicago/Turabian Stylevon Gastrow, Lucas, Marie-Noëlle Madec, Victoria Chuat, Stanislas Lubac, Clémence Morinière, Sébastien Lé, Sylvain Santoni, Delphine Sicard, and Florence Valence. 2020. "Microbial Diversity Associated with Gwell, a Traditional French Mesophilic Fermented Milk Inoculated with a Natural Starter" Microorganisms 8, no. 7: 982. https://doi.org/10.3390/microorganisms8070982
APA Stylevon Gastrow, L., Madec, M.-N., Chuat, V., Lubac, S., Morinière, C., Lé, S., Santoni, S., Sicard, D., & Valence, F. (2020). Microbial Diversity Associated with Gwell, a Traditional French Mesophilic Fermented Milk Inoculated with a Natural Starter. Microorganisms, 8(7), 982. https://doi.org/10.3390/microorganisms8070982






