Molecular Ecology of Isoprene-Degrading Bacteria
Abstract
:1. Isoprene and Climate
2. Global Isoprene Emissions
3. Biological Sinks for Isoprene
4. Diversity of Isoprene-degrading Bacteria
5. Isoprene Degradation Pathway
6. Molecular Techniques to Study the Ecology of Isoprene Degraders
6.1. isoA Probes
6.2. DNA Stable-Isotope Probing (DNA-SIP)
7. Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Atkinson, R.; Arey, J. Gas-phase tropospheric chemistry of biogenic volatile organic compounds: A review. Atmos. Environ. 2003, 37, 197–219. [Google Scholar] [CrossRef]
- Guenther, A.B.; Jiang, X.; Heald, C.L.; Sakulyanontvittaya, T.; Duhl, T.; Emmons, L.K.; Wang, X. The model of emissions of gases and aerosols from nature version 2.1 (MEGAN2.1): An extended and updated framework for modelling biogenic emissions. Geosci. Model Dev. 2012, 5, 1471–1492. [Google Scholar] [CrossRef] [Green Version]
- Pacifico, F.; Harrison, S.P.; Jones, C.D.; Sitch, S. Isoprene emissions and climate. Atmos. Environ. 2009, 43, 6121–6135. [Google Scholar] [CrossRef]
- Collins, W.J.; Derwent, R.G.; Johnson, C.E.; Stevenson, D.S. The oxidation of organic compounds in the troposphere and their global warming potentials. Clim. Chang. 2002, 52, 453–479. [Google Scholar] [CrossRef]
- Folberth, G.A.; Hauglustaine, D.A.; Lathière, J.; Brocheton, F. Interactive chemistry in the Laboratoire de Météorologie Dynamique general circulation model: Model description and impact analysis of biogenic hydrocarbons on tropospheric chemistry. Atmos. Chem. Phys. 2006, 6, 2273–2319. [Google Scholar] [CrossRef] [Green Version]
- Ashworth, K.; Wild, O.; Hewitt, C.N. Impacts of biofuel cultivation on mortality and crop yields. Nat. Clim. Change. 2013, 3, 492–496. [Google Scholar] [CrossRef]
- Carlton, A.G.; Wiedinmyer, C.; Kroll, J.H. A review of secondary organic aerosol (SOA) formation from isoprene. Atmos. Chem. Phys. 2009, 9, 4987–5005. [Google Scholar] [CrossRef] [Green Version]
- Engelhart, G.J.; Moore, R.H.; Nenes, A.; Pandis, S.N. Cloud condensation nuclei activity of isoprene secondary organic aerosol. J. Geophys. Res. 2011, 116, D02207. [Google Scholar] [CrossRef] [Green Version]
- Sharkey, T.D.; Wiberley, A.E.; Donohue, A.R. Isoprene emission from plants: Why and how. Ann. Bot. 2008, 101, 5–18. [Google Scholar] [CrossRef] [Green Version]
- Loreto, F.; Ciccioli, P.; Brancaleoni, E.; Valentini, R.; De Lillis, M.; Csiky, O.; Seufert, G. A hypothesis on the evolution of isoprenoid emission by oaks based on the correlation between emission type and Quercus taxonomoy. Oeacologia 1998, 115, 17430–17435. [Google Scholar]
- Monson, R.K.; Jones, R.T.; Rosenstiel, T.N.; Schnitzler, J.P. Why only some plants emit isoprene. Plant Cell Environ. 2013, 36, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D. Is it useful to ask why plants emit isoprene? Plant Cell Environ. 2013, 36, 517–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohmer, M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999, 16, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D.; Singsaas, E.L.; Vanderveer, P.J.; Geron, C. Field measurements of isoprene emission from trees in response to temperature and light. Tree Physiol. 1996, 16, 649–654. [Google Scholar] [CrossRef]
- Lantz, A.T.; Allman, J.; Weraduwage, S.M.; Sharkey, T.D. Isoprene: New insights into the control of emission and mediation of stress tolerance by gene expression. Plant Cell Environ. 2019, 42, 2808–2826. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, C.N.; MacKenzie, A.R.; Di Carlo, P.; Di Marco, C.F.; Dorsey, J.R.; Evans, M.; Fowler, D.; Gallagher, M.W.; Hopkins, J.R.; Jones, C.E.; et al. Nitrogen management is essential to prevent tropical oil palm plantations from causing ground-level ozone pollution. Proc. Natl. Acad. Sci. USA 2009, 106, 18447–18451. [Google Scholar] [CrossRef] [Green Version]
- Monson, R.K.; Winkler, B.; Rosenstiel, T.N.; Block, K.; Merl-Pham, J.; Strauss, S.H.; Ault, K.; Maxfield, J.; Moore, D.J.P.; Trahan, N.A.; et al. High productivity in hybrid poplar plantations without isoprene emission to the atmosphere. Proc. Natl. Acad. Sci. USA 2020, 117, 1596–1605. [Google Scholar] [CrossRef] [Green Version]
- Bäck, J.; Aaltonen, H.; Hellén, H.; Kajos, M.K.; Patokoski, J.; Taipale, R.; Pumpanen, J.; Heinonsalo, J. Variable emissions of microbial volatile organic compounds (MVOCs) from root-associated fungi isolated from Scots pine. Atmosph. Environ. 2010, 44, 3651–3659. [Google Scholar] [CrossRef]
- Dani, S.K.G.; Benavides, A.M.S.; Michelozzi, M.; Peluso, G.; Torzillo, G.; Loreto, F. Relationship between isoprene emission and photosynthesis in diatoms, and its implications for global marine isoprene estimates. Mar. Chem. 2017, 189, 17–24. [Google Scholar] [CrossRef]
- Exton, D.A.; McGenity, T.J.; Steinke, M.; Smith, D.J.; Suggett, D.J. Uncovering the volatile nature of tropical coastal marine ecosystems in a changing world. Glob. Chang. Biol. 2015, 21, 1383–1394. [Google Scholar] [CrossRef]
- Fall, R.; Copley, S.D. Bacterial sources and sinks of isoprene, a reactive atmospheric hydrocarbon. Environ. Microbiol. 2000, 2, 123–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelmont, D.; Stein, R.A.; Mead, J.F. Isoprene—the main hydrocarbon in human breath. Biochem. Biophys. Res. Commun. 1981, 99, 1456–1460. [Google Scholar] [CrossRef]
- Kuzma, J.; Nemecek-Marshall, M.; Pollock, W.; Fall, R. Bacteria produce the volatile hydrocarbon isoprene. Curr. Microbiol. 1995, 30, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Schöller, C.; Molin, S.; Wilkins, K. Volatile metabolites from some Gram-negative bacteria. Chemosphere 1997, 35, 1487–1495. [Google Scholar] [CrossRef]
- Schöller, C.E.G.; Gürtler, H.; Pederson, R.; Molin, S.; Wilkins, K. Volatile metabolites from Actinomycetes. J. Agric. Food Chem. 2002, 50, 2615–2621. [Google Scholar] [CrossRef]
- McGenity, T.J.; Crombie, A.T.; Murrell, J.C. Microbial cycling of isoprene, the most abundantly produced biological volatile organic compound on Earth. ISME J. 2018, 12, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Dani, S.K.G.; Loreto, F. Trade-off between dimethyl sulfide and isoprene emissions from marine phytoplankton. Trends Plant Sci. 2017, 22, 361–372. [Google Scholar] [CrossRef]
- Shaw, S.L.; Gantt, B.; Meskhidze, N. Production and emissions of marine isoprene and monoterpenes: A review. Adv. Meterol. 2010. [Google Scholar] [CrossRef]
- Exton, D.A.; Suggett, D.J.; Steinke, M.; McGenity, T.J. Spatial and temporal variability of biogenic isoprene emissions from a temperate estuary. Glob. Biogeochem. Cycl. 2012, 26, GB2012. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, S.C.; Andrews, S.J.; Airs, R.; Arnold, S.R.; Bouman, H.A.; Brewin, R.J.W.; Chance, R.J.; Cummings, D.; Dall’Omo, G.; Lewis, A.C.; et al. Potential controls of isoprene in the surface ocean. Glob. Biogeochem. Cycles 2017, 31, 644–662. [Google Scholar] [CrossRef]
- Meskhidze, N.; Sabolis, A.; Reed, R.; Kamykowski, D. Quantifying environmental stress-induced emissions of algal isoprene and monoterpenes using laboratory measurements. Biogeosciences 2015, 12, 637–651. [Google Scholar] [CrossRef] [Green Version]
- Ekberg, A.; Arneth, A.; Hakota, H.; Hayward, S.; Holst, T. Isoprene emission from wetland sedges. Biogeosciences 2009, 6, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Steinke, M.; Hodapp, B.; Subhan, R.; Bell, T.G.; Martin-Creuzburg, D. Flux of the biogenic volatiles isoprene and dimethyl sulfide from an oligotrophic lake. Sci. Rep. 2018, 8, 630. [Google Scholar] [CrossRef] [PubMed]
- Morais, A.R.C.; Dworakowska, S.; Reis, A.; Gouveia, L.; Matos, C.T.; Bogdal, D.; Bogel-Lukasik, R. Chemical and biological-based isoprene production: Green metrics. Catal. Today 2015, 239, 38–43. [Google Scholar] [CrossRef] [Green Version]
- Borbon, A.; Fontaine, H.; Veillerot, M.; Locoge, N.; Galloo, J.C.; Guillermo, R. An investigation into the traffic-related fraction of isoprene at an urban location. Atmos. Environ. 2001, 35, 3749–3760. [Google Scholar] [CrossRef]
- Chen, J.; Li, C.; Ristovski, Z.; Milic, A.; Gu, Y.; Islam, M.S.; Whang, S.; Hao, J.; Zhang, H.; He, C.; et al. A review of biomass burning: Emissions and impacts on air quality, health and climate in China. Sci. Total Environ. 2017, 579, 1000–1034. [Google Scholar] [CrossRef] [Green Version]
- Cleveland, C.C.; Yavitt, J.B. Consumption of atmospheric isoprene in soil. Geophys. Res. Lett. 1997, 24, 2379–2382. [Google Scholar] [CrossRef] [Green Version]
- Cleveland, C.C.; Yavitt, J.B. Microbial consumption of atmospheric isoprene in a temperate forest soil. Appl. Environ. Microbiol. 1998, 64, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Gray, C.M.; Helmig, D.; Fierer, N. Bacteria and fungi associated with isoprene consumption in soil. Elem. Sci. Anth. 2015, 3, 000053. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, E.; Abrell, L.; Van Haren, J.; Barron-Gafford, G.; Grieve, K.A.; Yadvinder, M.; Murthy, R.; Lin, G. The effect of elevated atmospheric CO2 and drought on sources and sinks of isoprene in a temperate and tropical rainforest mesocosm. Glob. Chang. Biol. 2005, 11, 1234–1246. [Google Scholar] [CrossRef]
- Acuña Alvarez, L.; Exton, D.A.; Timmis, K.N.; Suggett, D.J.; McGenity, T.J. Characterization of marine isoprene-degrading communities. Environ. Microbiol. 2009, 11, 3280–3291. [Google Scholar] [CrossRef] [PubMed]
- Carrión, O.; Gibson, L.; Elias, D.M.O.; McNamara, N.P.; van Alen, T.A.; Op den Camp, H.J.M.; Supramaniam, C.V.; McGenity, T.J.; Murrell, J.C. Diversity of isoprene-degrading bacteria in phyllosphere and soil communities from a high isoprene-emitting environment: A Malaysian oil palm plantation. Microbiome 2020, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Crombie, A.T.; Larke-Mejía, N.L.; Emery, H.; Dawson, R.; Pratscher, J.; Murphy, G.P.; McGenity, T.J.; Murrell, J.C. Poplar phyllosphere harbors disparate isoprene-degrading bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 13081–13086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, L.; Crombie, A.T.; Emery, H.; McGenity, T.J.; Murrell, J.C. Community profiling of isoprene-degrading bacteria associated with the phyllosphere of a Salix tree. Manuscript in preparation.
- Larke-Mejía, N.L.; Carrión, O.; Crombie, A.T.; McGenity, T.J.; Murrell, J.C. Sphingopyxis sp. OPL5, an isoprene-degrading bacterium from the Sphingomonadaceae family isolated from oil palm leaves. Manuscript in preparation.
- Ewers, J.; Freier-Schroder, D.; Knackmuss, H.J. Selection of trichloroethene (TCE) degrading bacteria that resist inactivation by TCE. Arch. Microbiol. 1990, 154, 410–413. [Google Scholar] [CrossRef]
- Van Ginkel, C.G.; de Jong, E.; Tilanus, J.W.R.; de Bont, J.A.M. Microbial oxidation of isoprene, a biogenic foliage volatile and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol. Lett. 1987, 45, 275–279. [Google Scholar] [CrossRef]
- Van Ginkel, C.G.; Welten, H.G.J.; de Bont, J.A.M. Oxidation of gaseous and volatile hydrocarbons by selected alkene-utilizing bacteria. Appl. Environ. Microb. 1987, 53, 2903–2907. [Google Scholar] [CrossRef] [Green Version]
- Srivastva, N.; Shukla, A.K.; Singh, R.S.; Upadhyay, S.N.; Dubey, S.K. Characterisation of bacterial isolates from rubber dump site and their use in biodegradation of isoprene in batch and continuous bioreactors. Bioresour. Technol. 2015, 188, 84–91. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Dubey, S.K. Molecular characterization and kinetics of isoprene degrading bacteria. Bioresour. Technol. 2019, 278, 51–56. [Google Scholar] [CrossRef]
- El Khawand, M.; Crombie, A.T.; Johnston, A.; Vavlline, D.V.; McAuliffe, J.C.; Latone, J.A.; Primak, Y.A.; Lee, S.K.; Whited, G.M.; McGenity, T.J.; et al. Isolation of isoprene degrading bacteria from soils, development of isoA gene probes and identification of the active isoprene-degrading soil community using DNA-stable isotope probing. Environ. Microbiol. 2016, 18, 2743–2753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crombie, A.T.; Emery, H.; McGenity, T.J.; Murrell, J.C. Draft genome sequences of three terrestrial isoprene-degrading Rhodococcus strains. Genome Announc. 2017, 5, e01256-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, R.A.; Larke-Mejía, N.L.; Crombie, A.T.; Ul Haque, M.F.; Murrell, J.C. Isoprene oxidation by the Gram-negative model bacterium Variovorax sp. WS11. Microorganisms 2020, 8, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, L.; Larke-Mejía, N.L.; Murrell, J.C. Complete genome of isoprene degrading Nocardioides sp. WS12. Microorganisms 2020, 8, 889. [Google Scholar] [CrossRef]
- Larke-Mejía, N.L. Microbial Ecology of Isoprene degraders in the Terrestrial Environment. Ph.D. Thesis, University of East Anglia, Norwich, UK, 2018. [Google Scholar]
- Larke-Mejía, N.L.; Crombie, A.T.; Pratscher, J.; McGenity, T.J.; Murrell, J.C. Novel isoprene-degrading Proteobacteria from soil and leaves identified by cultivation and metagenomics analysis of stable isotope probing experiments. Front. Microbiol. 2019, 10, 2700. [Google Scholar] [CrossRef]
- Johnston, A.; Crombie, A.T.; El Khawand, E.; Sims, L.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Environ. Microbiol. 2017, 19, 3526–3537. [Google Scholar] [CrossRef] [Green Version]
- Van Hylckama Vlieg, J.E.T.; Kingma, J.; van den Wijngaard, A.J.; Janssen, D.B. A glutathione S-transferase with activity towards cis-1, 2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 1998, 64, 2800–2805. [Google Scholar] [CrossRef] [Green Version]
- Van Hylckama Vlieg, J.E.T.; Kingma, J.; Kruizinga, W.; Janssen, D.B. Purification of a glutathione S-transferase and a conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 1999, 181, 2094–2101. [Google Scholar] [CrossRef] [Green Version]
- Van Hylckama Vlieg, J.E.T.; Leemhuis, H.; Spelberg, J.H.L.; Janssen, D.B. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 2000, 182, 1956–1963. [Google Scholar] [CrossRef] [Green Version]
- Crombie, A.T.; El Khawand, M.; Rhodius, V.A.; Fengler, K.A.; Miller, M.C.; Whited, G.M.; McGenity, T.J.; Murrell, J.C. Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle. Environ. Microbiol. 2015, 17, 3314–3329. [Google Scholar] [CrossRef] [Green Version]
- Murrell, J.C.; McGenity, T.J.; Crombie, A.T. Microbial metabolism of isoprene: A much-neglected climate-active gas. Microbiology 2020. [Google Scholar] [CrossRef]
- Kronen, M.; Lee, M.; Jones, Z.L.; Manefield, M.J. Reductive metabolism of the important atmospheric gas isoprene by homoacetogens. ISME J. 2019, 13, 1168–1182. [Google Scholar] [CrossRef] [PubMed]
- Leahy, J.G.; Batchelor, P.J.; Morcomb, S.M. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 2003, 27, 449–479. [Google Scholar] [CrossRef]
- Dumont, M.G.; Murrell, J.C. Community-level analysis: Key genes of aerobic methane oxidation. Methods Enzymol. 2005, 397, 413–427. [Google Scholar] [PubMed]
- McDonald, I.R.; Kenna, E.M.; Murrell, J.C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl. Environ. Microbiol. 1995, 61, 116–121. [Google Scholar] [CrossRef] [Green Version]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef] [Green Version]
- Farhan Ul Haque, M.; Crombie, A.T.; Ensminger, S.A.; Baciu, C.; Murrell, J.C. Facultative methanotrophs are abundant at terrestrial natural gas seeps. Microbiome 2018, 6, 118. [Google Scholar] [CrossRef]
- Carrión, O.; Larke-Mejía, N.L.; Gibson, L.; Farhan Ul Haque, M.; Ramiro-García, J.; McGenity, T.J.; Murrell, J.C. Gene probing reveals the widespread distribution, diversity and abundance of isoprene-degrading bacteria in the environment. Microbiome 2018, 6, 219. [Google Scholar] [CrossRef]
- Kesselmeier, J.; Staudt, M. Biogenic volatile organic compounds (VOC): An overview on emission, physiology and ecology. J. Atmos. Chem. 1999, 33, 23–88. [Google Scholar] [CrossRef]
- Radajewski, S.; Ineson, P.; Parekh, N.R.; Murrell, J.C. Stable-isotope probing as a tool in microbial ecology. Nature 2000, 403, 646–649. [Google Scholar] [CrossRef]
- Neufeld, J.D.; Vohra, J.; Dumont, M.G.; Lueders, T.; Manefield, M.; Friedrich, M.W.; Murrell, J.C. DNA stable-isotope probing. Nat. Protoc. 2007, 2, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Jehmlich, N.; Schmidt, F.; Taubert, M.; Seifert, J.; Bastida, F.; von Bergen, M.; Richnow, H.H.; Vogt, C. Protein-based stable isotope probing. Nat. Protoc. 2010, 5, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu. Rev. Microbiol. 2009, 63, 411–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, W.E.; Cui, L.; Wagner, M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 2016, 41, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemlich, N.; Vogt, C.; Lüsmann, V.; Richnow, H.H.; von Bergen, M. Protein-SIP in environmental studies. Curr. Opinion. Biotechnol. 2016, 41, 26–33. [Google Scholar]
- Lünsmann, V.; Kappelmeyer, U.; Benndorf, R.; Martinez-Lavanchy, P.M.; Taubert, A.; Adrian, L.; Duarte, M.; Pieper, D.H.; von Bergen, M.; Müller, J.A.; et al. In situ protein-SIP highlights Burkholderiaceae as key players degrading toluene by para ring hydroxylation in a constructed wetland model. Environ. Microbiol. 2016, 18, 1176–1186. [Google Scholar] [CrossRef]
- Ouyang, W.Y.; Su, J.Q.; Richnow, H.H.; Adrian, L. Identification of dominant sulfamethoxazole-degraders in pig farm-impacted soil by DNA and protein stable isotope probing. Environ. Int. 2019, 126, 118–126. [Google Scholar] [CrossRef]
- Hatzenpichler, R.; Krukenberg, V.; Spietz, R.L.; Jay, Z.J. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Microbiol. 2020, 18, 241–256. [Google Scholar] [CrossRef]
- Singer, E.; Wagner, M.; Woyke, T. Capturing the genetic makeup of the active microbiome in situ. ISME J. 2017, 11, 1949–1963. [Google Scholar] [CrossRef]
- Berry, D.; Mader, E.; Lee, T.K.; Woebken, D.; Wang, Y.; Zhu, D.; Palatinszky, M.; Schintlmeister, A.; Schmid, M.C.; Hanson, B.T.; et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. USA 2015, 112, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.E.; Ward, A.D.; Whiteley, A.S. Raman tweezers sorting of single microbial cells. Environ. Microbiol. 2009, 1, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Gou, H.; Gong, Y.; Su, X.; Xu, L.; Yuetong, J.; Song, Y.; Thompson, I.P.; Xu, J.; Huang, W.E. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean. Environ. Microbiol. 2018, 20, 2241–2255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichorst, S.A.; Strasser, F.; Woyke, T.; Schintlmeister, A.; Wagner, M.; Woebken, D. Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol. Ecol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, M.G.; Pommerenke, B.; Casper, P. Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment. Environ. Microbiol. Rep. 2013, 5, 757–764. [Google Scholar] [CrossRef]
- Fortunato, C.S.; Huber, J.A. Coupled RNA-SIP and metatranscriptomics of active chemolithoautotrophic communities at a deep-sea hydrothermal vent. ISME J. 2015, 10, 1925–1938. [Google Scholar] [CrossRef]
- Murphy, G.P.; Uttarotai, T.; Crombie, A.T.; Lawson, T.; Chitov, T.; Murrell, J.C.; Steinke, M.; McGenity, T.J. Soil as a sink for isoprene: The effects of isoprene concentration on consumption, and the identification of isoprene-degrading bacteria. Manuscript in preparation.
- Griggs, D.; Stafford-Smith, M.; Gaffney, O.; Rockström, J.; Öhman, M.C.; Shyamsundar, P.; Steffen, W.; Glaser, G.; Kanie, N.; Noble, I. Sustainable development goals for people and planet. Nature 2013, 495, 305–307. [Google Scholar] [CrossRef]
- Janson, R.; De Serves, C. Isoprene emissions from boreal wetlands in Scandinavia. J. Geosphys. Res. 1998, 103, 513–517. [Google Scholar] [CrossRef]
- Hanson, D.T.; Swanson, S.; Graham, L.E.; Sharkey, T.D. Evolutionary significance of isoprene emission from mosses. Am. J. Bot. 1999, 86, 634–639. [Google Scholar] [CrossRef]
- Lindwall, F.; Svendsen, S.S.; Nielsen, C.S.; Michelsen, A.; Rinnan, R. Warming increases isoprene emissions from an arctic fen. Sci. Total Environ. 2016, 553, 297–304. [Google Scholar] [CrossRef]
- Rinnan, R.; Albers, C. Soil uptake of volatile organic compounds: Ubiquitous and underestimated? JGR Biogeosciences 2020. [Google Scholar] [CrossRef]
- Bender, M.; Conrad, R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Lett. 1992, 101, 261–269. [Google Scholar] [CrossRef]
- Crombie, A.T. Metabolism of Methane and Propane and the Role of the Glyoxylate Bypass Enzymes in Methylocella silvestris BL2. Ph.D. Thesis, University of Warwick, Coventry, UK, 2011. [Google Scholar]

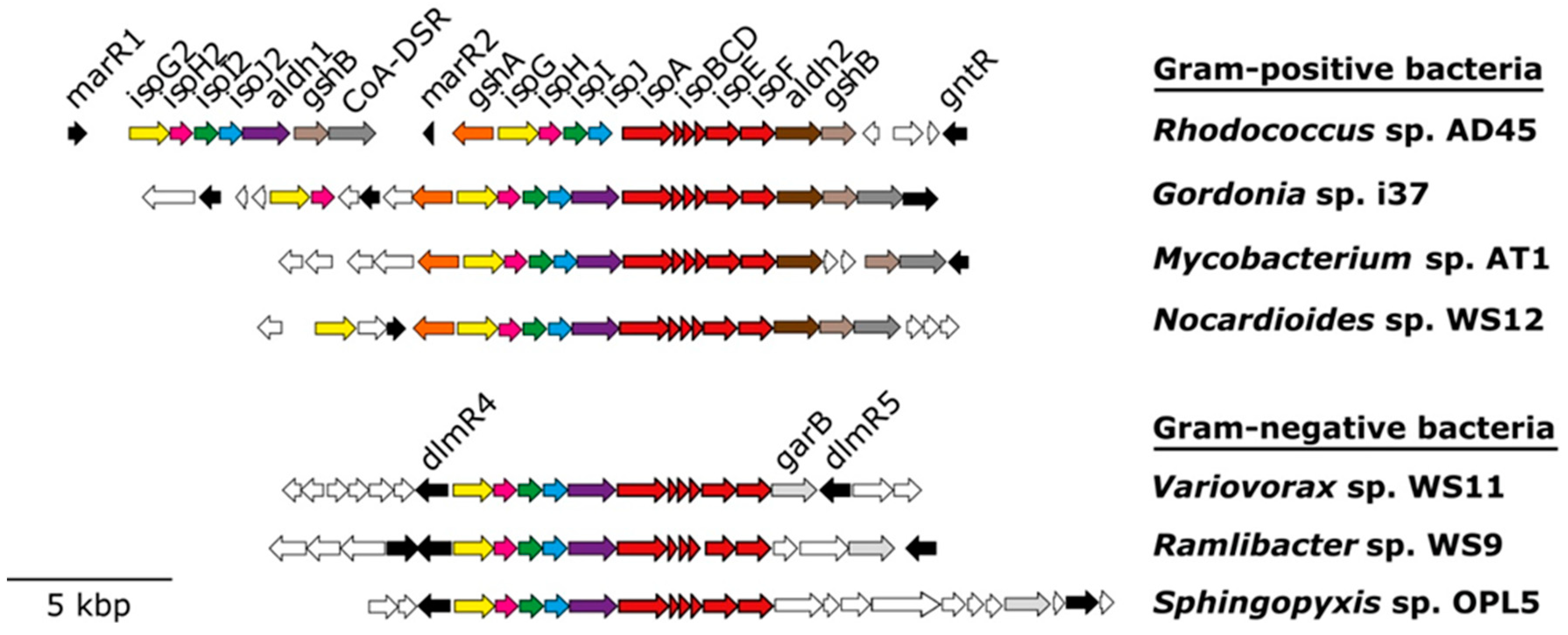
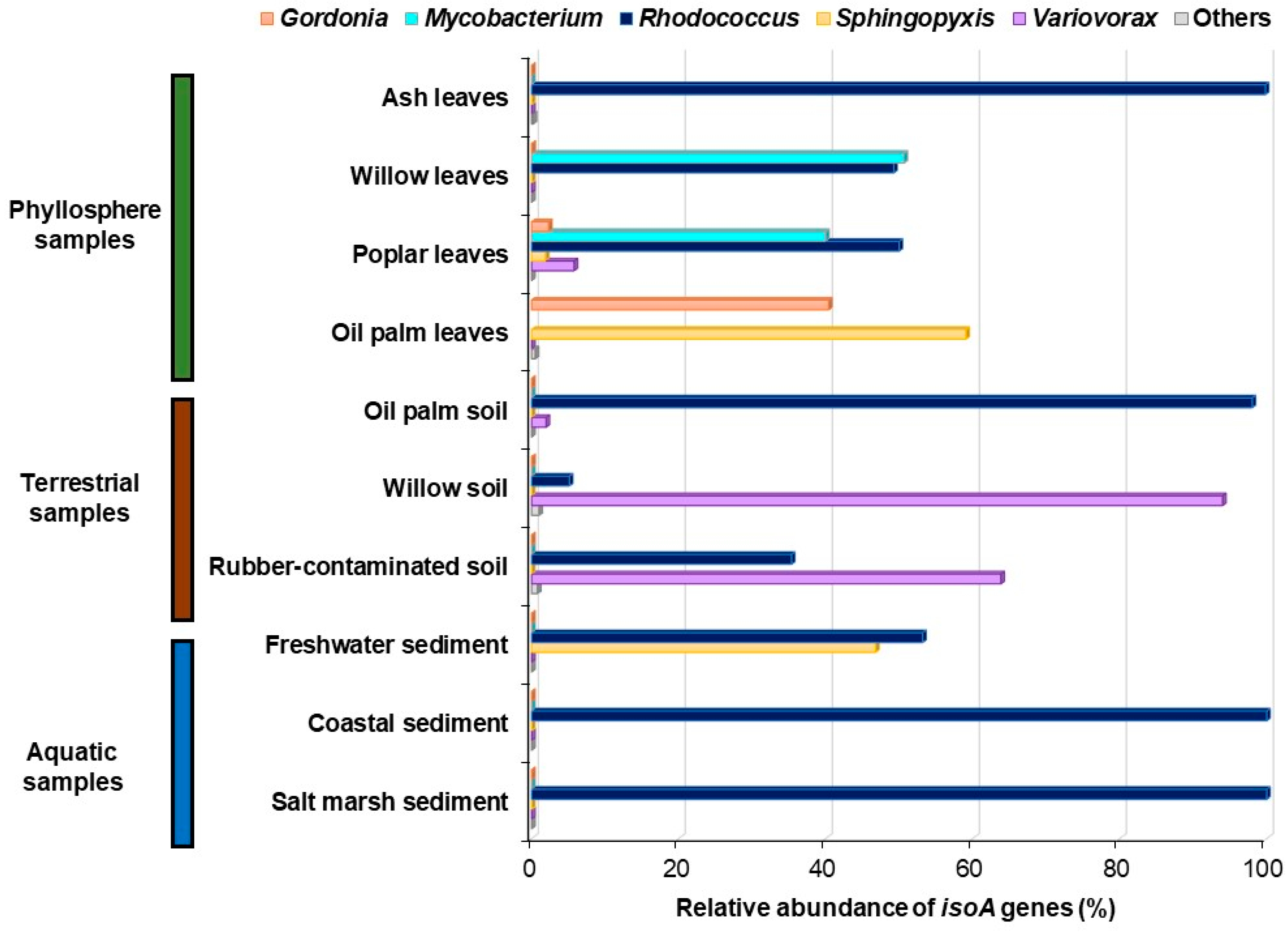
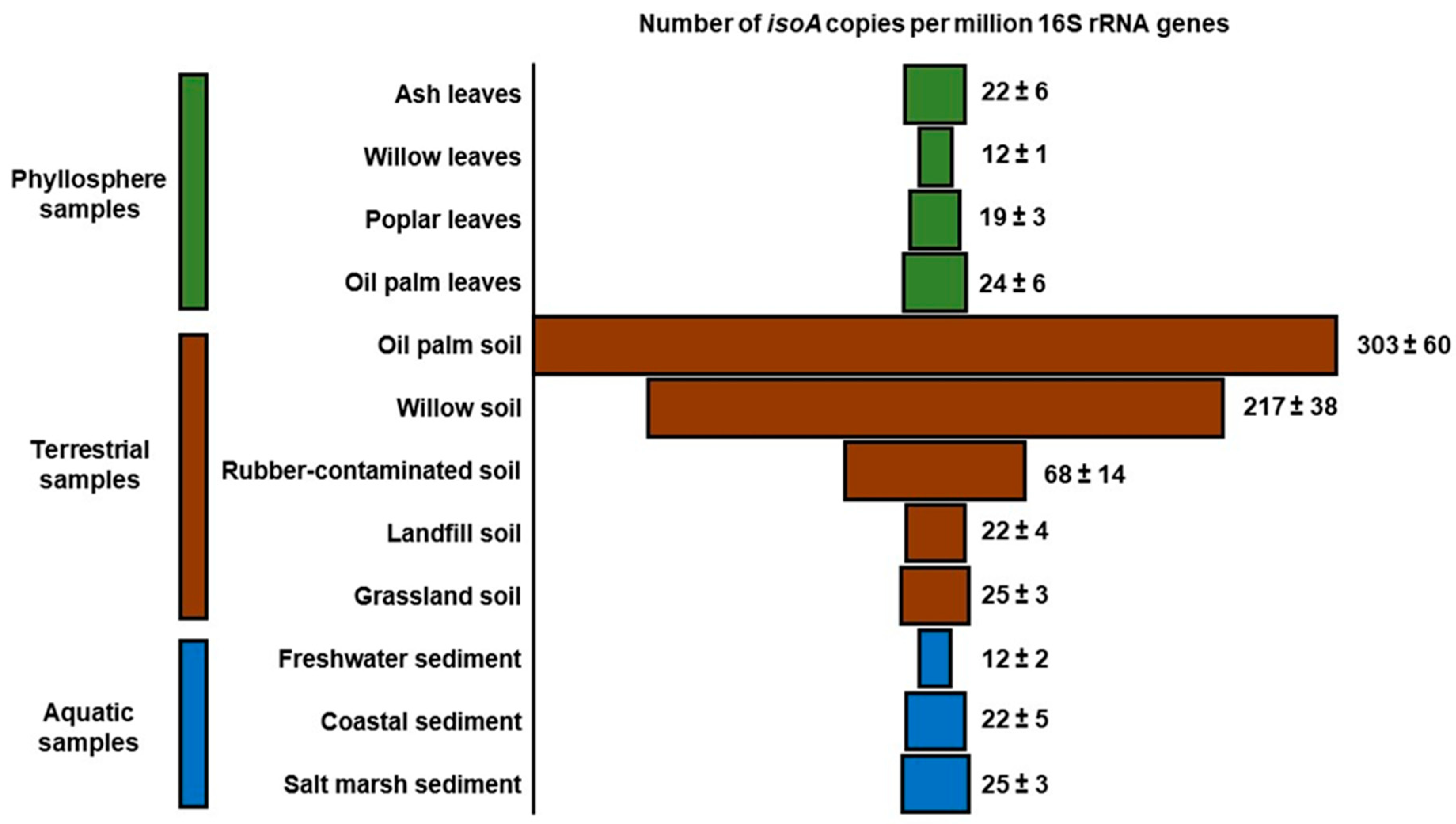
| Study | Environment | Isoprene Concentration (ppm) | Active Isoprene Degraders |
|---|---|---|---|
| El Khawand et al., 2016 [51] | Willow soil | 5000 | Rhodococcus, Variovorax, Comamonas |
| Johnston et al., 2017 [57] | Estuarine water and sediment | 2000 | Microbacterium, Rhodococcus, Mycobacterium, Gordonia |
| Crombie et al., 2018 [43] | Poplar leaves | 500 | Rhodococcus, Xanthomonadaceae, Comamonadaceae |
| 150 | Rhodococcus, Variovorax | ||
| Larke-Mejía et al., 2019 [56] | Willow soil | 25 | Ramlibacter, Variovorax, Rhodococcus |
| Carrión et al., 2020 [42] | Oil palm leaves | 25 | Gordonia, Zoogloea |
| Oil palm soil | 25 | Pelomonas, Novosphingobium, Rhodoblastus, Sphingomonas | |
| Larke-Mejía et al., unpublished [45] | Oil palm leaves | 25 | Gordonia, Sphingomonas, Aquincola |
| Oil palm soil | 25 | Aquabacterium, Rhodococcus, Saccharibacter |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrión, O.; McGenity, T.J.; Murrell, J.C. Molecular Ecology of Isoprene-Degrading Bacteria. Microorganisms 2020, 8, 967. https://doi.org/10.3390/microorganisms8070967
Carrión O, McGenity TJ, Murrell JC. Molecular Ecology of Isoprene-Degrading Bacteria. Microorganisms. 2020; 8(7):967. https://doi.org/10.3390/microorganisms8070967
Chicago/Turabian StyleCarrión, Ornella, Terry J. McGenity, and J. Colin Murrell. 2020. "Molecular Ecology of Isoprene-Degrading Bacteria" Microorganisms 8, no. 7: 967. https://doi.org/10.3390/microorganisms8070967
APA StyleCarrión, O., McGenity, T. J., & Murrell, J. C. (2020). Molecular Ecology of Isoprene-Degrading Bacteria. Microorganisms, 8(7), 967. https://doi.org/10.3390/microorganisms8070967





