Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. Cocoyam Tissue Culture, Plant Experiment, Bacterial Isolation and Culture Conditions
2.2. LC-MS and Nuclear Magnetic Resonance (NMR) Analysis
2.3. Genome Sequencing, Annotation, Mining, Bioinformatics and Phylogenetic Analyses
2.4. Swarming Motility and White Line-in-Agar Assay
2.5. Antimicrobial Activity Assays
3. Results
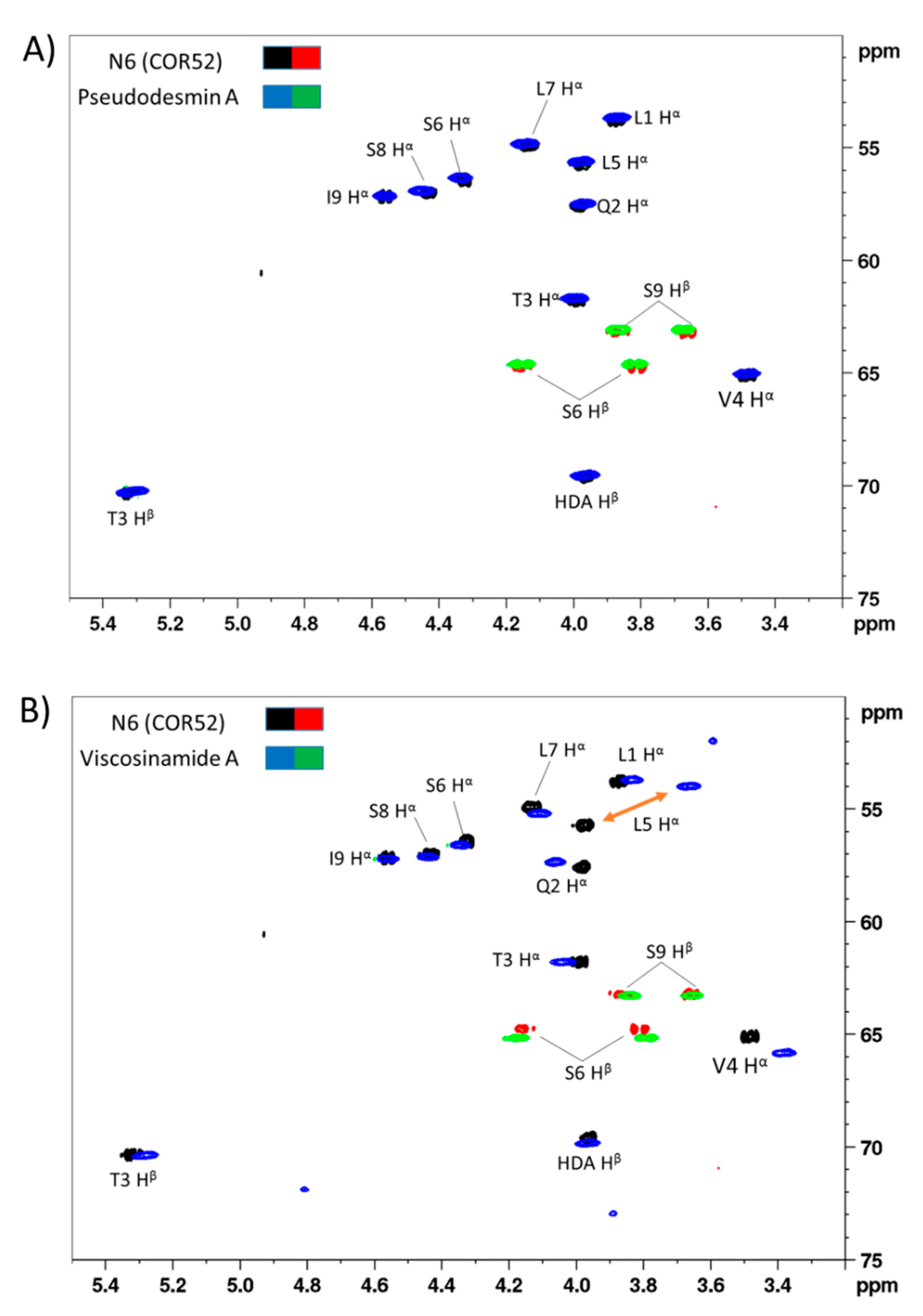
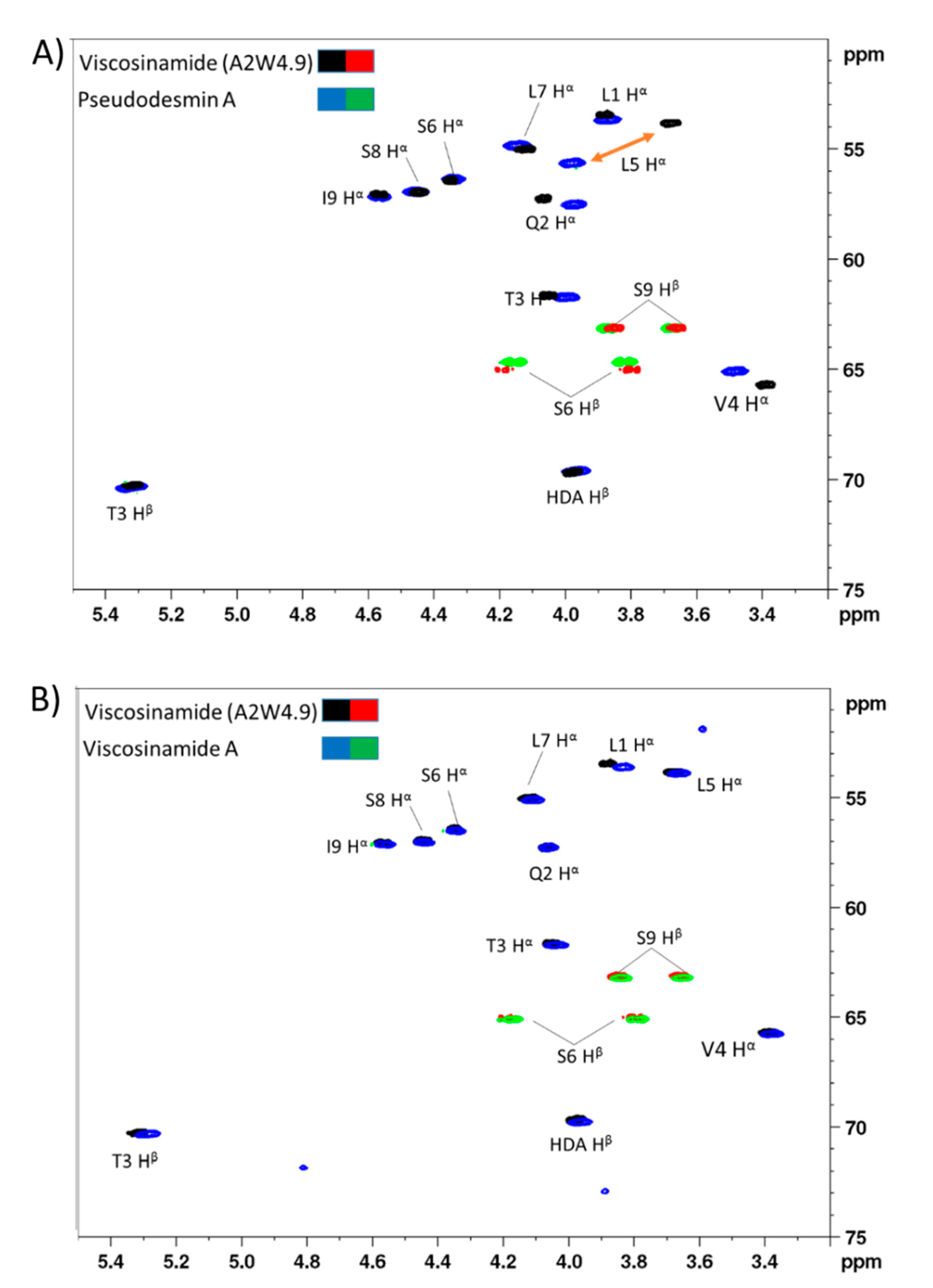
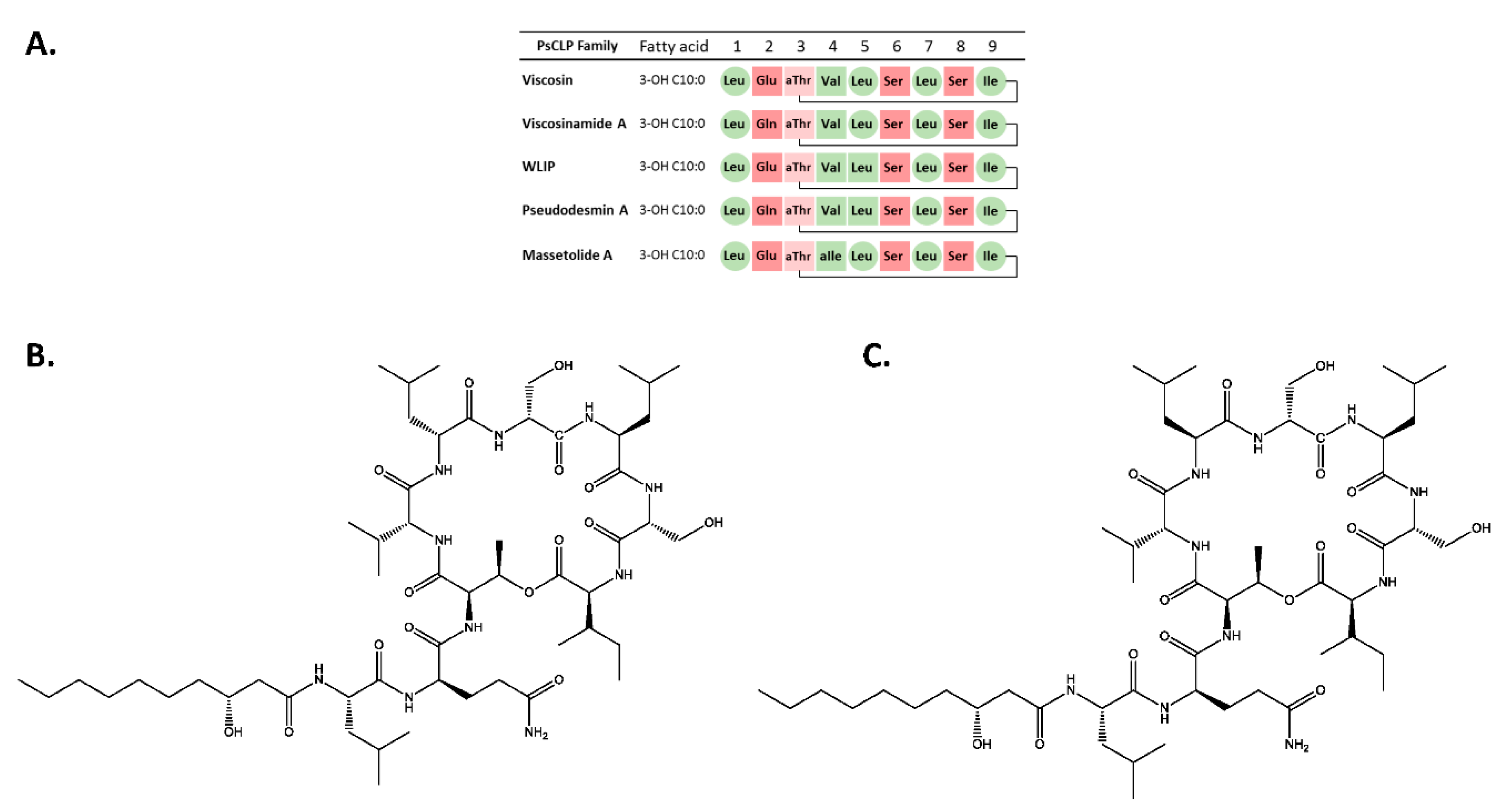
3.1. Chemical Structural Elucidation of Isolated CLPs
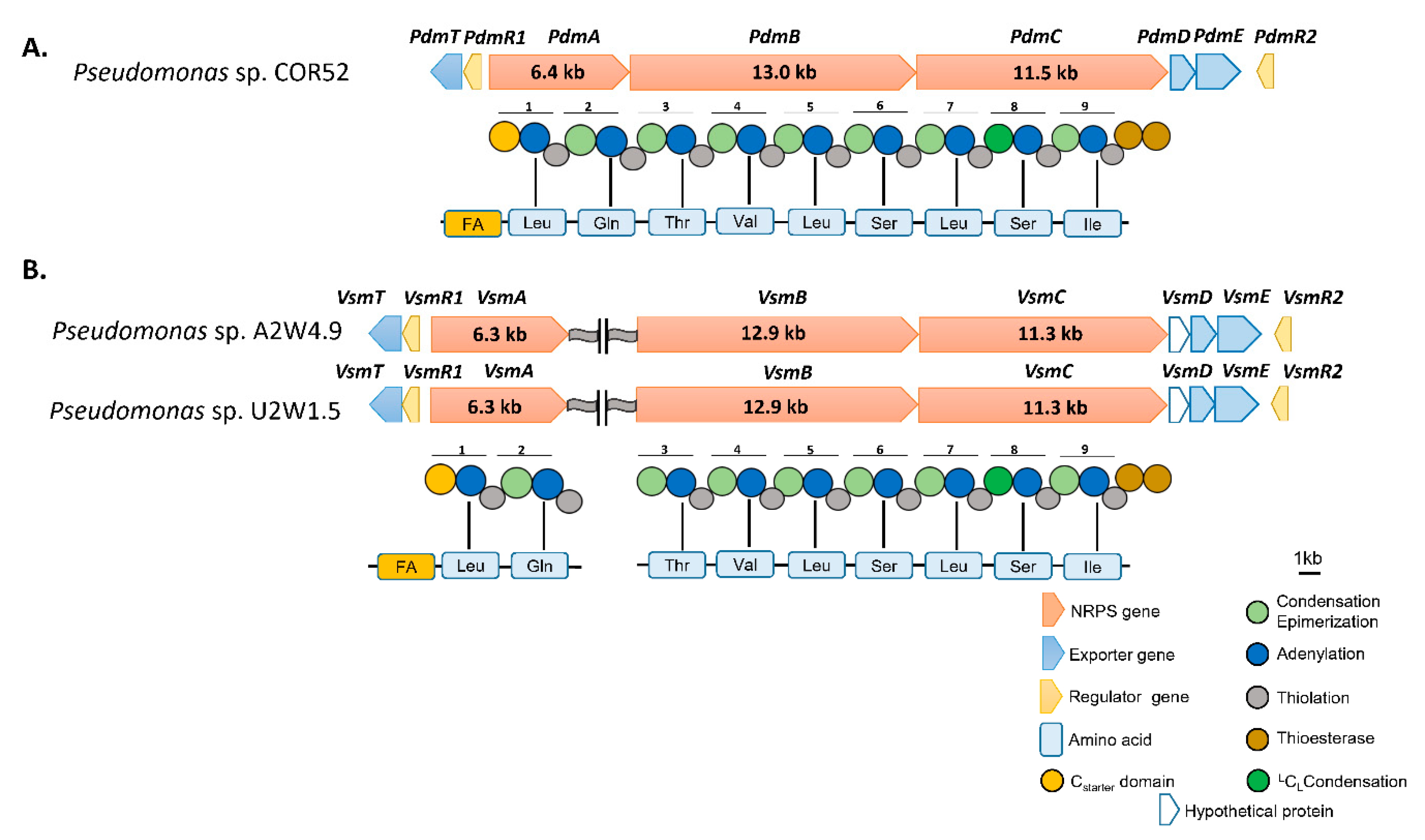
3.2. In Silico Analysis of the Pseudodesmin and Viscosinamide Biosynthetic Gene Cluster
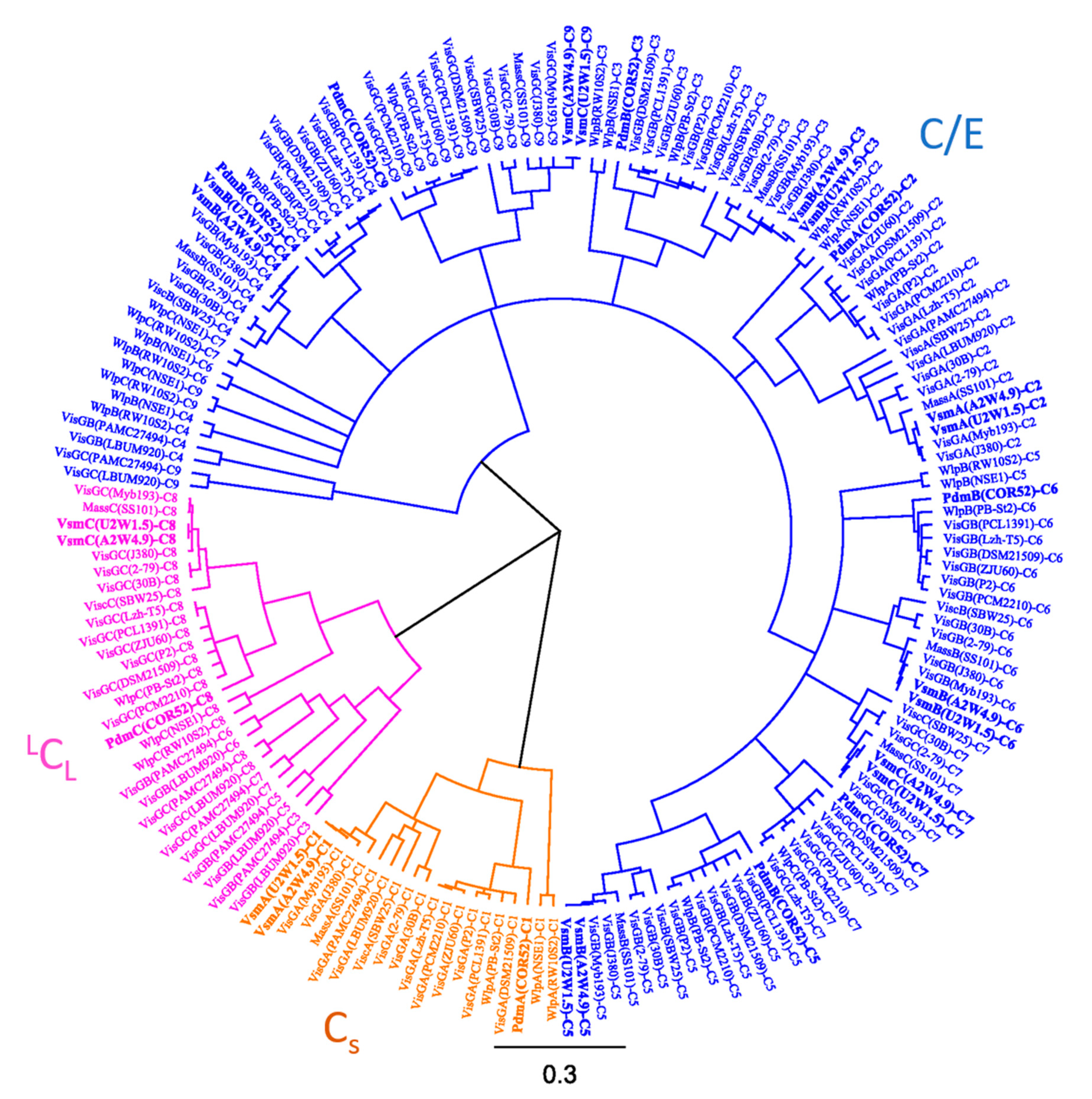
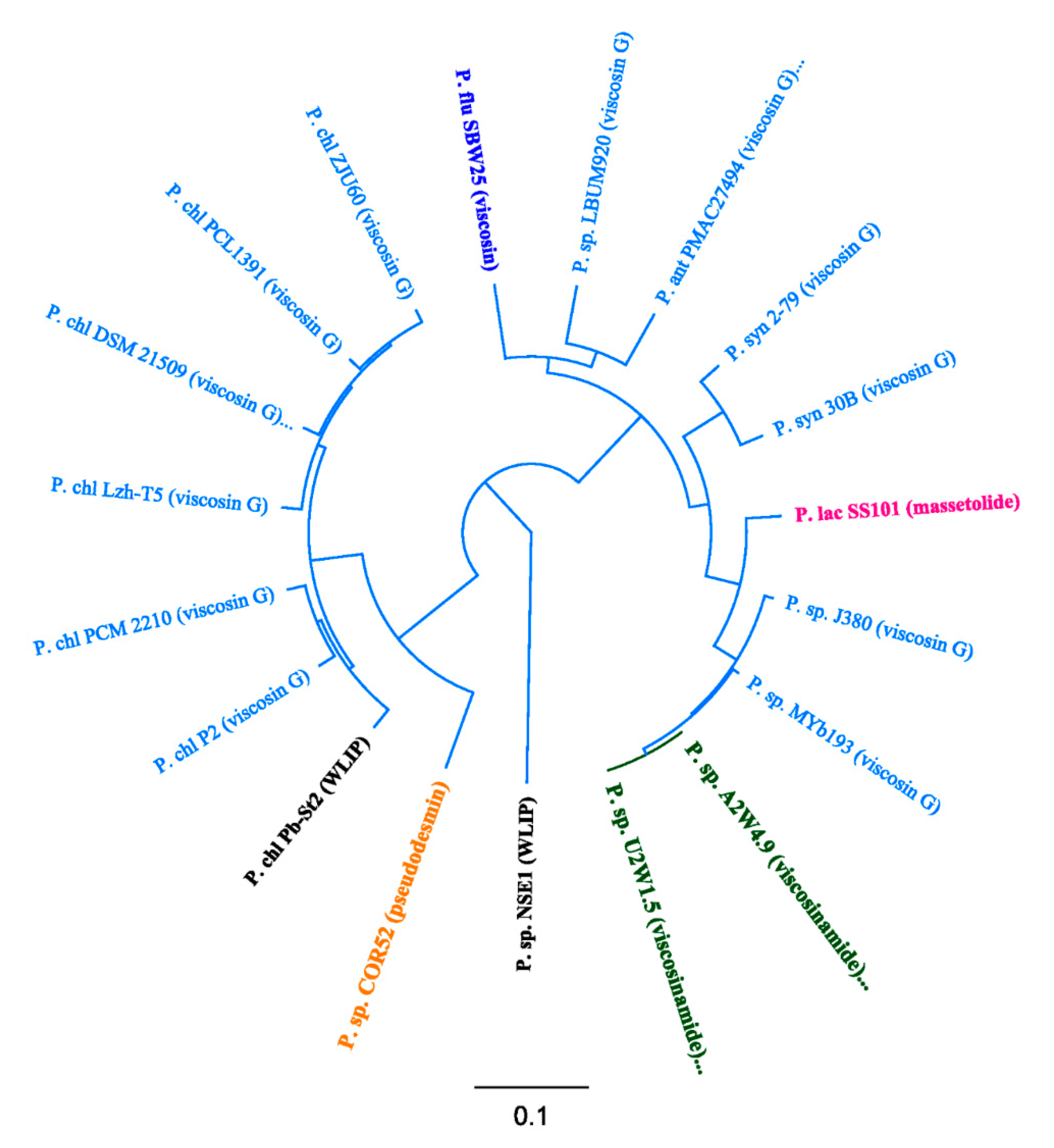
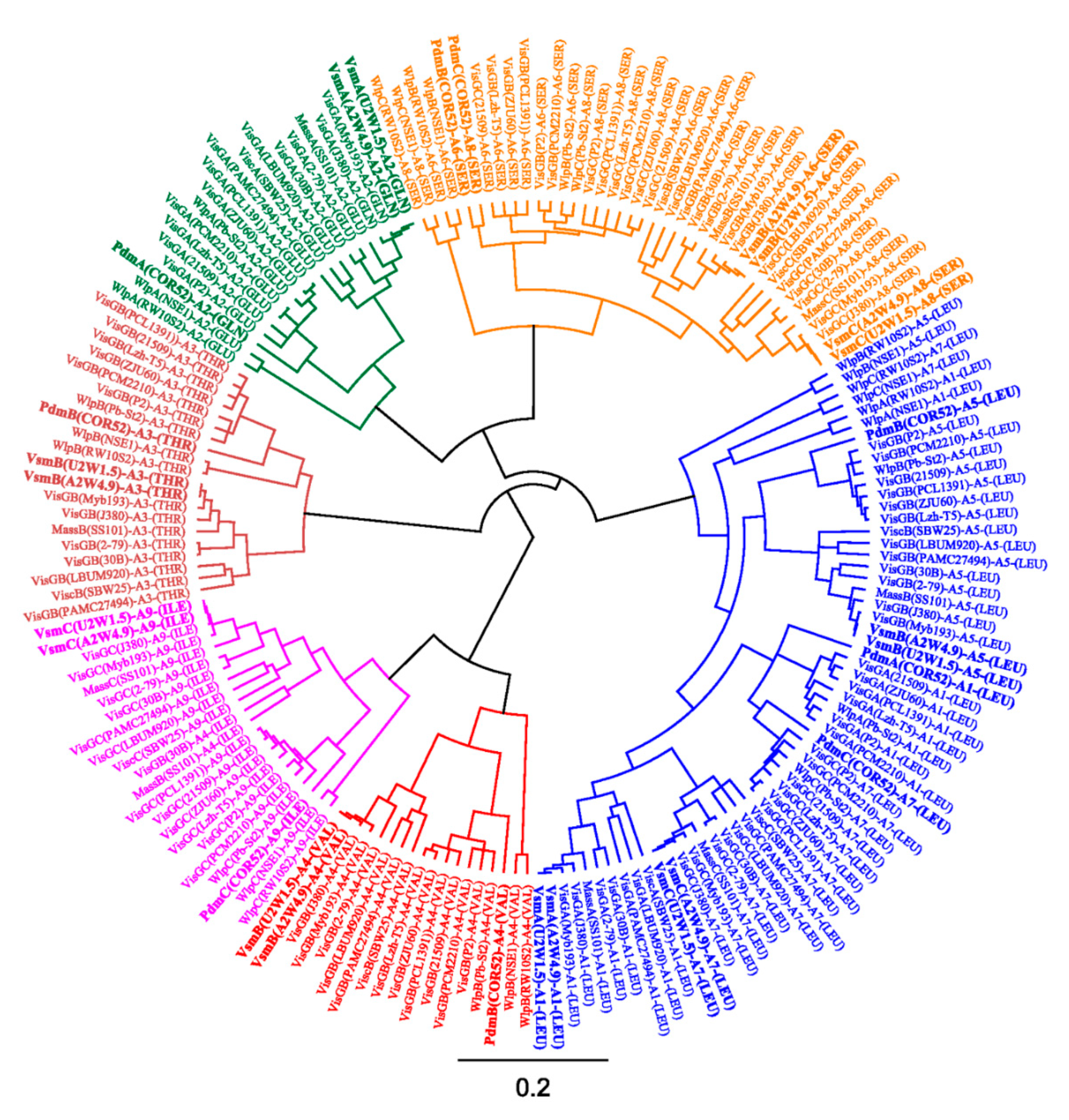
3.3. Phylogenetic Analyses of CLP-Producing Strains within the Viscosin Group
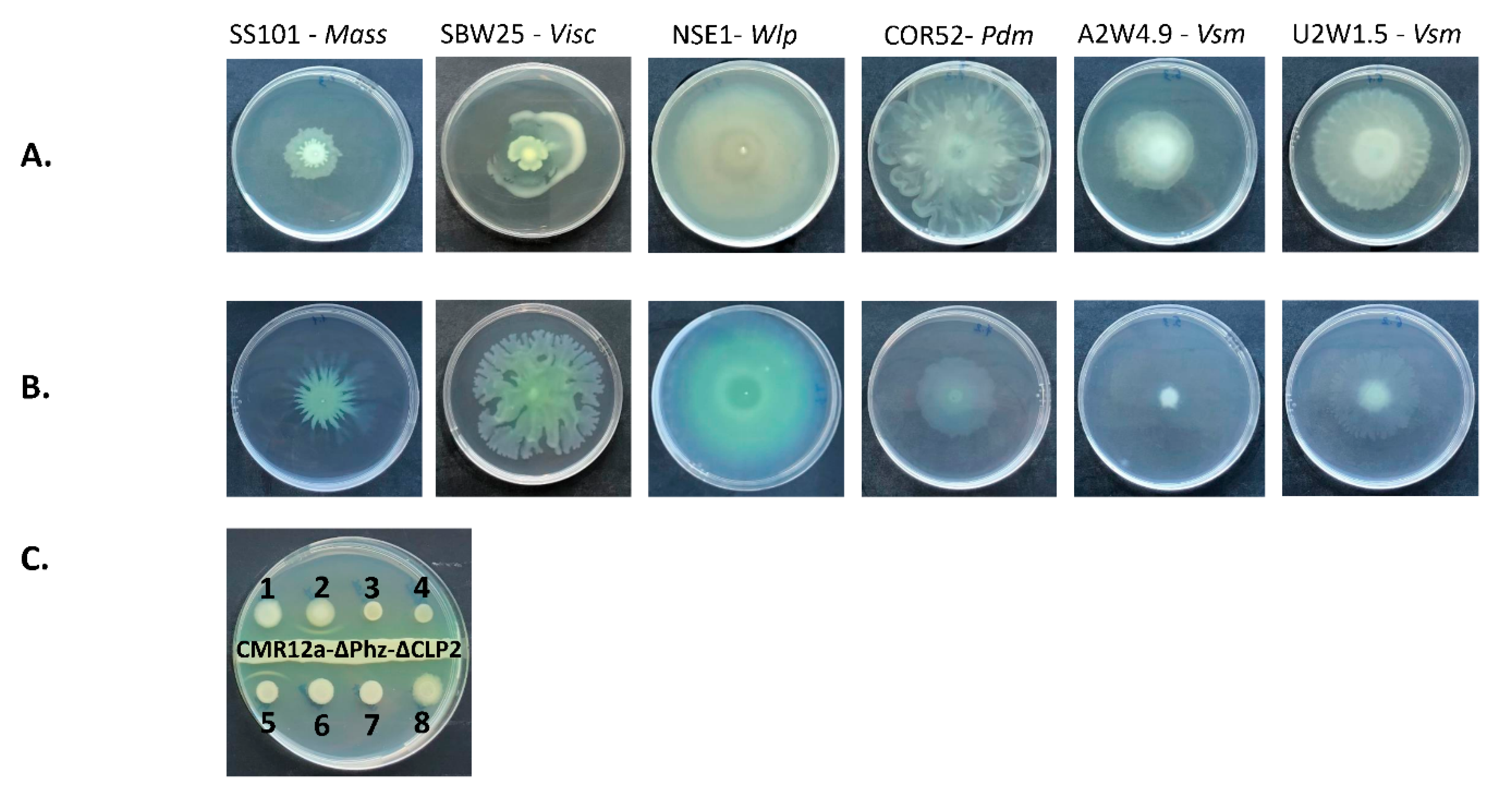
3.4. Swarming Motility and White Line-in-Agar Assay
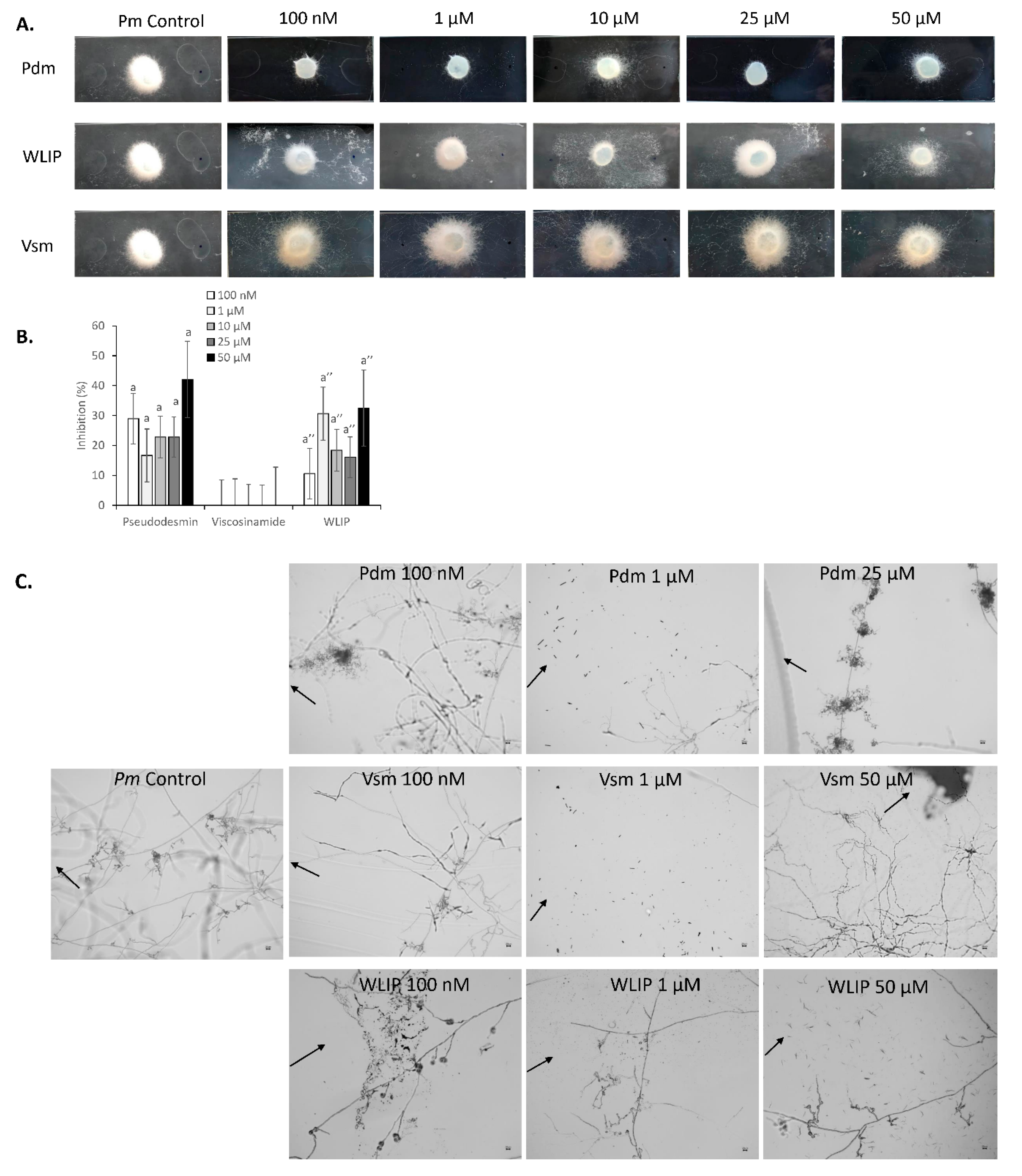
3.5. Inhibition of P. myriotylum by Pseudodesmin, Viscosinamide and WLIP
3.6. P. myriotylum-CLP Perception and Interaction
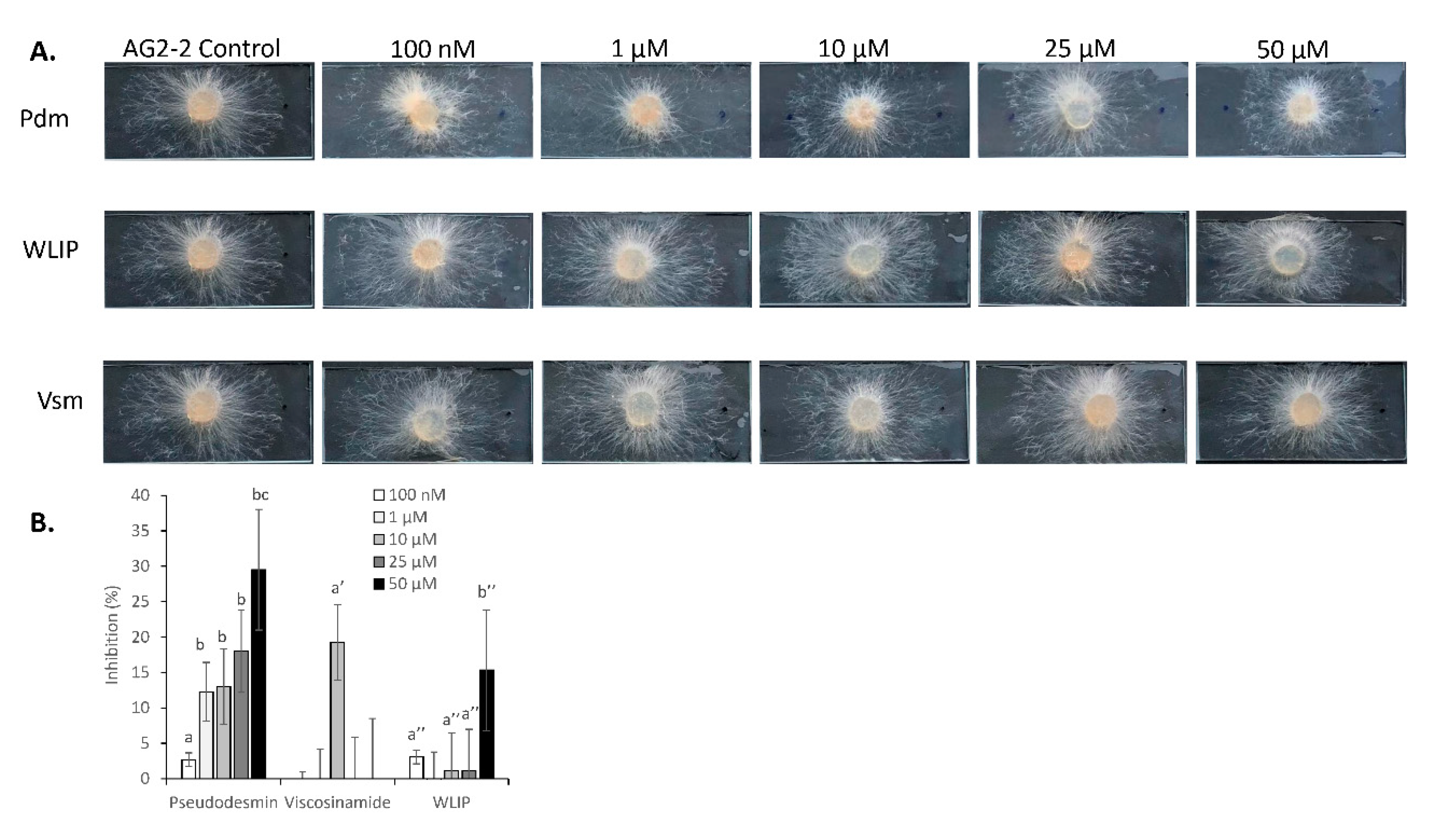
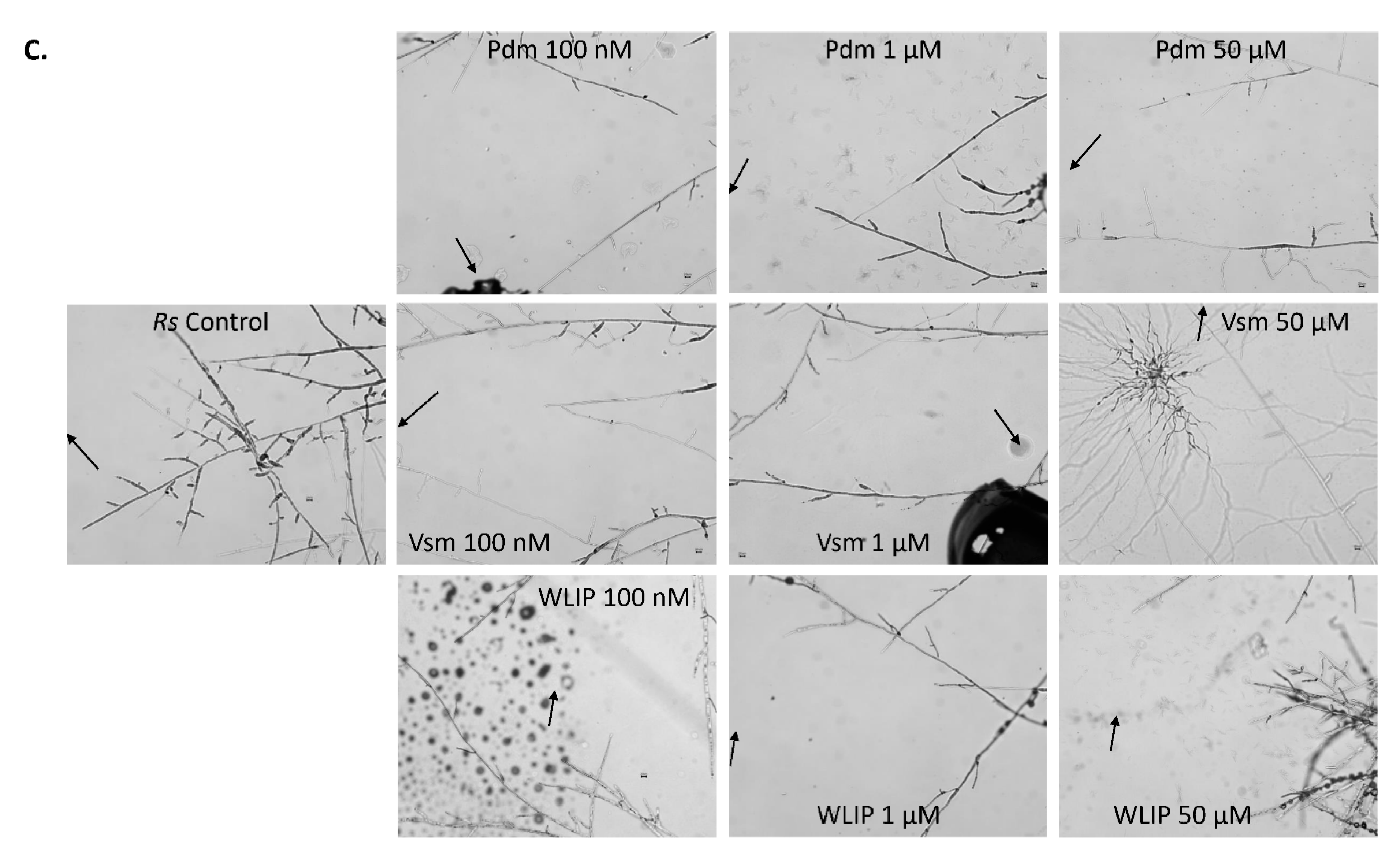
3.7. Inhibition of R. solani by Pseudodesmin, Viscosinamide and WLIP
3.8. R. solani-CLP Perception and Interaction
4. Discussion
4.1. CLP Identification and in silico Analysis of Pseudodesmin and Viscosinamide Genes
4.2. Taxonomic Congruence and Functional Analyses of Pseudodesmin- and Viscosinamide-Producing Strains
4.3. Swarming Motility Rates in Different Media
4.4. CLP Rarity vs. Functional Relevance
4.5. Biological Activity of Pseudodesmin, Viscosinamide and WLIP against P. myriotylum—Mechanism of Action?
4.6. Perception of CLPs by P. myriotylum—Implications for CRRD Suppression
4.7. Differential Responses of R. solani to Pseudodesmin, Viscosinamide and WLIP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Species | Strain Name | CLP Produced * | Origin/Source | Host | Accession | Country | Reference |
|---|---|---|---|---|---|---|---|
| P. chlororaphis subsp. piscium | PCL1391 | viscosin G 1 | Tomato rhizosphere | Solanum lycopersicum | CP027736.1 | Spain | [50] |
| P. chlororaphis subsp. piscium | DSM 21509 | viscosin G 1 | European perch intestine | Perca fluviatilis | CP027707.1 | Switzerland | [50] |
| P. chlororaphis subsp. aureofaciens | P2 | viscosin G 1 | Potato roots | Solanum tuberosum | CP027719.1 | Algeria | [50] |
| P. chlororaphis | Pb-St2 | WLIP | Sugarcane stem | Saccharum sp. | CP027716.1 | Pakistan | [50] |
| P. chlororaphis subsp. aurantiaca | PCM 2210 | viscosin G 1 | Sugar beet roots | Beta vulgaris | CP027717.1 | Poland | [50] |
| P. chlororaphis subsp. piscium | ZJU60 | viscosin G 1 | - | Wheat | CP027656.1 | China | Chen and Ma, unpublished |
| P. chlororaphis | Lzh-T5 | viscosin G 1 | Soil | - | CP025309.1 | China | Li et al., unpublished |
| Pseudomonas sp. | MYb193 | viscosin G 2 | Compost | - | CP023269.1 | Germany | [59] |
| Pseudomonas sp. | J380 | viscosin G 2 | - | Cunner fish | CP043060.1 | Canada | Santander et al., unpublished |
| P. synxantha | 2-79 | viscosin G 3 | Wheat rhizosphere | Triticum sp. | CP027755.1 | USA | [50] |
| Pseudomonas sp. | LBUM920 | viscosin G 3 | White spruce rhizosphere | Picea glauca | CP027762.1 | Canada | [50] |
| P. antarctica | PAMC 27494 | viscosin G 3 | Freshwater | - | CP015600.1 | Antarctica | Lee, unpublished |
| P. lactis | SS101 | massetolide | Wheat rhizosphere | Triticum sp. | CM001513.1 | Netherlands | [23] |
| P. synxantha | 30B | viscosin G 3 | Wheat rhizosphere | Triticum sp. | CP027754.1 | Iran | [50] |
| P. putida | RW10S2 | WLIP | Rice rhizosphere | Oryza sativa | JN982332.1 | Sri Lanka | [24] |
| (wlipA) | |||||||
| JN982333.1 (wlipBC) | |||||||
| Pseudomonas sp. | NSE1 | WLIP | white cocoyam rhizosphere | X. sagittifolium | MK534106.1 | Nigeria | [25] |
| (wlipA) | |||||||
| MK650230.2 | |||||||
| (wlipBC) | |||||||
| P. fluorescens | SBW25 | viscosin | Wheat rhizosphere | Triticum sp. | NC_012660.1 | Netherlands | [60] |
| Pseudomonas sp. | COR52 | pseudodesmin | tissue cultured red cocoyam | X.. sagittifolium | MT577358 | Cameroon | This study |
| Pseudomonas sp. | U2W1.5 | viscosinamide | tissue cultured white cocoyam | X. sagittifolium | MT771986 | Nigeria | This study |
| (vsmA) | |||||||
| MT749673 | |||||||
| (vsmBC) | |||||||
| Pseudomonas sp. | A2W4.9 | viscosinamide | tissue cultured white cocoyam | X. sagittifolium | MT749674 | Nigeria | This study |
| (vsmA) | |||||||
| MT771985 | |||||||
| (vsmBC) |
| CLP | Concentration | Growth Inhibition | Hyphal Blockage | Hypal Distortion | Hyphal Branching | Hyper Branching | Hyphal Disintegration | Hyphal Lysis | CLP Evasion | Convergent Structures * |
|---|---|---|---|---|---|---|---|---|---|---|
| pseudodesmin | 100 nM | +/+ | −/+ | ++/+ | ++/− | ++/− | −/− | −/+ | −/+ | −/− |
| 1 µM | +/+ | −/+ | ++/+ | ++/− | −/− | −/− | ++/+ | −/+ | −/− | |
| 10 µM | +/+ | −/+ | ++/+ | ++/− | −/− | −/− | −/+ | +/+ | −/− | |
| 25 µM | +/+ | −/+ | ++/+ | +/− | ++/− | −/− | −/+ | +/+ | −/− | |
| 50 µM | ++/++ | −/+ | ++/+ | −/− | −/− | −/− | −/+ | −/+ | −/− | |
| viscosinamide | 100 nM | −/− | −/+ | +/+ | ++/− | −/− | −/− | −/− | −/+ | −/− |
| 1 µM | −/− | −/+ | +/+ | ++/− | −/− | −/− | +/− | −/+ | −/− | |
| 10 µM | −/+ | −/+ | +/+ | +/− | −/− | −/− | −/− | +/+ | −/− | |
| 25 µM | −/− | −/+ | +/+ | ++/− | −/− | −/− | +/− | −/+ | −/− | |
| 50 µM | −/− | −/+ | +/+ | ++/− | −/− | −/− | −/− | +/+ | ++/++ | |
| WLIP | 100 nM | +/+ | +/+ | +/+ | −/− | −/− | +/+ | −/− | +/− | −/− |
| 1 µM | +/− | +/+ | +/+ | −/− | −/− | +/+ | −/− | +/− | −/− | |
| 10 µM | +/+ | +/+ | +/+ | −/− | −/− | ++/+ | −/− | +/− | −/− | |
| 25 µM | +/+ | +/+ | +/+ | −/− | −/− | ++/++ | −/− | +/− | −/− | |
| 50 µM | ++/++ | +/+ | +/+ | −/− | −/− | ++/++ | −/− | +/− | −/− |
References
- Raaijmakers, J.M.; de Bruijn, I.; de Kock, M.J.D. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: Diversity, activity, biosynthesis, and regulation. Mol. Plant. Microbe Interact. 2006, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Geudens, N.; Martins, J.C. Cyclic lipodepsipeptides from Pseudomonas spp.–biological swiss-army knives. Front. Microbiol. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Ann. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Yang, S.Y.; Kim, Y.C.; Lee, C.W.; Park, M.S.; Kim, J.C.; Kim, I.S. Identification of orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. J. Agric. Food Chem 2013, 61, 6786–6791. [Google Scholar] [CrossRef] [PubMed]
- Cautain, B.; de Pedro, N.; Schulz, C.; Pascual, J.; Sousa, T.D.; Martin, J.; Perez-Victoria, I.; Asensio, F.; Gonzalez, I.; Bills, G.F.; et al. Identification of the lipodepsipeptide MDN-0066, a novel inhibitor of vhl/hif pathway produced by a new Pseudomonas species. PLoS ONE 2015, 10, e0125221. [Google Scholar] [CrossRef]
- Olorunleke, F.E.; Kieu, N.P.; Höfte, M. Recent advances in Pseudomonas biocontrol. In Bacteria-Plant Interactions: Advance Research and Future Trends; Murillo, J., Vinatzer, B.A., Jackson, R.W., Eds.; Caister Academic Press: Cambridge, UK, 2015; pp. 167–198. [Google Scholar]
- Flury, P.; Vesga, P.; Pechy-Tarr, M.; Aellen, N.; Dennert, F.; Hofer, N.; Kupferschmied, K.P.; Kupferschmied, P.; Metla, Z.; Ma, Z.W.; et al. Antimicrobial and insecticidal: Cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 2017, 8, 100. [Google Scholar] [CrossRef]
- Oni, F.E.; Geudens, N.; Onyeka, J.T.; Olorunleke, O.F.; Salami, A.E.; Omoboye, O.O.; Arias, A.A.; Adiobo, A.; De Neve, S.; Ongena, M.; et al. Cyclic lipopeptide-producing Pseudomonas koreensis group strains dominate the cocoyam rhizosphere of a Pythium root rot suppressive soil contrasting with P. putida prominence in conducive soils. Environ. Microbiol. 2020, in press. [Google Scholar] [CrossRef]
- Gotze, S.; Stallforth, P. Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads. Nat. Prod. Rep. 2020, 37, 29–54. [Google Scholar] [CrossRef]
- Jahanshah, G.; Yang, Q.; Gerhardt, H.; Pataj, Z.; Lammerhofer, M.; Pianet, I.; Josten, M.; Sahl, H.G.; Silby, M.W.; Loper, J.E.; et al. Discovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective gacA regulator in Pseudomonas fluorescens Pf0-1. J. Nat. Prod. 2019, 82, 301–308. [Google Scholar] [CrossRef]
- Oni, F.E.; Geudens, N.; Omoboye, O.O.; Bertier, L.; Hua, H.G.K.; Adiobo, A. Fluorescent Pseudomonas and cyclic lipopeptide diversity in the rhizosphere of cocoyam (Xanthosoma sagittifolium) (vol 21, pg 1019, 2019). Environ. Microbiol. 2019, 21, 4887. [Google Scholar]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes-a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Roongsawang, N.; Washio, K.; Morikawa, M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 2011, 12, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Jensen, P.R. Phylogenetic approaches to natural product structure prediction. Methods Enzymol. 2012, 517, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Laycock, M.V.; Hildebrand, P.D.; Thibault, P.; Walter, J.A.; Wright, J.L.C. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of pseudomonas-fluorescens. J. Agric. Food Chem 1991, 39, 483–489. [Google Scholar] [CrossRef]
- Nielsen, T.H.; Christophersen, C.; Anthoni, U.; Sorensen, J. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 1999, 87, 80–90. [Google Scholar] [CrossRef]
- Sinnaeve, D.; Michaux, C.; Van Hemel, J.; Vandenkerckhove, J.; Peys, E.; Borremans, F.A.M.; Sas, B.; Wouters, J.; Martins, J.C. Structure and x-ray conformation of pseudodesmins A and B, two new cyclic lipodepsipeptides from Pseudomonas bacteria. Tetrahedron 2009, 65, 4173–4181. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Ismail, N.; Quail, J.W.; Boyetchko, S.M. Structure, chemistry, and biological activity of pseudophomins A and B, new cyclic lipodepsipeptides isolated from the biocontrol bacterium Pseudomonas fluorescens. Phytochemistry 2003, 62, 1105–1114. [Google Scholar] [CrossRef]
- Gerard, J.; Lloyd, R.; Barsby, T.; Haden, P.; Kelly, M.T.; Andersen, R.J. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 1997, 60, 223–229. [Google Scholar] [CrossRef]
- Mortishiresmith, R.J.; Nutkins, J.C.; Packman, L.C.; Brodey, C.L.; Rainey, P.B.; Johnstone, K.; Williams, D.H. Determination of the structure of an extracellular peptide produced by the mushroom saprotroph Pseudomonas-reactans. Tetrahedron 1991, 47, 3645–3654. [Google Scholar] [CrossRef]
- De Bruijn, I.; de Kock, M.J.D.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 2777–2789. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, I.; de Kock, M.J.D.; Yang, M.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 2007, 63, 417–428. [Google Scholar] [CrossRef]
- Rokni-Zadeh, H.; Li, W.; Sanchez-Rodriguez, A.; Sinnaeve, D.; Rozenski, J.; Martins, J.C.; De Mot, R. Genetic and functional characterization of cyclic lipopeptide white-line-inducing principle (WLIP) production by rice rhizosphere isolate Pseudomonas putida RW10S2. Appl. Environ. Microbiol. 2012, 78, 4826–4834. [Google Scholar] [CrossRef] [PubMed]
- Omoboye, O.O.; Oni, F.E.; Batool, H.; Yimer, H.Z.; De Mot, R.; Hofte, M. Pseudomonas cyclic lipopeptides suppress the rice blast fungus Magnaporthe oryzae by induced resistance and direct antagonism. Front. Plant. Sci. 2019, 10, 901. [Google Scholar] [CrossRef] [PubMed]
- Vlassak, K.; Vanholm, L.; Duchateau, L.; Vanderleyden, J.; Demot, R. Isolation and characterization of fluorescent Pseudomonas associated with the roots of rice and banana grown in Sri-lanka. Plant. Soil 1992, 145, 51–63. [Google Scholar] [CrossRef]
- De Souza, J.T.; Mazzola, M.; Raaijmakers, J.M. Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 2003, 5, 1328–1340. [Google Scholar] [CrossRef]
- Mehnaz, S.; Saleem, R.S.Z.; Yameen, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.G.; Franzblau, S.G.; et al. Lahorenoic acids a-c, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef]
- Martin, C.; Ibanez, R.; Nothias, L.F.; Boya, C.A.; Reinert, L.K.; Rollins-Smithv, L.A.; Dorrestein, P.C.; Gutierrez, M. Viscosin-like lipopeptides from frog skin bacteria inhibit Aspergillus fumigatus and Batrachochytrium dendrobatidis detected by imaging mass spectrometry and molecular networking. Sci. Rep. 2019, 9, 3019. [Google Scholar] [CrossRef]
- Geudens, N.; Nasir, M.N.; Crowet, J.M.; Raaijmakers, J.M.; Feher, K.; Coenye, T.; Martins, J.C.; Lins, L.; Sinnaeve, D.; Deleu, M. Membrane interactions of natural cyclic lipodepsipeptides of the viscosin group. BBA-Biomembranes 2017, 1859, 331–339. [Google Scholar] [CrossRef]
- De Vleeschouwer, M.; Van Kersavond, T.; Verleysen, Y.; Sinnaeve, D.; Coenye, T.; Martins, J.C.; Madder, A. Identification of the molecular determinants involved in antimicrobial activity of pseudodesmin a, a cyclic lipopeptide from the viscosin group. Front. Microbiol. 2020, 11, 646. [Google Scholar] [CrossRef]
- Thrane, C.; Olsson, S.; Nielsen, T.H.; Sorensen, J. Vital fluorescent stains for detection of stress in Pythium ultimum and Rhizoctonia solani challenged with viscosinamide from Pseudomonas fluorescens DR54. FEMS Microbiol. Ecol. 1999, 30, 11–23. [Google Scholar] [CrossRef]
- Olorunleke, F.E. Cyclic Lipopeptides Produced by Pseudomonas spp. Associated with the Cocoyam (Xanthosoma sagittifolium (l.) Schott) Rhizosphere: Diversity, Regulation, Secretion and Biological Activity. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2017. [Google Scholar]
- Omoboye, O.O. Cyclic lipopeptide Diversity and Biocontrol Versatility of Pseudomonas species Associated with the Cocoyam Rhizosphere. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2019. [Google Scholar]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Tambong, J.T.; Sapra, V.T.; Garton, S. In vitro induction of tetraploids in colchicine-treated cocoyam plantlets. Euphytica 1998, 104, 191–197. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Nielsen, T.H.; Sorensen, D.; Tobiasen, C.; Andersen, J.B.; Christophersen, C.; Givskov, M.; Sorensen, J. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. From the sugar beet rhizosphere. Appl. Environ. Microbiol. 2002, 68, 3416–3423. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Rottig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef]
- D’aes, J.; Kieu, N.P.; Leclere, V.; Tokarski, C.; Olorunleke, F.E.; De Maeyer, K.; Jacques, P.; Hofte, M.; Ongena, M. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 2014, 16, 2282–2300. [Google Scholar] [CrossRef]
- Perneel, M.; Tambong, J.T.; Adiobo, A.; Floren, C.; Saborio, F.; Levesque, A.; Hofte, M. Intraspecific variability of Pythium myriotylum isolated from cocoyam and other host crops. Mycol. Res. 2006, 110, 583–593. [Google Scholar] [CrossRef]
- Nerey, Y.; Pannecoucque, J.; Hernandez, H.P.; Diaz, M.; Espinosa, R.; De Vos, S.; Van Beneden, S.; Herrera, L.; Hofte, M. Rhizoctonia spp. causing root and hypocotyl rot in Phaseolus vulgaris in cuba. J. Phytopathol. 2010, 158, 236–243. [Google Scholar] [CrossRef]
- Olorunleke, F.E.; Hua, G.K.H.; Kieu, N.P.; Ma, Z.W.; Hofte, M. Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ. Microbiol. Rep. 2015, 7, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Balibar, C.J.; Vaillancourt, F.H.; Walsh, C.T. Generation of d amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 2005, 12, 1189–1200. [Google Scholar] [CrossRef]
- De Bruijn, I.; Raaijmakers, J.M. Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the Clpp protease. J. Bacteriol. 2009, 191, 1910–1923. [Google Scholar] [CrossRef]
- Rokni-Zadeh, H.; Li, W.; Yilma, E.; Sanchez-Rodriguez, A.; De Mot, R. Distinct lipopeptide production systems for WLIP (white line-inducing principle) in Pseudomonas fluorescens and Pseudomonas putida. Environ. Microbiol. Rep. 2013, 5, 160–169. [Google Scholar] [CrossRef]
- Sajben, E.; Manczinger, L.; Nagy, A.; Kredics, L.; Vagvolgyi, C. Characterization of pseudomonads isolated from decaying sporocarps of oyster mushroom. Microbiol. Res. 2011, 166, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Mulet, M.; Gomila, M.; Garcia-Valdes, E. Genomics in bacterial taxonomy: Impact on the genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef]
- Biessy, A.; Novinscak, A.; Blom, J.; Leger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Osborn, R.W. Plant defensins-novel antimicrobial peptides as components of the host-defense system. Plant. Physiol. 1995, 108, 1353–1358. [Google Scholar] [CrossRef]
- Thomma, B.; Cammue, B.P.A.; Thevissen, K. Plant defensins. Planta 2002, 216, 193–202. [Google Scholar] [CrossRef]
- Spelbrink, R.G.; Dilmac, N.; Allen, A.; Smith, T.J.; Shah, D.M.; Hockerman, G.H. Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant. Physiol. 2004, 135, 2055–2067. [Google Scholar] [CrossRef]
- Pombeiro-Sponchiado, S.R.; Sousa, G.S.; Andrade, J.C.R.; Lisboa, H.F.; Goncalves, R.C.R. Production of melanin pigment by fungi and its biotechnological applications. Melanin 2017. [Google Scholar] [CrossRef]
- Ipcho, S.; Sundelin, T.; Erbs, G.; Kistler, H.C.; Newman, M.A.; Olsson, S. Fungal innate immunity induced by bacterial microbe-associated molecular patterns (MAMPs). G3 (Bethesda) 2016, 6, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Adiobo, A.; Oumar, O.; Perneel, M.; Zok, S.; Hofte, M. Variation of Pythium-induced cocoyam root rot severity in response to soil type. Soil Biol. Biochem. 2007, 39, 2915–2925. [Google Scholar] [CrossRef]
- Schroeder, K.L.; Martin, F.N.; de Cock, A.; Levesque, C.A.; Spies, C.F.J.; Okubara, P.A.; Paulitz, T.C. Molecular detection and quantification of Pythium species: Evolving taxonomy, new tools, and challenges. Plant. Dis. 2013, 97, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Thrane, C.; Olsson, S.; Sorensen, J. Confocal imaging of living fungal hyphae challenged with the fungal antagonist viscosinamide. Mycologia 2000, 92, 216–221. [Google Scholar] [CrossRef]
- Dirksen, P.; Marsh, S.A.; Braker, I.; Heitland, N.; Wagner, S.; Nakad, R.; Mader, S.; Petersen, C.; Kowallik, V.; Rosenstiel, P.; et al. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol. 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Silby, M.W.; Cerdeno-Tarraga, A.M.; Vernikos, G.S.; Giddens, S.R.; Jackson, R.W.; Preston, G.M.; Zhang, X.X.; Moon, C.D.; Gehrig, S.M.; Godfrey, S.A.; et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009, 10, R51. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oni, F.E.; Geudens, N.; Adiobo, A.; Omoboye, O.O.; Enow, E.A.; Onyeka, J.T.; Salami, A.E.; De Mot, R.; Martins, J.C.; Höfte, M. Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere. Microorganisms 2020, 8, 1079. https://doi.org/10.3390/microorganisms8071079
Oni FE, Geudens N, Adiobo A, Omoboye OO, Enow EA, Onyeka JT, Salami AE, De Mot R, Martins JC, Höfte M. Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere. Microorganisms. 2020; 8(7):1079. https://doi.org/10.3390/microorganisms8071079
Chicago/Turabian StyleOni, Feyisara E., Niels Geudens, Amayana Adiobo, Olumide O. Omoboye, Elsie A. Enow, Joseph T. Onyeka, Ayodeji E. Salami, René De Mot, José C. Martins, and Monica Höfte. 2020. "Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere" Microorganisms 8, no. 7: 1079. https://doi.org/10.3390/microorganisms8071079
APA StyleOni, F. E., Geudens, N., Adiobo, A., Omoboye, O. O., Enow, E. A., Onyeka, J. T., Salami, A. E., De Mot, R., Martins, J. C., & Höfte, M. (2020). Biosynthesis and Antimicrobial Activity of Pseudodesmin and Viscosinamide Cyclic Lipopeptides Produced by Pseudomonads Associated with the Cocoyam Rhizosphere. Microorganisms, 8(7), 1079. https://doi.org/10.3390/microorganisms8071079





